The Inactivation of LPS Biosynthesis Genes in E. coli Cells Leads to Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
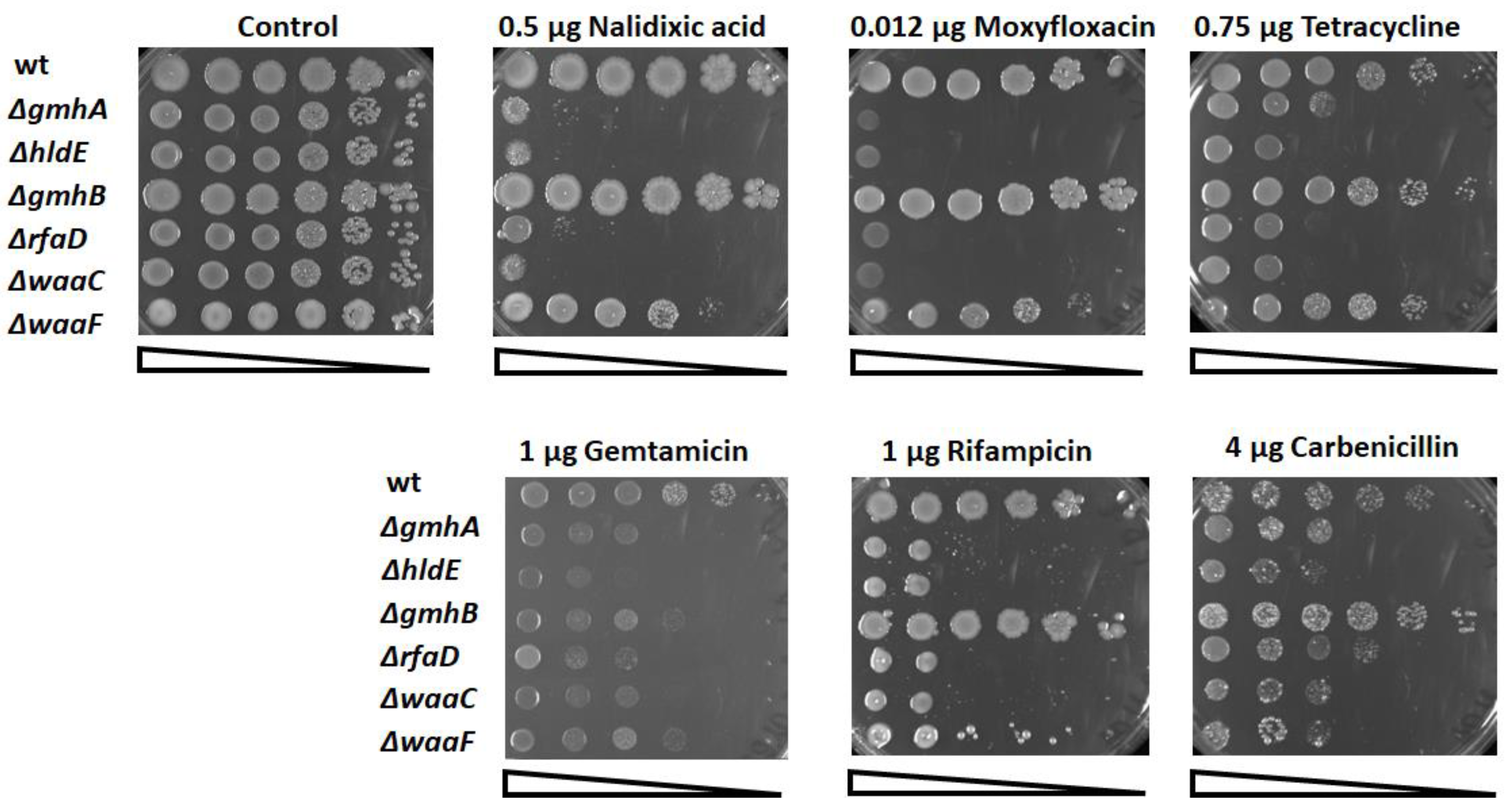
2.2. Determination of Sensitivity to Antibiotics
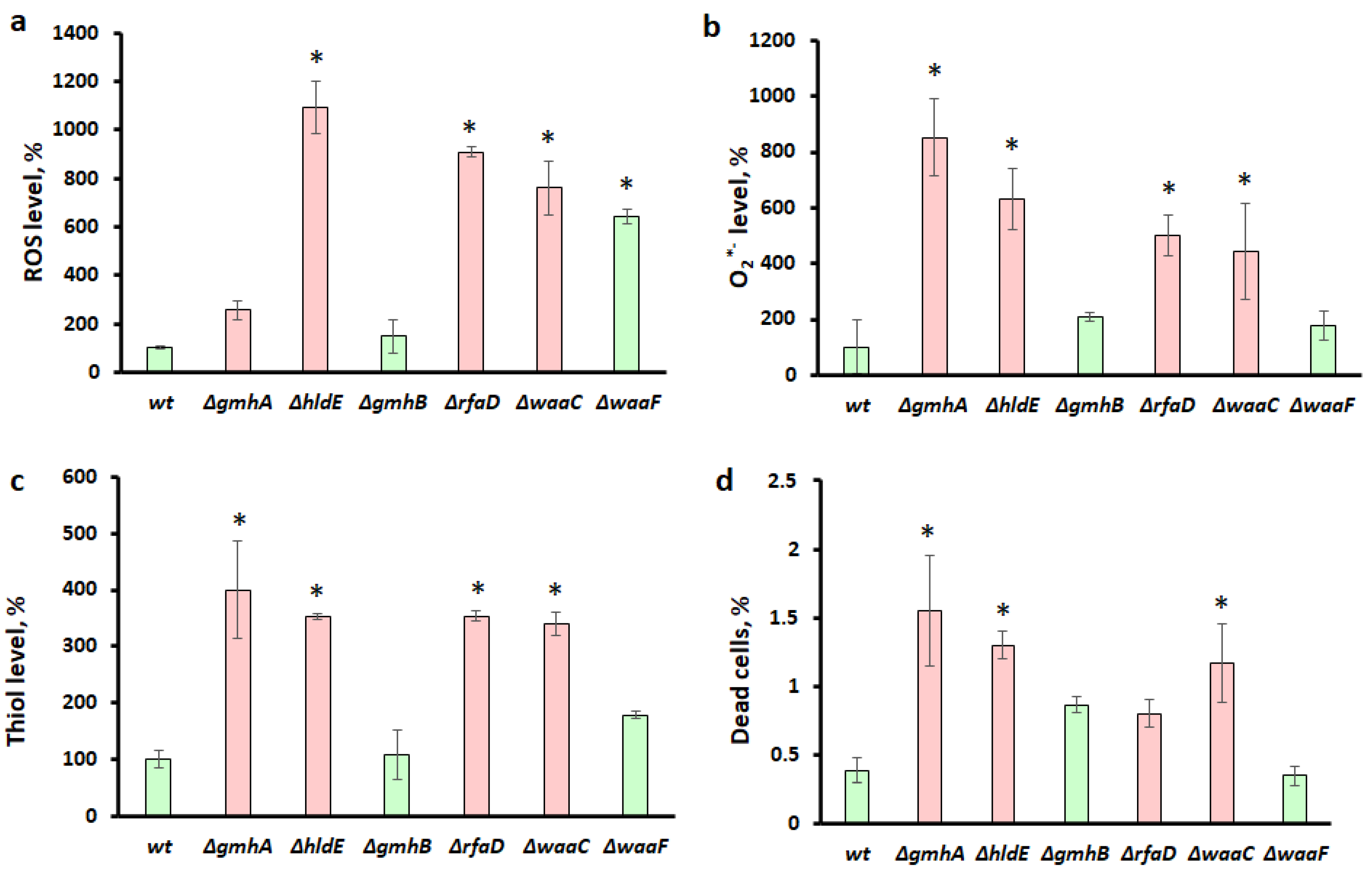
2.3. Determination of Viability and Redox Status of Cells Using Flow Cytometry
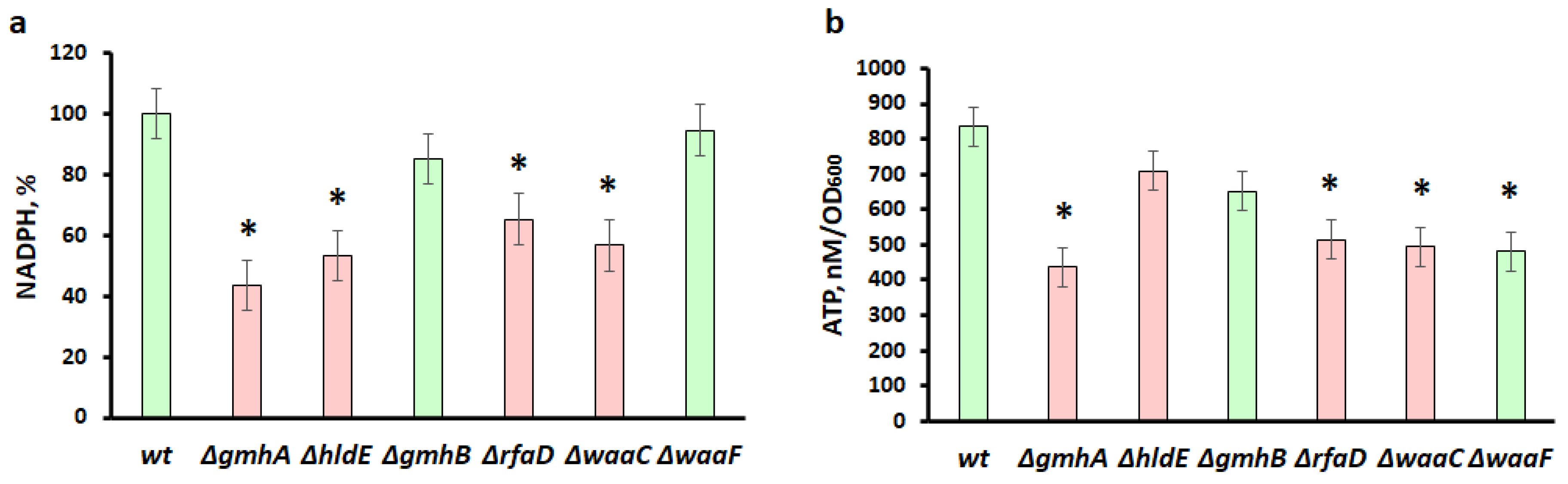
2.4. Measurement of NADPH
2.5. Measurement of ATP
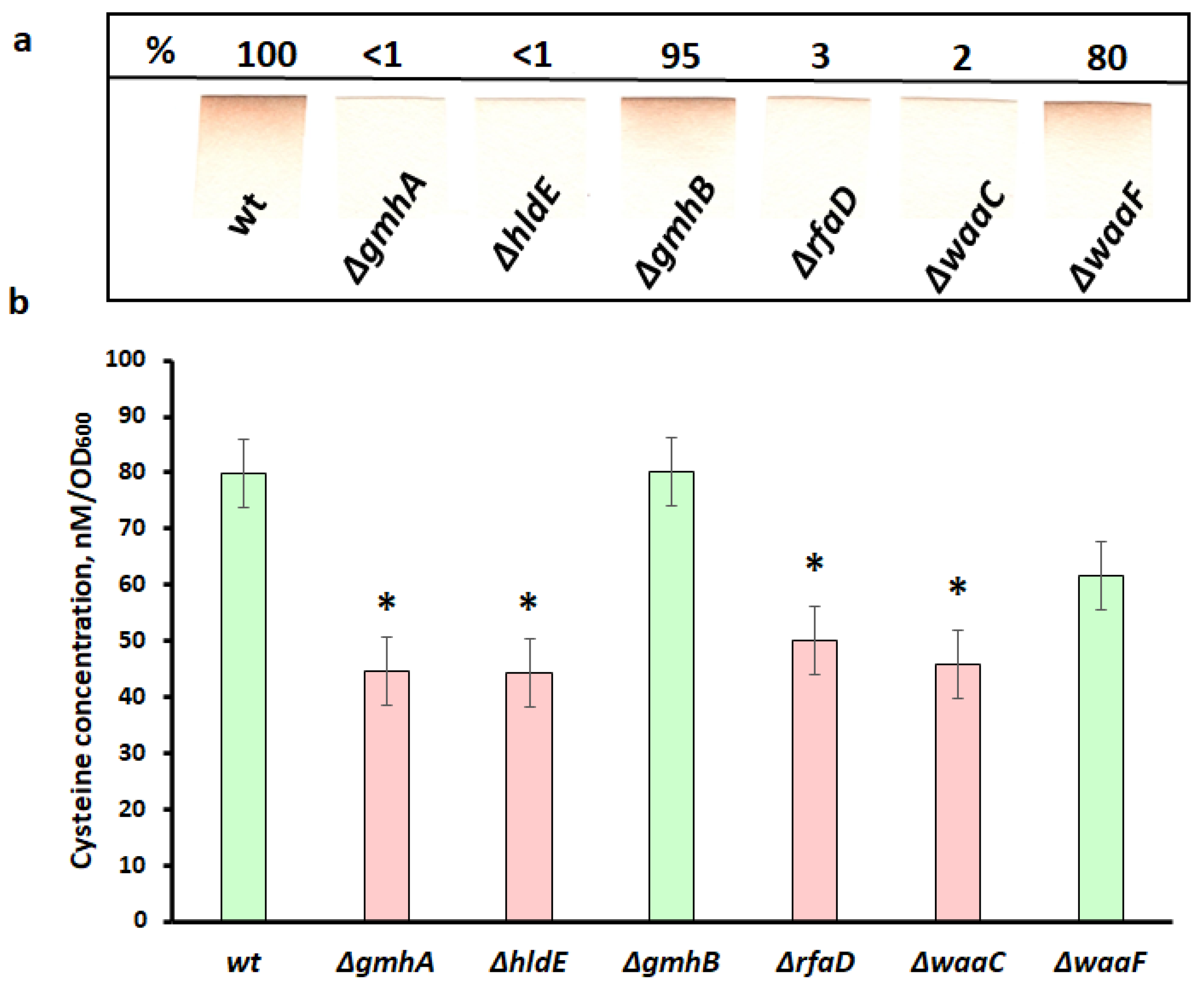
2.6. H2S Detection
2.7. Determination of Intracellular Cysteine
2.8. Statistical Analysis
3. Results
3.1. Oxidative Stress and Antibiotic Sensitivity
3.2. Reducing NADPH Equivalents and the ATP Pool
3.3. H2S Generation and Cysteine Deficiency
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kneidinger, B.; Marolda, C.; Graninger, M.; Zamyatina, A.; McArthur, F.; Kosma, P.; Valvano, M.A.; Messner, P. Biosynthesis Pathway of ADP-l-Glycero-β-d-Manno-Heptose in Escherichia coli. J. Bacteriol. 2002, 184, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.L.; Blakely, K.M.; de Leon, G.P.; Walker, J.R.; McArthur, F.; Evdokimova, E.; Zhang, K.; Valvano, M.A.; Wright, G.D.; Junop, M.S. Structure and Function of Sedoheptulose-7-Phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants *. J. Biol. Chem. 2008, 283, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Durka, M.; Tikad, A.; Périon, R.; Bosco, M.; Andaloussi, M.; Floquet, S.; Malacain, E.; Moreau, F.; Oxoby, M.; Gerusz, V.; et al. Systematic Synthesis of Inhibitors of the Two First Enzymes of the Bacterial Heptose Biosynthetic Pathway: Towards Antivirulence Molecules Targeting Lipopolysaccharide Biosynthesis. Chem. A Eur. J. 2011, 17, 11305–11313. [Google Scholar] [CrossRef]
- Huang, K.C.; Mukhopadhyay, R.; Wen, B.; Gitai, Z.; Wingreen, N.S. Cell Shape and Cell-Wall Organization in Gram-Negative Bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 19282–19287. [Google Scholar] [CrossRef] [PubMed]
- Schnaitman, C.A.; Klena, J.D. Genetics of Lipopolysaccharide Biosynthesis in Enteric Bacteria. Microbiol. Rev. 1993, 57, 655–682. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.-M.; Krüger, A.; Tauqeer Alam, M.; et al. The Return of Metabolism: Biochemistry and Physiology of the Pentose Phosphate Pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed]
- De Leon, G.P.; Elowe, N.H.; Koteva, K.P.; Valvano, M.A.; Wright, G.D. An In Vitro Screen of Bacterial Lipopolysaccharide Biosynthetic Enzymes Identifies an Inhibitor of ADP-Heptose Biosynthesis. Chem. Biol. 2006, 13, 437–441. [Google Scholar] [CrossRef]
- Valvano, M.A. Biosynthesis and Genetics of ADP-Heptose. J. Endotoxin Res. 1999, 5, 90–95. [Google Scholar] [CrossRef]
- Desroy, N.; Moreau, F.; Briet, S.; Fralliec, G.L.; Floquet, S.; Durant, L.; Vongsouthi, V.; Gerusz, V.; Denis, A.; Escaich, S. Towards Gram-Negative Antivirulence Drugs: New Inhibitors of HldE Kinase. Tetrahedron Young Investig. Award. 2006 Jonathan Ellman 2009, 17, 1276–1289. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Cockerill, F.R.; Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; ISBN 1-56238-784-7. [Google Scholar]
- Mitkevich, V.A.; Kretova, O.V.; Petrushanko, I.Y.; Burnysheva, K.M.; Sosin, D.V.; Simonenko, O.V.; Ilinskaya, O.N.; Tchurikov, N.A.; Makarov, A.A. Ribonuclease Binase Apoptotic Signature in Leukemic Kasumi-1 Cells. Biochimie 2013, 95, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- Mitkevich, V.A.; Petrushanko, I.Y.; Yegorov, Y.E.; Simonenko, O.V.; Vishnyakova, K.S.; Kulikova, A.A.; Tsvetkov, P.O.; Makarov, A.A.; Kozin, S.A. Isomerization of Asp7 Leads to Increased Toxic Effect of Amyloid-Β42 on Human Neuronal Cells. Cell Death Dis. 2013, 4, e939. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Smirnova, O.A.; Petrushanko, I.Y.; Ivanova, O.N.; Karpenko, I.L.; Alekseeva, E.; Sominskaya, I.; Makarov, A.A.; Bartosch, B.; Kochetkov, S.N.; et al. HCV Core Protein Uses Multiple Mechanisms to Induce Oxidative Stress in Human Hepatoma Huh7 Cells. Viruses 2015, 7, 2745–2770. [Google Scholar] [CrossRef] [PubMed]
- Tille, P.M. Bailey & Scott’s Diagnostic Microbiology, 13th ed.; Elsevier: St. Louis, MS, USA, 2014. [Google Scholar]
- Gaitonde, M. A Spectrophotometric Method for the Direct Determination of Cysteine in the Presence of Other Naturally Occurring Amino Acids. Biochem. J. 1967, 104, 627–633. [Google Scholar] [CrossRef]
- Coleman, W.G.; Leive, L. Two Mutations Which Affect the Barrier Function of the Escherichia Coli K-12 Outer Membrane. J. Bacteriol. 1979, 139, 899–910. [Google Scholar] [CrossRef]
- Dwyer Daniel, J.; Belenky Peter, A.; Yang Jason, H.; MacDonald, I.C.; Martell Jeffrey, D.; Takahashi, N.; Chan Clement, T.Y.; Lobritz Michael, A.; Braff, D.; Schwarz Eric, G.; et al. Antibiotics Induce Redox-Related Physiological Alterations as Part of Their Lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef]
- Shatalin, K.; Shatalina, E.; Mironov, A.; Nudler, E. H2S: A Universal Defense Against Antibiotics in Bacteria. Science 2011, 334, 986–990. [Google Scholar] [CrossRef]
- Mironov, A.; Seregina, T.; Nagornykh, M.; Luhachack, L.G.; Korolkova, N.; Lopes, L.E.; Kotova, V.; Zavilgelsky, G.; Shakulov, R.; Shatalin, K.; et al. Mechanism of H2S-Mediated Protection against Oxidative Stress in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 6022–6027. [Google Scholar] [CrossRef]
- Colnaghi, R.; Cassinelli, G.; Drummond, M.; Forlani, F.; Pagani, S. Properties of the Escherichia Coli Rhodanese-like Protein SseA: Contribution of the Active-Site Residue Ser240 to Sulfur Donor Recognition. FEBS Lett. 2001, 500, 153–156. [Google Scholar] [CrossRef]
- Linkevicius, M.; Sandegren, L.; Andersson, D.I. Mechanisms and Fitness Costs of Tigecycline Resistance in Escherichia coli. J. Antimicrob. Chemother. 2013, 68, 2809–2819. [Google Scholar] [CrossRef] [Green Version]
- Coleman, W.G., Jr. The RfaD Gene Codes for ADP-L-Glycero-D-Mannoheptose-6-Epimerase. An Enzyme Required for Lipopolysaccharide Core Biosynthesis. J. Biol. Chem. 1983, 258, 1985–1990. [Google Scholar] [CrossRef]
- Fraenkel, D.G.; Horecker, B.L. Fructose-1,6-Diphosphatase and Acid Hexose Phosphatase of Escherichia coli. J. Bacteriol. 1965, 90, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel, D.G.; Pontremoli, S.; Horecker, B.L. The Specific Fructose Diphosphatase of Escherichia coli: Properties and Partial Purification. Arch. Biochem. Biophys. 1966, 114, 4–12. [Google Scholar] [CrossRef]
- Alzayn, M.; Dulyayangkul, P.; Satapoomin, N.; Heesom Kate, J.; Avison Matthew, B. OmpF Downregulation Mediated by Sigma E or OmpR Activation Confers Cefalexin Resistance in Escherichia Coli in the Absence of Acquired β-Lactamases. Antimicrob. Agents Chemother. 2021, 65, e01004-21. [Google Scholar] [CrossRef] [PubMed]
- Meredith, T.C.; Mamat, U.; Kaczynski, Z.; Lindner, B.; Holst, O.; Woodard, R.W. Modification of Lipopolysaccharide with Colanic Acid (M-Antigen) Repeats in Escherichia coli*. J. Biol. Chem. 2007, 282, 7790–7798. [Google Scholar] [CrossRef] [PubMed]
- Joloba Moses, L.; Clemmer Katy, M.; Sledjeski Darren, D.; Rather Philip, N. Activation of the Gab Operon in an RpoS-Dependent Manner by Mutations That Truncate the Inner Core of Lipopolysaccharide in Escherichia coli. J. Bacteriol. 2004, 186, 8542–8546. [Google Scholar] [CrossRef]
- Ren, G.; Wang, Z.; Li, Y.; Hu, X.; Wang, X. Effects of Lipopolysaccharide Core Sugar Deficiency on Colanic Acid Biosynthesis in Escherichia coli. J. Bacteriol. 2016, 198, 1576–1584. [Google Scholar] [CrossRef]
- Samain, E. 1.23—Production of Oligosaccharides in Microbes. In Comprehensive Glycoscience; Kamerling, H., Ed.; Elsevier: Oxford, UK, 2007; pp. 923–947. ISBN 978-0-444-51967-2. [Google Scholar]
- Lobritz Michael, A.; Belenky, P.; Porter Caroline, B.M.; Gutierrez, A.; Yang Jason, H.; Schwarz Eric, G.; Dwyer Daniel, J.; Khalil Ahmad, S.; Collins James, J. Antibiotic Efficacy Is Linked to Bacterial Cellular Respiration. Proc. Natl. Acad. Sci. USA 2015, 112, 8173–8180. [Google Scholar] [CrossRef]
- Canonaco, F.; Hess, T.A.; Heri, S.; Wang, T.; Szyperski, T.; Sauer, U. Metabolic Flux Response to Phosphoglucose Isomerase Knock-out in Escherichia coli and Impact of Overexpression of the Soluble Transhydrogenase UdhA. FEMS Microbiol. Lett. 2001, 204, 247–252. [Google Scholar] [CrossRef]
- Reitzer, L.; DiRita, V.J. Death by Cystine: An Adverse Emergent Property from a Beneficial Series of Reactions. J. Bacteriol. 2015, 197, 3626–3628. [Google Scholar] [CrossRef] [Green Version]
- Ohtsu, I.; Wiriyathanawudhiwong, N.; Morigasaki, S.; Nakatani, T.; Kadokura, H.; Takagi, H. The L-Cysteine/l-Cystine Shuttle System Provides Reducing Equivalents to the Periplasm in Escherichia coli*. J. Biol. Chem. 2010, 285, 17479–17487. [Google Scholar] [CrossRef] [PubMed]
- Mironov, A.; Seregina, T.; Shatalin, K.; Nagornykh, M.; Shakulov, R. Nudler, E. CydDC Functions as a Cytoplasmic Cystine Reductase to Sensitize Escherichia Coli to Oxidative Stress and Aminoglycosides. Proc. Natl. Acad. Sci. USA 2020, 117, 23565–23570. [Google Scholar] [CrossRef]
- Park, S.; Imlay, J. High Levels of Intracellular Cysteine Promote Oxidative DNA Damage by Driving the Fenton Reaction. J. Bacteriol. 2003, 185, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Lopatkin, A.J.; Lobritz, M.A.; Collins, J.J. Bacterial Metabolism and Antibiotic Efficacy. Cell Metab. 2019, 30, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Moreau, F.; Desroy, N.; Genevard, J.M.; Vongsouthi, V.; Gerusz, V.; Le Fralliec, G.; Oliveira, C.; Floquet, S.; Denis, A.; Escaich, S.; et al. Discovery of New Gram-Negative Antivirulence Drugs: Structure and Properties of Novel, E. coli WaaC Inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 4022–4026. [Google Scholar] [CrossRef]
- Chiu, S.-F.; Teng, K.-W.; Wang, P.-C.; Chung, H.-Y.; Wang, C.-J.; Cheng, H.-C.; Kao, M.-C. Helicobacter Pylori GmhB Enzyme Involved in ADP-Heptose Biosynthesis Pathway Is Essential for Lipopolysaccharide Biosynthesis and Bacterial Virulence. Virulence 2021, 12, 1610–1628. [Google Scholar] [CrossRef] [PubMed]
- Karan, S.; Pratap, B.; Yadav, S.P.; Ashish, F.; Saxena, A.K. Structural and Functional Characterization of M. tuberculosis Sedoheptulose- 7-Phosphate Isomerase, a Critical Enzyme Involved in Lipopolysaccharide Biosynthetic Pathway. Sci. Rep. 2020, 10, 20813. [Google Scholar] [CrossRef]



| Genotype | Nature of Mutation | Reference |
|---|---|---|
| wt | F- wild type | Laboratory collection |
| ∆gmhA | Deletion of the gmhA D-sedoheptulose 7-phosphate isomerase | This work |
| ∆hldE | Deletion of the hldE gene encoding heptose-7-phosphate kinase | “ |
| ∆gmhB | Deletion of the gmhB gene encoding | “ |
| D,D-heptose 1,7-bisphosphate phosphatase | ||
| ∆rfaD | Deletion of the rfaD gene encoding | “ |
| ADP-L-glycero-D-mannoheptose-6-epimerase | ||
| ∆waaC | Deletion of the waaC gene encoding | “ |
| ADP-heptose:LPS heptosyltransferase I | ||
| ∆waaF | Deletion of the waaF gene encoding | “ |
| ADP-heptose:LPS heptosyltransferase II |
| Name | Sequence |
|---|---|
| GmhA1 | 5′–atg-tac-cag-gat-ctt-att-cgt-acc-gaa-ctg-aac-gac-gct-caa-gtt-agt-ata-aaa-aag-ct–3′ |
| GmhA2 | 5′–tta-ctt-aac-cat-ctc-ttt-ttc-aat-taa-ctg-gat-cag-tga-agc-ctg-ctt-ttt-tat-act-aag–3′ |
| GmhA3 | 5′–ccg-tac-ttc-tcg-ctt-ttg-gc–3′ |
| GmhA4 | 5′–taa-gac-gcg-tca-gca-tcg-ca–3′ |
| HldE1 | 5′–atg-aaa-gta-acg-ctg-cca-gag-ttt-gaa-cgt-gca-ggc-gct-caa-gtt-agt-ata-aaa-aag-ct–3′ |
| HldE2 | 5′–tta-gcc-ttt-ttt-atc-ctg-ttg-gat-ctt-ctt-gat-gat-tga-agc-ctg-ctt-ttt-tat-act-aag–3′ |
| HldE3 | 5′–ggt-gga-aga-atg-aag-tat-gg–3′ |
| HldE4 | 5′–gtt-gaa-aaa-aca-aca-gcg-tca–3′ |
| GmhB1 | 5′–gtg-gcg-aag-agc-gta-ccc-gca-att-ttt-ctt-gac-cgc-gct-caa-gtt-agt-ata-aaa-aag-ct–3′ |
| GmhB2 | 5′–tca-ttg-tgc-cgg-ttt-ttg-ctg-ctt-ttt-tat-cgc-ttg-tga-agc-ctg-ctt-ttt-tat-act-aag–3′ |
| GmhB3 | 5′–ttc-ttg-cag-gtc-gaa-aca-tg–3′ |
| GmhB4 | 5′–c-tca-gga-aga-caa-gcg-gaa–3′ |
| RfaD1 | 5′–atg-atc-atc-gtt-acc-ggc-ggc-gcg-ggc-ttt-atc-ggc-gct-caa-gtt-agt-ata-aaa-aag-ct–3′ |
| RfaD2 | 5′–tta-tgc-gtc-gcg-att-cag-cca-ggc-cat-gta-ttc-cgt-tga-agc-ctg-ctt-ttt-tat-act-aag–3′ |
| RfaD3 | 5′–atg-att-aca-gac-att-cgt-gtc–3′ |
| RfaD4 | 5′–ga-ctt-tgc-gac-atc-atc-atg–3′ |
| WaaC1 | 5′–atg-cgg-gtt-ttg-atc-gtt-aaa-aca-tcg-tcg-atg-ggc-gct-caa-gtt-agt-ata-aaa-aag-ct–3′ |
| WaaC2 | 5′–tta-taa-tga-tga-taa-ctt-ttc-caa-aac-tgc-ttg-act-tga-agc-ctg-ctt-ttt-tat-act-aag–3′ |
| WaaC3 | 5′–gcg-tac-tgg-aag-aac-tca-ac–3′ |
| WaaC4 | 5′–gat-ttc-aga-gtg-taa-ggt-ttc–3′ |
| WaaF1 | 5′–atg-aaa-ata-ctg-gtg-atc-ggc-ccg-tct-tgg-gtt-ggc-gct-caa-gtt-agt-ata-aaa-aag-ct–3′ |
| WaaF2 | 5′–tca-ggc-ttc-ctc-ttg-taa-caa-tag-cgc-gtt-gag-ttc-tga-agc-ctg-ctt-ttt-tat-act-aag–3′ |
| WaaF3 | 5′–gtt-gct-gaa-ggt-gta-acg-ga–3′ |
| WaaF4 | 5′–ggc-aac-gta-tgg-aga-aca-tc–3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seregina, T.A.; Petrushanko, I.Y.; Shakulov, R.S.; Zaripov, P.I.; Makarov, A.A.; Mitkevich, V.A.; Mironov, A.S. The Inactivation of LPS Biosynthesis Genes in E. coli Cells Leads to Oxidative Stress. Cells 2022, 11, 2667. https://doi.org/10.3390/cells11172667
Seregina TA, Petrushanko IY, Shakulov RS, Zaripov PI, Makarov AA, Mitkevich VA, Mironov AS. The Inactivation of LPS Biosynthesis Genes in E. coli Cells Leads to Oxidative Stress. Cells. 2022; 11(17):2667. https://doi.org/10.3390/cells11172667
Chicago/Turabian StyleSeregina, Tatiana A., Irina Yu. Petrushanko, Rustem S. Shakulov, Pavel I. Zaripov, Alexander A. Makarov, Vladimir A. Mitkevich, and Alexander S. Mironov. 2022. "The Inactivation of LPS Biosynthesis Genes in E. coli Cells Leads to Oxidative Stress" Cells 11, no. 17: 2667. https://doi.org/10.3390/cells11172667
APA StyleSeregina, T. A., Petrushanko, I. Y., Shakulov, R. S., Zaripov, P. I., Makarov, A. A., Mitkevich, V. A., & Mironov, A. S. (2022). The Inactivation of LPS Biosynthesis Genes in E. coli Cells Leads to Oxidative Stress. Cells, 11(17), 2667. https://doi.org/10.3390/cells11172667






