Cryopreservation and Cryobanking of Cells from 100 Coral Species
Abstract
:1. Introduction
2. Materials and Methods
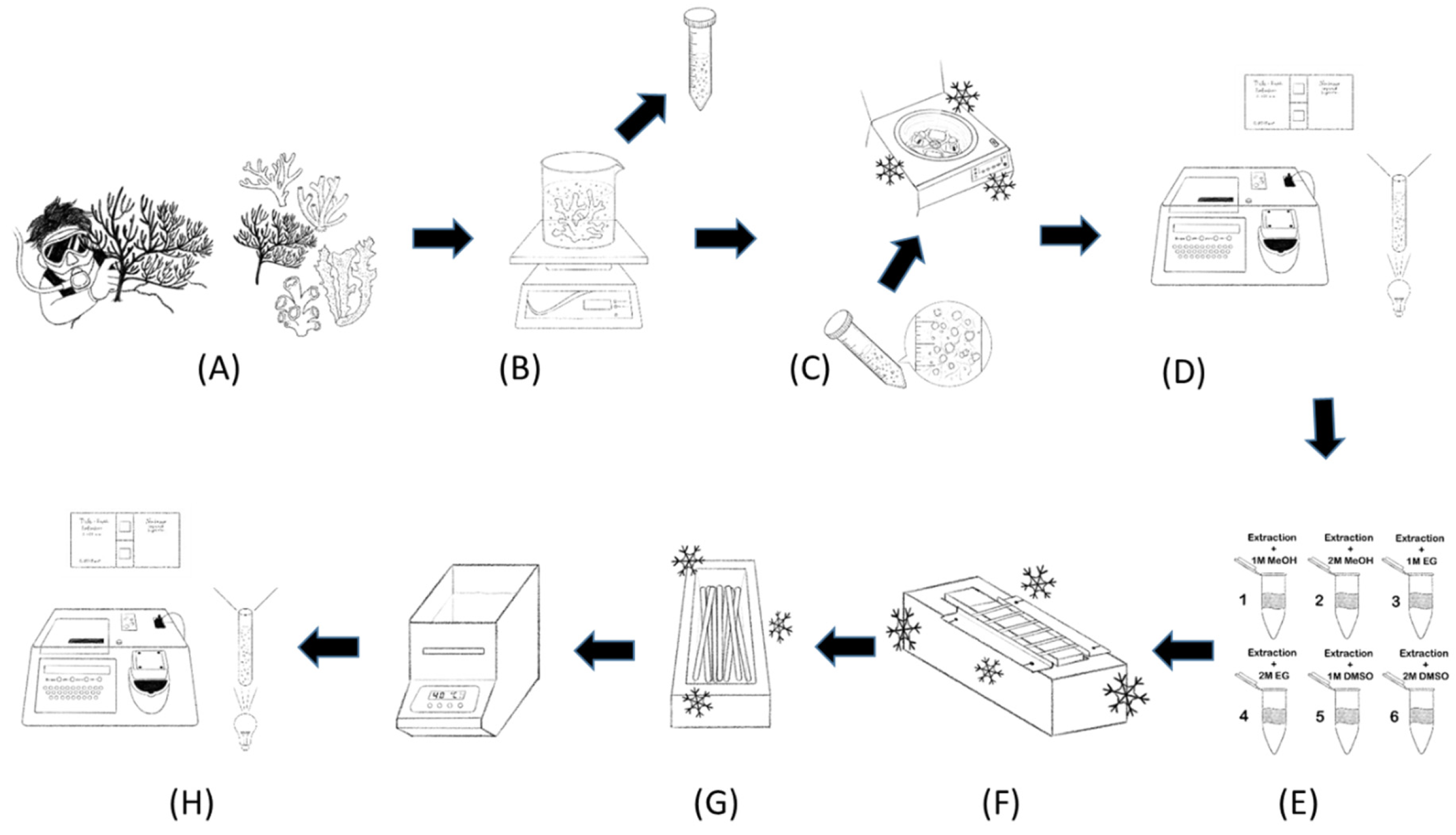
2.1. Coral Collection
2.2. Coral Identification
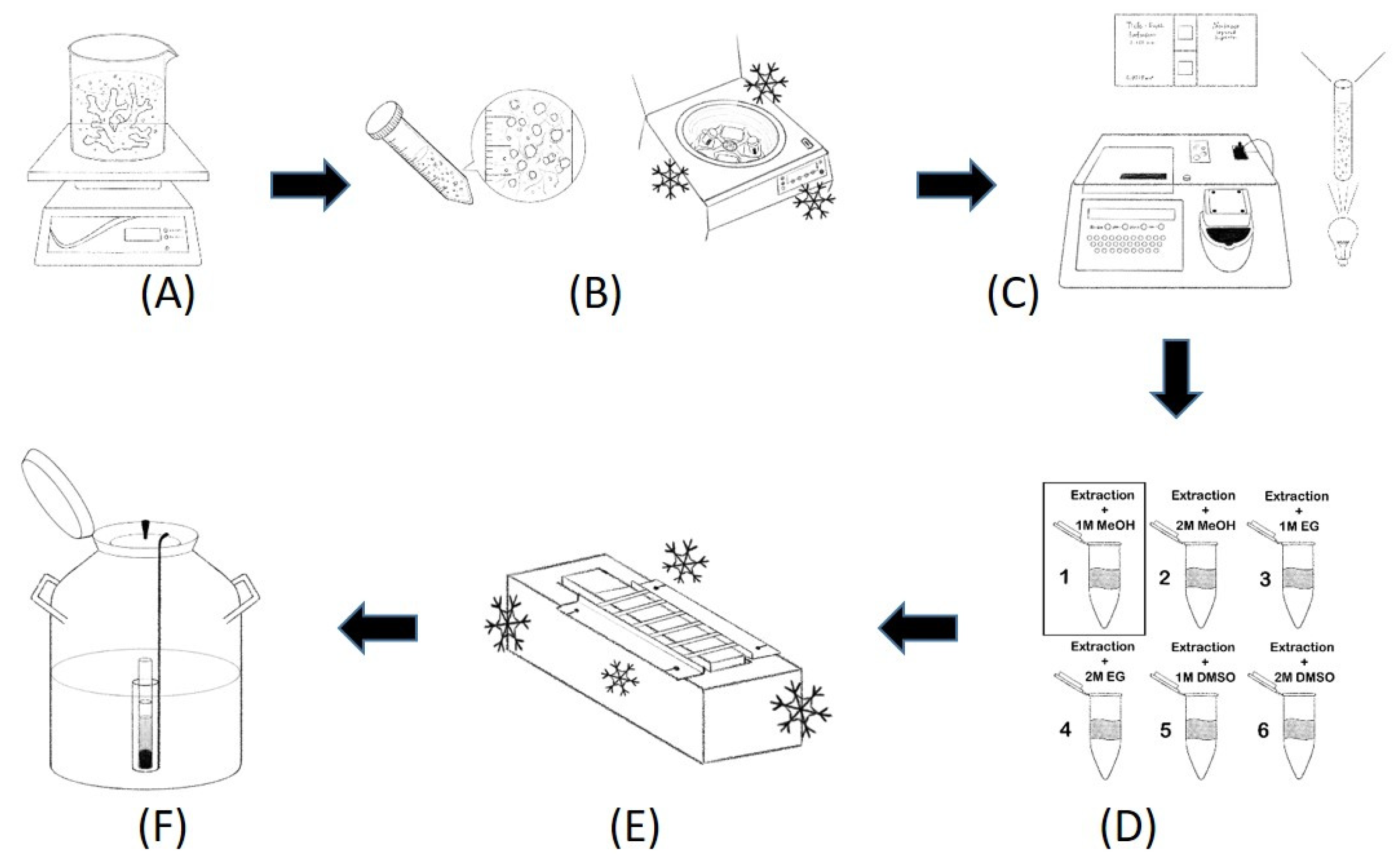
2.3. Coral Host Cell and Symbiodiniaceae Extraction
2.4. Cryopreservation
2.5. Viability Assay
2.6. Coral Cryobanking
2.7. Statistical Analysis
3. Results
3.1. Coral Cell Types
3.2. Cryopreservation of Coral Cells
3.3. Identification of Cell Types
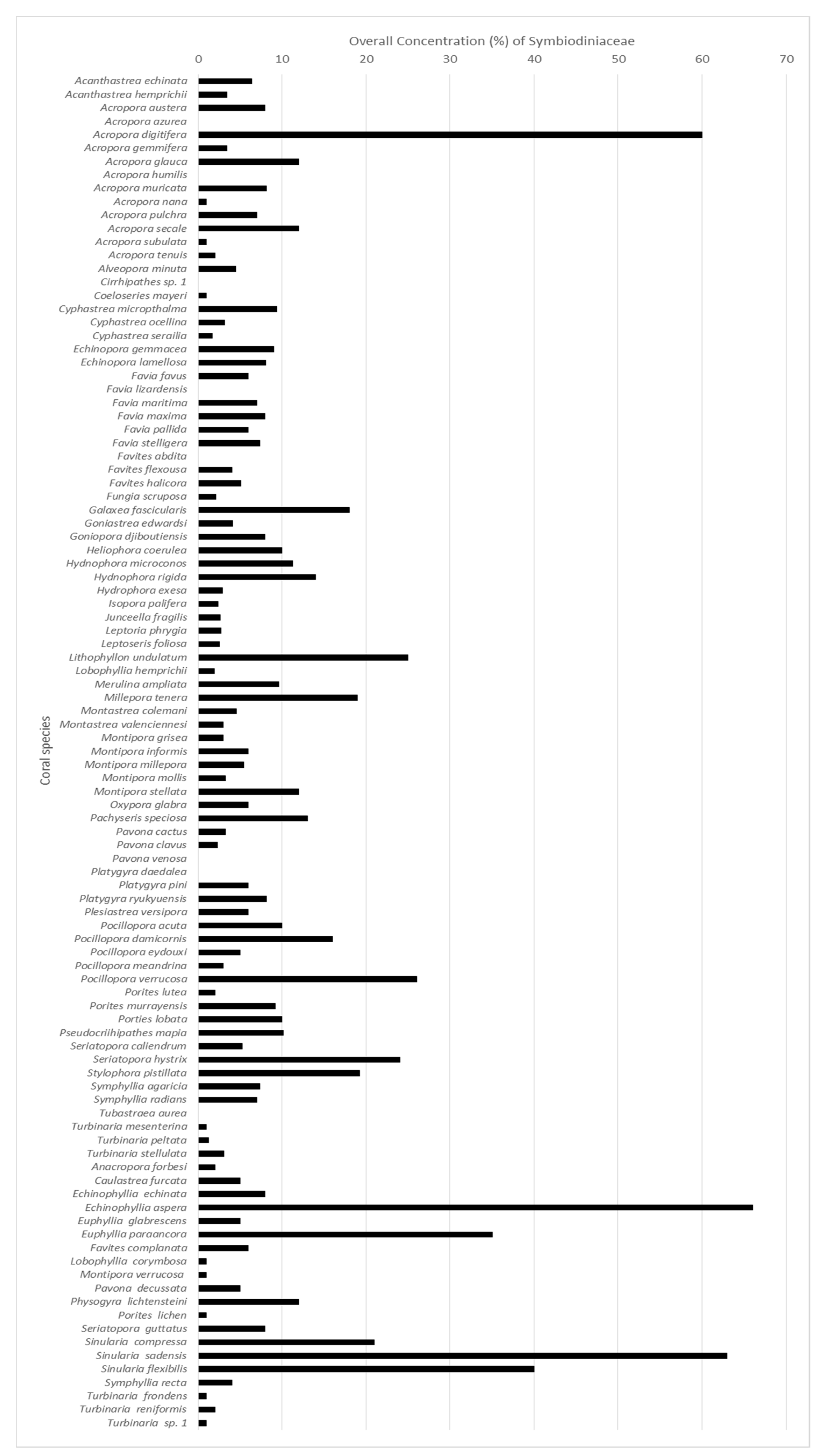
3.4. Symbiodiniaceae Concentration in Cells of Coral Species
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- McCook, L.J.; Ayling, T.; Cappo, M.; Choat, J.H.; Evans, R.D.; De Freitas, D.M.; Heupel, M.; Hughes, T.P.; Jones, G.P.; Mapstone, B.; et al. Adaptive management of the Great Barrier Reef: A globally significant demonstration of the benefits of networks of marine reserves. Proc. Natl. Acad. Sci. 2010, 107, 18278–18285. [Google Scholar] [CrossRef] [PubMed]
- Shulman, M.J. Recruitment of Coral Reef Fishes: Effects of Distribution of Predators and Shelter. Ecology 1985, 66, 1056–1066. [Google Scholar] [CrossRef]
- Lesser, M.P.; Farrell, J.H. Exposure to solar radiation increases damage to both host tissues and algal symbionts of corals during thermal stress. Coral Reefs 2004, 23, 367–377. [Google Scholar] [CrossRef]
- Nitrogen fixation in coral reef sponges with symbiotic cyanobacteria. Nature 1979, 279, 527–529. [CrossRef]
- Carr, L.; Mendelsohn, R. Valuing Coral Reefs: A Travel Cost Analysis of the Great Barrier Reef. Ambio 2003, 32, 353–357. [Google Scholar] [CrossRef]
- Cooper, E.L.; Hirabayashi, K.; Strychar, K.B.; Sammarco, P.W. Corals and Their Potential Applications to Integrative Medicine. Evidence-Based Complement. Altern. Med. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Bellwood, D.R.; Hughes, T.; Folke, C.; Nyström, M. Confronting the coral reef crisis. Nature 2004, 429, 827–833. [Google Scholar] [CrossRef]
- Feely, R.A.; Sabine, C.L.; Lee, K.; Berelson, W.; Kleypas, J.; Fabry, V.J.; Millero, F.J. Impact of Anthropogenic CO 2 on the CaCO 3 System in the Oceans. Science 2004, 305, 362–366. [Google Scholar] [CrossRef]
- Hughes, T.P.; Kerry, J.T.; Álvarez-Noriega, M.; Álvarez-Romero, J.G.; Anderson, K.D.; Baird, A.H.; Babcock, R.C.; Beger, M.; Bellwood, D.R.; Berkelmans, R.; et al. Global warming and recurrent mass bleaching of corals. Nature 2017, 543, 373–377. [Google Scholar] [CrossRef]
- Viyakarn, V.; Chavanich, S.; Chong, G.; Tsai, S.; Lin, C. Cryopreservation of sperm from the coral Acropora humilis. Cryobiology 2018, 80, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Chong, G.; Tsai, S.; Wang, L.-H.; Huang, C.-Y.; Lin, C. Cryopreservation of the gorgonian endosymbiont Symbiodinium. Sci. Rep. 2016, 6, 18816. [Google Scholar] [CrossRef]
- Comizzoli, P.; Wildt, D.E. Cryobanking biomaterials from wild animal species to conserve genes and bio-diversity: Relevance to human biobanking and biomedical research. In Biobanking of Human Biospecimens; Springer: Berlin/Heidelberg, Germany, 2017; pp. 217–235. [Google Scholar]
- Cressey, D. Coral crisis: Great Barrier Reef bleaching is “the worst we’ve ever seen”. Nature 2016. [Google Scholar] [CrossRef]
- Di Genio, S.; Wang, L.-H.; Meng, P.-J.; Tsai, S.; Lin, C. “Symbio-Cryobank”: Toward the Development of a Cryogenic Archive for the Coral Reef Dinoflagellate Symbiont Symbiodiniaceae. Biopreservation Biobanking 2021, 19, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, K. Can cloning save endangered species? Curr. Biol. 2001, 11, R245–R246. [Google Scholar] [CrossRef] [PubMed]
- A Odintsova, N.; Boroda, A. Cryopreservation of the cells and larvae of marine organisms. Russ. J. Mar. Biol. 2012, 38, 101–111. [Google Scholar] [CrossRef]
- Lin, C.; Wang, L.-H.; Meng, P.-J.; Chen, C.-S.; Tsai, S. Lipid Content and Composition of Oocytes from Five Coral Species: Potential Implications for Future Cryopreservation Efforts. PLoS ONE 2013, 8, e57823. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Tsai, S.; Lin, C. The Coral Hospital. Biopreservation Biobanking 2019, 17, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Hagedorn, M.; van Oppen, M.J.; Carter, V.; Henley, M.; Abrego, D.; Puill-Stephan, E.; Negri, A.; Heyward, A.; MacFarlane, D.; Spindler, R. First frozen repository for the Great Barrier Reef coral created. Cryobiology 2012, 65, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Li, H.-H.; Tsai, S.; Lin, C. Tissue Cryopreservation and Cryobanking: Establishment of a Cryogenic Resource for Coral Reefs. Biopreservation Biobanking 2021. [Google Scholar] [CrossRef]
- Clarke, A.G. The Frozen Ark Project: The role of zoos and aquariums in preserving the genetic material of threatened animals. Int. Zoo Yearb. 2009, 43, 222–230. [Google Scholar] [CrossRef]
- Droege, G.; Barker, K.; Seberg, O.; Coddington, J.A.; Benson, E.; Berendsohn, W.; Bunk, B.; Butler, C.; Cawsey, E.M.; Deck, J.; et al. The Global Genome Biodiversity Network (GGBN) Data Standard specification. Database 2016, 2016. [Google Scholar] [CrossRef]
- Koepfli, K.-P.; Paten, B.; O’Brien, S.J.; The Genome 10K Community of Scientists. The Genome 10K Project: A Way Forward. Annu. Rev. Anim. Biosci. 2015, 3, 57–111. [Google Scholar] [CrossRef]
- Kawamura, K.; Nishitsuji, K.; Shoguchi, E.; Fujiwara, S.; Satoh, N. Establishing Sustainable Cell Lines of a Coral, Acropora tenuis. Mar. Biotechnol. 2021, 23, 373–388. [Google Scholar] [CrossRef]
- Higuchi, R.; Bowman, B.; Freiberger, M.; Ryder, O.A.; Wilson, A.C. DNA sequences from the quagga, an extinct member of the horse family. Nature 1984, 312, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.H.; Chen, C.-K.; Ravikrishnan, A.; Rane, S.; Pfeifer, B.A. Overcoming Nonviral Gene Delivery Barriers: Perspective and Future. Mol. Pharm. 2013, 10, 4082–4098. [Google Scholar] [CrossRef] [PubMed]
- Bak, R.O.; Gomez-Ospina, N.; Porteus, M.H. Gene Editing on Center Stage. Trends Genet. 2018, 34, 600–611. [Google Scholar] [CrossRef]
- Wang, Y.; Shyy, J.Y.-J.; Chien, S. Fluorescence Proteins, Live-Cell Imaging, and Mechanobiology: Seeing Is Believing. Annu. Rev. Biomed. Eng. 2008, 10, 1–38. [Google Scholar] [CrossRef]
- Mass, T.; Drake, J.L.; Haramaty, L.; Rosenthal, Y.; Schofield, O.M.E.; Sherrell, R.M.; Falkowski, P.G. Aragonite Precipitation by “Proto-Polyps” in Coral Cell Cultures. PLoS ONE 2012, 7, e35049. [Google Scholar] [CrossRef]
- Khalesi, M.K. Cell cultures from the symbiotic soft coral Sinularia flexibilis. Vitr. 2008, 44, 330–338. [Google Scholar] [CrossRef]
- Tsai, S.; Jhuang, Y.; Spikings, E.; Sung, P.-J.; Lin, C. Ultrastructural observations of the early and late stages of gorgonian coral (Junceella juncea) oocytes. Tissue Cell 2014, 46, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Tsai, S.; Chang, W.-C.; Chavanich, S.; Viyakarn, V.; Lin, C. Ultrastructural observation of oocytes in six types of stony corals. Tissue Cell 2016, 48, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Cirino, L.; Tsai, S.; Wang, L.-H.; Chen, C.-S.; Hsieh, W.-C.; Huang, C.-L.; Wen, Z.-H.; Lin, C. Supplementation of exogenous lipids via liposomes improves coral larvae settlement post-cryopreservation and nano-laser warming. Cryobiology 2020, 98, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Ryder, O.A.; Onuma, M. Viable Cell Culture Banking for Biodiversity Characterization and Conservation. Annu. Rev. Anim. Biosci. 2018, 6, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Frank, U.; Rabinowitz, C.; Rinkevich, B. In vitro establishment of continuous cell cultures and cell lines from ten colonial cnidarians. Mar. Biol. 1994, 120, 491–499. [Google Scholar] [CrossRef]
- Kopecky, E.J.; Ostrander, G.K. Isolation and primary culture of viable multicellular endothelial isolates from hard corals. Vitr. 1999, 35, 616–624. [Google Scholar] [CrossRef]
- Domart-Coulon, I.J.; Elbert, D.C.; Scully, E.P.; Calimlim, P.S.; Ostrander, G.K. Aragonite crystallization in primary cell cultures of multicellular isolates from a hard coral, Pocillopora damicornis. Proc. Natl. Acad. Sci. USA 2001, 98, 11885–11890. [Google Scholar] [CrossRef]
- Domart-Coulon, I.J.; Sinclair, C.S.; Hill, R.T.; Puverel, S.; Ostrander, G.K. A basidiomycete isolated from the skeleton of Pocillopora damicornis (Scleractinia) selectively stimulates short-term survival of coral skeletogenic cells. Mar. Biol. 2003, 144, 583–592. [Google Scholar] [CrossRef]
- Domart-Coulon, I.; Tambutté, S.; Tambutté, E.; Allemand, D. Short term viability of soft tissue detached from the skeleton of reef-building corals. J. Exp. Mar. Biol. Ecol. 2004, 309, 199–217. [Google Scholar] [CrossRef]
- Helman, Y.; Natale, F.; Sherrell, R.M.; LaVigne, M.; Starovoytov, V.; Gorbunov, M.Y.; Falkowski, P.G. Extracellular matrix production and calcium carbonate precipitation by coral cells in vitro. Proc. Natl. Acad. Sci. USA 2008, 105, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Nesa, B.; Hidaka, M. High zooxanthella density shortens the survival time of coral cell aggregates under thermal stress. J. Exp. Mar. Biol. Ecol. 2009, 368, 81–87. [Google Scholar] [CrossRef]
- Reyes-Bermudez, A.; Miller, D.J. In vitro culture of cells derived from larvae of the staghorn coral Acropora millepora. Coral Reefs 2009, 28, 859–864. [Google Scholar] [CrossRef]
- Downs, C.A.; Fauth, J.E.; Downs, V.D.; Ostrander, G.K. In vitro cell-toxicity screening as an alternative animal model for coral toxicology: Effects of heat stress, sulfide, rotenone, cyanide, and cuprous oxide on cell viability and mitochondrial function. Ecotoxicology 2009, 19, 171–184. [Google Scholar] [CrossRef]
- Auzoux-Bordenave, S.; Domart-Coulon, I. Short review marine invertebrate cell cultures as tools for biomineralization studies. J. Sci. Hal. Aquat. 2010, 2, 42–47. [Google Scholar]
- Vizel, M.; Loya, Y.; Downs, C.A.; Kramarsky-Winter, E. A Novel Method for Coral Explant Culture and Micropropagation. Mar. Biotechnol. 2010, 13, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-L.; Chang, C.-F.; Shikina, S. Development of an in vitro tissue culture system for hammer coral (Fimbriaphyllia ancora) ovaries. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Roger, L.M.; Reich, H.G.; Lawrence, E.; Li, S.; Vizgaudis, W.; Brenner, N.; Kumar, L.; Klein-Seetharaman, J.; Yang, J.; Putnam, H.M.; et al. Applying model approaches in non-model systems: A review and case study on coral cell culture. PLoS ONE 2021, 16, e0248953. [Google Scholar] [CrossRef]
- Levy, S.; Elek, A.; Grau-Bové, X.; Menéndez-Bravo, S.; Iglesias, M.; Tanay, A.; Mass, T.; Sebé-Pedrós, A. A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell 2021, 184, 2973–2987.e18. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.J. Lipid Bodies in the Marine Endosymbiosis. Master’s thesis, National Dong Hwa University, Hualien, Taiwan, 2007; pp. 1–188. [Google Scholar]
- Östman, C.; Kultima, J.R.; Roat, C.; Rundblom, K. Acontia and mesentery nematocysts of the sea anemone Metridium senile (Linnaeus, 1761) (Cnidaria: Anthozoa). Sci. Mar. 2010, 74, 483–497. [Google Scholar] [CrossRef]
- Lin, C.; Tsai, S. Fifteen years of coral cryopreservation. Platax 2020, 2020, 53–75. [Google Scholar] [CrossRef]
- Wildt, D.E.; Rall, W.F.; Critser, J.K.; Monfort, S.L.; Seal, U.S. Genome Resource Banks. BioScience 1997, 47, 689–698. [Google Scholar] [CrossRef]
- Gwo, J.-C. Cryopreservation of aquatic invertebrate semen: A review. Aquac. Res. 2000, 31, 259–271. [Google Scholar] [CrossRef]
- Ohki, S.; Morita, M.; Kitanobo, S.; Kowalska, A.A.; Kowalski, R.K. Cryopreservation of Acropora digitifera sperm with use of sucrose and methanol based solution. Cryobiology 2014, 69, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.; Kuit, V.; Lin, Z.G.; Lin, C. Application of a functional marker for the effect of cryoprotectant agents on gorgonian coral (Junceella juncea and J. fragilis) sperm sacs. Cryoletters 2014, 35. [Google Scholar]
- Lin, C.; Zhang, T.; Kuo, F.W.; Tsai, S. Gorgonian coral (Junceella juncea and Junceella fragilis) oocyte chilling sensitivity in the context of adenosine triphosphate response (ATP). Cryoletters 2011, 32. [Google Scholar]
- Tsai, S.; Lin, C. Advantages and Applications of Cryopreservation in Fisheries Science. Braz. Arch. Biol. Technol. 2012, 55, 425–434. [Google Scholar] [CrossRef]
- Tsai, S.; Chen, J.-C.; Spikings, E.; Li, J.-J.; Lin, C. Degradation of mitochondrial DNA in cryoprotectant-treated hard coral (Echinopora spp.) oocytes. Mitochondrial DNA 2013, 26, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.; Yang, V.; Lin, C. Comparison of the cryo-tolerance of vitrified gorgonian oocytes. Sci. Rep. 2016, 6, 23290. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.; Zuchowicz, N.; Lendo, C.I.N.; Khosla, K.; Lager, C.; Henley, E.M.; Bischof, J.; Kleinhans, F.W.; Lin, C.; Peters, E.C.; et al. Successful cryopreservation of coral larvae using vitrification and laser warming. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Cirino, L.; Wen, Z.-H.; Hsieh, K.; Huang, C.-L.; Leong, Q.L.; Wang, L.-H.; Chen, C.-S.; Daly, J.; Tsai, S.; Lin, C. First instance of settlement by cryopreserved coral larvae in symbiotic association with dinoflagellates. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Cirino, L.; Tsai, S.; Wen, Z.-H.; Wang, L.-H.; Chen, H.-K.; Cheng, J.-O.; Lin, C. Lipid profiling in chilled coral larvae. Cryobiology 2021, 102, 56–67. [Google Scholar] [CrossRef]
- Tsai, S.; Spikings, E.; Kuo, F.; Lin, N.; Lin, C. Use of an adenosine triphosphate assay, and simultaneous staining with fluorescein diacetate and propidium iodide, to evaluate the effects of cryoprotectants on hard coral (Echinopora spp.) oocytes. Theriogenology 2010, 73, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.; Spikings, E.; Huang, I.-C.; Lin, C. Study on the mitochondrial activity and membrane potential after exposing later stage oocytes of two gorgonian corals (Junceella juncea and Junceella fragilis) to cryoprotectants. Cryoletters 2011, 32. [Google Scholar]
- Feuillassier, L.; Romans, P.; Engelmann-Sylvestre, I.; Masanet, P.; Barthélémy, D.; Engelmann, F. Tolerance of apexes of coral Pocillopora damicornis L. to cryoprotectant solutions. Cryobiology 2014, 68, 96–106. [Google Scholar] [CrossRef]
- Feuillassier, L.; Masanet, P.; Romans, P.; Barthélémy, D.; Engelmann, F. Towards a vitrification-based cryopreservation protocol for the coral Pocillopora damicornis L.: Tolerance of tissue balls to 4.5M cryoprotectant solutions. Cryobiology 2015, 71, 224–235. [Google Scholar] [CrossRef]
- Tsai, S.; Chong, G.; Meng, P.-J.; Lin, C. Sugars as supplemental cryoprotectants for marine organisms. Rev. Aquac. 2017, 10, 703–715. [Google Scholar] [CrossRef]
- Tsai, S.; Lin, C. Effects of cryoprotectant on the embryos of banded coral shrimp (Stenopus hispidus); preliminary studies to establish freezing protocols. Cryoletters 2009, 30. [Google Scholar]
- Zheng, Y.-J.; Ornstein, R.L. A Molecular Dynamics and Quantum Mechanics Analysis of the Effect of DMSO on Enzyme Structure and Dynamics: Subtilisin. J. Am. Chem. Soc. 1996, 118, 4175–4180. [Google Scholar] [CrossRef]
- Gurtovenko, A.A.; Anwar, J. Modulating the Structure and Properties of Cell Membranes: The Molecular Mechanism of Action of Dimethyl Sulfoxide. J. Phys. Chem. B 2007, 111, 10453–10460. [Google Scholar] [CrossRef] [PubMed]
- Thongpoo, P.; Tsai, S.; Lin, C. Assessing the impacts of cryopreservation on the mitochondria of a thermotolerant Symbiodinium lineage: Implications for reef coral conservation. Cryobiology 2019, 89, 96–99. [Google Scholar] [CrossRef]
- Lin, C.; Kuo, F.-W.; Chavanich, S.; Viyakarn, V. Membrane Lipid Phase Transition Behavior of Oocytes from Three Gorgonian Corals in Relation to Chilling Injury. PLoS ONE 2014, 9, e92812. [Google Scholar] [CrossRef]
- Desai, K.; Spikings, E.; Zhang, T. Use of methanol as cryoprotectant and its effect on sox genes and proteins in chilled zebrafish embryos. Cryobiology 2015, 71, 1–11. [Google Scholar] [CrossRef]
- Vian, A.M.; Higgins, A.Z. Membrane permeability of the human granulocyte to water, dimethyl sulfoxide, glycerol, propylene glycol and ethylene glycol. Cryobiology 2013, 68, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Hughes, S.; Fang, C.; Huang, J.; Fu, B.; Zhao, G.; Fialkow, M.; Lentz, G.; Hladik, F.; Gao, D. A study of the osmotic characteristics, water permeability, and cryoprotectant permeability of human vaginal immune cells. Cryobiology 2016, 72, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Sydykov, B.; Oldenhof, H.; Barros, L.D.O.; Sieme, H.; Wolkers, W.F. Membrane permeabilization of phosphatidylcholine liposomes induced by cryopreservation and vitrification solutions. Biochim. et Biophys. Acta (BBA) - Biomembr. 2018, 1860, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Bruckner, A.W. History of Coral Disease Research. Dis. Coral 2015, 52–84. [Google Scholar] [CrossRef]
- Hernandez-Agreda, A.; Gates, R.D.; Ainsworth, T.D. Defining the Core Microbiome in Corals’ Microbial Soup. Trends Microbiol. 2017, 25, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.E.; Bythell, J.C. Perspectives on mucus secretion in reef corals. Mar. Ecol. Prog. Ser. 2005, 296, 291–309. [Google Scholar] [CrossRef]
- Goldberg, W.M. Feeding behavior, epidermal structure and mucus cytochemistry of the scleractinian Mycetophyllia reesi, a coral without tentacles. Tissue Cell 2002, 34, 232–245. [Google Scholar] [CrossRef]
- Sleigh, M.A.; Blake, J.R.; Liron, N. The Propulsion of Mucus by Cilia. Am. Rev. Respir. Dis. 1988, 137, 726–741. [Google Scholar] [CrossRef]
- Bythell, J.C.; Wild, C. Biology and ecology of coral mucus release. J. Exp. Mar. Biol. Ecol. 2011, 408, 88–93. [Google Scholar] [CrossRef]
- Wild, C.; Huettel, M.; Klueter, A.; Kremb, S.G.; Rasheed, M.; Jorgensen, B.B. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 2004, 428, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.C. Diseases of Coral Reef Organisms. In Coral reefs in the Anthropocene; Springer: Dordrecht, The Netherland, 2015; pp. 147–178. [Google Scholar] [CrossRef]
- Guo, Q.; Whipps, C.M.; Zhai, Y.; Li, D.; Gu, Z. Quantitative Insights into the Contribution of Nematocysts to the Adaptive Success of Cnidarians Based on Proteomic Analysis. Biology 2022, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, A.; Özbek, S. The Nematocyst: A molecular map of the Cnidarian stinging organelle. Int. J. Dev. Biol. 2012, 56, 577–582. [Google Scholar] [CrossRef]
- Tardent, P. The cnidarian cnidocyte, a hightech cellular weaponry. BioEssays 1995, 17, 351–362. [Google Scholar] [CrossRef]
- Jouiaei, M.; Yanagihara, A.A.; Madio, B.; Nevalainen, T.J.; Alewood, P.F.; Fry, B.G. Ancient Venom Systems: A Review on Cnidaria Toxins. Toxins 2015, 7, 2251–2271. [Google Scholar] [CrossRef]
- Schmidt, C.A.; Daly, N.L.; Wilson, D.T. Coral Venom Toxins. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef]
- Glider, W.V.; Phipps, D.W.; Pardy, R.L. Localization of Symbiotic Dinoflagellate Cells within Tentacle Tissue of Aiptasia pallida (Coelenterata, Anthozoa). Trans. Am. Microsc. Soc. 1980, 99, 426. [Google Scholar] [CrossRef]
- Gates, R.D.; Baghdasarian, G.; Muscatine, L. Temperature Stress Causes Host Cell Detachment in Symbiotic Cnidarians: Implications for Coral Bleaching. Biol. Bull. 1992, 182, 324–332. [Google Scholar] [CrossRef]
- Westfall, J.A. Ultrastructure of synapses in the first-evolved nervous systems. J. Neurocytol. 1996, 25, 735–746. [Google Scholar] [CrossRef]
- Berzins, I.K.; Yanong, R.P.E.; LaDouceur, E.E.; Peters, E.C. Cnidaria. Invertebr. Histol. 2021, 55–86. [Google Scholar] [CrossRef]
- Hu, M.; Zheng, X.; Fan, C.-M.; Zheng, Y. Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia. Nature 2020, 582, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Rosental, B.; Kozhekbaeva, Z.; Fernhoff, N.; Tsai, J.M.; Traylor-Knowles, N. Coral cell separation and isolation by fluorescence-activated cell sorting (FACS). BMC Cell Biol. 2017, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Snyder, G.A.; Browne, W.; Traylor-Knowles, N.; Rosental, B. Fluorescence-Activated Cell Sorting for the Isolation of Scleractinian Cell Populations. J. Vis. Exp. 2020. [Google Scholar] [CrossRef]
- Giese, W.; Eigel, M.; Westerheide, S.; Engwer, C.; Klipp, E. Influence of cell shape, inhomogeneities and diffusion barriers in cell polarization models. Phys. Biol. 2015, 12, 066014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelstein-Keshet, L.; Holmes, W.R.; Zajac, M.; Dutot, M. From simple to detailed models for cell polarization. Philos. Trans. R. Soc. B: Biol. Sci. 2013, 368, 20130003. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.-L.; Pasaribu, B.; Chen, C.-S. Nitrogen-Deprivation Elevates Lipid Levels in Symbiodinium spp. by Lipid Droplet Accumulation: Morphological and Compositional Analyses. PLoS ONE 2014, 9, e87416. [Google Scholar] [CrossRef]
- Weng, L.-C.; Pasaribu, B.; Lin, I.-P.; Tsai, C.-H.; Chen, C.-S.; Jiang, P.-L. Nitrogen Deprivation Induces Lipid Droplet Accumulation and Alters Fatty Acid Metabolism in Symbiotic Dinoflagellates Isolated from Aiptasia pulchella. Sci. Rep. 2014, 4, srep05777. [Google Scholar] [CrossRef]
- Elkhateeb, A.; El-Beih, A.A.; Gamal-Eldeen, A.M.; Alhammady, M.A.; Ohta, S.; Paré, P.W.; Hegazy, M.-E.F. New Terpenes from the Egyptian Soft Coral Sarcophyton ehrenbergi. Mar. Drugs 2014, 12, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.-E.F.; Gamal-Eldeen, A.M.; Mohamed, T.A.; Alhammady, M.A.; Hassanien, A.A.; Shreadah, M.A.; Abdelgawad, I.I.; Elkady, E.M.; Paré, P.W. New cytotoxic constituents from the Red Sea soft coral Nephthea sp. Nat. Prod. Res. 2015, 30, 1266–1272. [Google Scholar] [CrossRef]
- Lin, C.; Chong, G.; Wang, L.; Kuo, F.; Tsai, S. Use of luminometry and flow cytometry for evaluating the effects of cryoprotectants in the gorgonian coral endosymbiont Symbiodinium. Phycol. Res. 2019, 67, 320–326. [Google Scholar] [CrossRef]
- Wang, S.-C.; Li, R.-N.; Lin, L.-C.; Tang, J.-Y.; Su, J.-H.; Sheu, J.-H.; Chang, H.-W. Comparison of Antioxidant and Anticancer Properties of Soft Coral-Derived Sinularin and Dihydrosinularin. Molecules 2021, 26, 3853. [Google Scholar] [CrossRef]
- Nowotny, J.D.; Connelly, M.T.; Traylor-Knowles, N. Novel methods to establish whole-body primary cell cultures for the cnidarians Nematostella vectensis and Pocillopora damicornis. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Grosso-Becerra, M.V.; Mendoza-Quiroz, S.; Maldonado, E.; Banaszak, A.T. Cryopreservation of sperm from the brain coral Diploria labyrinthiformis as a strategy to face the loss of corals in the Caribbean. Coral Reefs 2021, 40, 937–950. [Google Scholar] [CrossRef]
- Karl, D.; Holm-Hansen, O. Effects of luciferin concentration of the quantitative assay of ATP using crude luciferase preparations. Anal. Biochem. 1976, 75, 100–112. [Google Scholar] [CrossRef]
- Qureshi, A.A.; Patel, J. Adenosine Triphosphate (ATP) Levels in Microbial Cultures and a Review of the ATP Biomass Estima-tion Technique; Scientific Series; Environment Canada, Inland Waters Directorate: Burlington, ON, Canada, 1976; Volume 63, pp. 1–33. [Google Scholar]
- Sharon, G.; Rosenberg, E. Bacterial Growth on Coral Mucus. Curr. Microbiol. 2008, 56, 481–488. [Google Scholar] [CrossRef]
- Keshavmurthy, S.; Kuo, C.-Y.; Huang, Y.-Y.; Carballo-Bolaños, R.; Meng, P.-J.; Wang, J.-T.; Chen, C.A. Coral Reef Resilience in Taiwan: Lessons from Long-Term Ecological Research on the Coral Reefs of Kenting National Park (Taiwan). J. Mar. Sci. Eng. 2019, 7, 388. [Google Scholar] [CrossRef]
- Strychar, K.B.; Sammarco, P.W. Exaptation in corals to high seawater temperatures: Low concentrations of apoptotic and necrotic cells in host coral tissue under bleaching conditions. J. Exp. Mar. Biol. Ecol. 2009, 369, 31–42. [Google Scholar] [CrossRef]
- Yakovleva, I.M.; Baird, A.H.; Yamamoto, H.H.; Bhagooli, R.; Nonaka, M.; Hidaka, M. Algal symbionts increase oxidative damage and death in coral larvae at high temperatures. Mar. Ecol. Prog. Ser. 2009, 378, 105–112. [Google Scholar] [CrossRef]
- Jones, R.J.; A Ward, S.; Amri, A.Y.; Hoegh-Guldberg, O. Changes in quantum efficiency of Photosystem II of symbiotic dinoflagellates of corals after heat stress, and of bleached corals sampled after the 1998 Great Barrier Reef mass bleaching event. Mar. Freshw. Res. 2000, 51, 63–71. [Google Scholar] [CrossRef]

| (a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No | Genus | Species | Shape | Suitable CPA | Viability Rate (%) | Equilibrium Time (min) | Extraction Duration (min) | No of Straws | ANOVA |
| 1 | Acanthastrea | echinata | Massive | 1M EG | 6 ± 0.8 | 20 | 30 | 31 | F12,26 = 2.259, p < 0.05 |
| 2 | Acanthastrea | hemprichii | Massive | 1M DMSO | 9 ± 1.7 | 10 | 30 | 16 | F12,26 = 29.722, p < 0.001 |
| 3 | Acropora | austera | Branching | 1 M MeOH | 15 ± 1.3 | 10 | 30 | 16 | F12,26 = 50.236, p < 0.001 |
| 4 | Acropora | azurea | Branching | 1M EG | 8 ± 0.5 | 10 | 30 | 15 | F12,26 = 102.923, p < 0.001 |
| 5 | Acropora | digitifera | Branching | 2 M DMSO | 16 ± 1.0 | 10 | 30 | 15 | F12,26 = 9.342, p < 0.001 |
| 6 | Acropora | gemnifera | Branching | 1M EG | 25 ± 7.7 | 10 | 35 | 15 | F12,26 = 60.145, p < 0.001 |
| 7 | Acropora | glauca | Branching | 1M DMSO | 29 ± 0.0 | 20 | 30 | 16 | F12,26 = 43.432, p < 0.001 |
| 8 | Acropora | humilis | Branching | 2M DMSO | 18 ± 4.8 | 10 | 30 | 15 | F12,26 = 84.850, p < 0.001 |
| 9 | Acropora | muricata | Branching | 2M MeOH | 41 ± 12.6 | 20 | 30 | 47 | F12,26 = 5.144, p < 0.001 |
| 10 | Acropora | nana | Branching | 2M MeOH | 38 ± 8.3 | 20 | 30 | 47 | F12,26 = 79.298, p < 0.001 |
| 11 | Acropora | pulchra | Branching | 1M MeOH | 22 ± 0.3 | 20 | 30 | 16 | F12,26 = 239.681, p < 0.001 |
| 12 | Acropora | secale | Branching | 1M EG | 33 ± 0.1 | 20 | 30 | 16 | F12,26 = 25.654, p < 0.001 |
| 13 | Acropora | subulata | Branching | 1M EG | 7 ± 0.4 | 20 | 30 | 16 | F12,26 = 749.775, p < 0.001 |
| 14 | Acropora | tenuis | Branching | 1M DMSO | 49 ± 1.2 | 10 | 30 | 16 | F12,26 = 70.120, p < 0.001 |
| 15 | Alveopora | minuta | Massive | 2M MeOH | 25 ± 9.5 | 20 | 30 | 15 | F12,26 = 6.152, p < 0.001 |
| 16 | Cirrhipathes | sp 1 | Spines | 1M DMSO | 13 ± 2.4 | 20 | 30 | 0 | F12,26 = 554.545, p < 0.001 |
| 17 | Coeloseries | mayeri | Massive | 2M MeOH | 7 ± 1.5 | 20 | 30 | 15 | F12,26 = 5.749, p < 0.001 |
| 18 | Cyphastrea | micropthalma | Encrusting | 1M DMSO | 28 ± 9.6 | 20 | 30 | 16 | F12,26 = 2.377, p < 0.05 |
| 19 | Cyphastrea | Ocellina | Encrusting | 2M EG or 1M DMSO | 56 ± 12.0 | 20 | 30 | 16 | F12,26 = 21.547, p < 0.001 |
| 20 | Cyphastrea | serailia | Massive | 2M DMSO | 75 ± 15.9 | 20 | 30 | 16 | F12,26 = 3.037, p < 0.05 |
| 21 | Echinopora | gemmacea | Encrusting | 1M MeOH | 8 ± 1.5 | 10 | 60 | 16 | F12,26 = 103.324, p < 0.001 |
| 22 | Echinopora | lamellosa | Foliaceous | 2M EG | 13 ± 4.7 | 20 | 60 | 16 | F12,26 = 62.706, p < 0.001 |
| 23 | Favia | favus | Massive | 1M DMSO | 16 ± 1.8 | 10 | 30 | 15 | F12,26 = 25.545, p < 0.001 |
| 24 | Favia | lizardensis | Massive | 1M DMSO | 27 ± 13.6 | 20 | 30 | 31 | F12,26 = 8.990, p < 0.001 |
| 25 | Favia | maritima | Massive | 1M DMSO | 63 ± 15.4 | 10 | 30 | 15 | F12,26 = 21.565, p < 0.001 |
| 26 | Favia | maxima | Massive | 1M DMSO | 60 ± 24.7 | 20 | 30 | 16 | F12,26 = 76.453, p < 0.001 |
| 27 | Favia | pallida | Massive | 1M DMSO | 53 ± 21.5 | 20 | 30 | 15 | F12,26 = 2.511, p < 0.05 |
| 28 | Favia | stelligera | Massive | 2M DMSO | 96 ± 15.0 | 10 | 30 | 15 | F12,26 = 111.196, p < 0.001 |
| 29 | Favites | abdita | Massive | 1M DMSO | 6 ± 0.5 | 10 | 30 | 30 | F12,26 = 3209.843, p < 0.001 |
| 30 | Favites | flexousa | Massive | 1M EG | 73 ± 32.5 | 20 | 30 | 16 | F12,26 = 27.451, p < 0.001 |
| 31 | Favites | halicora | Massive | 1M DMSO | 34 ± 8.6 | 10 | 30 | 16 | F12,26 = 21.884, p < 0.001 |
| 32 | Fungia | scruposa | Massive | 1M MeOH | 10 ± 1.0 | 20 | 30 | 30 | F12,26 = 458.454, p < 0.001 |
| 33 | Galaxea | fascicularis | Massive | 2M DMSO | 10 ± 5.7 | 10 | 30 | 14 | F12,26 = 13.064, p < 0.001 |
| 34 | Goniastrea | edwardsi | Massive | 1M DMSO | 9 ± 0.2 | 10 | 30 | 47 | F12,26 = 4818.927, p < 0.001 |
| 35 | Goniopora | djiboutiensis | Massive | 1M MeOH | 39 ± 0.1 | 10 | 30 | 16 | F12,26 = 6.654, p < 0.001 |
| 36 | Heliopora | coerulea | Massive | 1M EG | 49 ± 5.0 | 10 | 76 | 16 | F12,26 = 49.548, p < 0.001 |
| 37 | Hydnophora | exesa | Massive | 1M DMSO | 50 ± 10.5 | 20 | 30 | 16 | F12,26 = 47.386, p < 0.001 |
| 38 | Hydnophora | microconos | Branching | 2M MeOH | 19 ± 5.8 | 10 | 30 | 15 | F12,26 = 2.295, p < 0.05 |
| 39 | Hydnophora | rigida | Encrusting | 1M EG | 12 ± 1.6 | 10 | 30 | 15 | F12,26 = 203.269, p < 0.001 |
| 40 | Isopora | palifera | Laminar | 2M DMSO | 18 ± 6.7 | 10 | 35 | 16 | F12,26 = 53.385, p < 0.001 |
| 41 | Junceella | fragilis | Columnar | 1M EG | 12 ± 1.8 | 20 | 30 | 15 | F12,26 = 517.381, p < 0.001 |
| 42 | Leptoria | phrygia | Massive | 1M DMSO | 35 ± 8.3 | 20 | 30 | 16 | F12,26 = 8.106, p < 0.001 |
| 43 | Leptoseries | foliosa | Encrusting | 1M DMSO | 50 ± 3.5 | 20 | 30 | 15 | F12,26 = 22.000, p < 0.001 |
| 44 | Lithophyllon | undulatum | Encrusting | 1M DMSO | 66 ± 17.3 | 20 | 35 | 16 | F12,26 = 54.412, p < 0.001 |
| 45 | Lobophyllia | hemprichii | Massive | 2M DMSO | 55 ± 17.5 | 20 | 30 | 16 | F12,26 = 10.507, p < 0.001 |
| 46 | Merulina | ampliata | Foliaceous | 1M DMSO | 40 ± 11.6 | 10 | 30 | 15 | F12,26 = 140.370, p < 0.001 |
| 47 | Millepora | tenera | Branching | 1M MeOH | 21 ± 1.8 | 20 | 35 | 30 | F12,26 = 28.980, p < 0.001 |
| 48 | Montastrea | colemani | Encrusting | 1M EG | 41 ± 7.0 | 10 | 30 | 30 | F12,26 = 49.303, p < 0.001 |
| 49 | Montastrea | valenciennesi | Massive | 1M MeOH | 57 ± 16.8 | 10 | 30 | 16 | F12,26 = 4.168, p = 0.001 |
| 50 | Montipora | grisea | Foliaceous | 1M DMSO | 8 ± 2.8 | 10 | 30 | 16 | F12,26 = 26.440, p < 0.001 |
| 51 | Montipora | informis | Encrusting | 1M DMSO | 2 ± 0.4 | 10 | 30 | 15 | F12,26 = 48.325, p < 0.001 |
| 52 | Montipora | millepora | Foliaceous | 1M MeOH | 10 ± 4.7 | 20 | 30 | 15 | F12,26 = 18.521, p < 0.001 |
| 53 | Montipora | mollis | Foliaceous | 1M DMSO | 25 ± 3.2 | 10 | 30 | 16 | F12,26 = 66.964, p < 0.001 |
| 54 | Montipora | stellata | Branching | 1M DMSO | 12 ± 2.3 | 10 | 30 | 15 | F12,26 = 15.343, p < 0.001 |
| 55 | Oxypora | glabra | Encrusting | 1M DMSO | 28 ± 4.3 | 10 | 30 | 15 | F12,26 = 53.656, p < 0.001 |
| 56 | Pachyseris | speciosa | Encrusting | 1M DMSO | 23 ± 6.2 | 10 | 30 | 15 | F12,26 = 15.573, p < 0.001 |
| 57 | Pavona | cactus | Massive | 1M DMSO | 16 ± 7.7 | 20 | 40 | 46 | F12,26 = 127.857, p < 0.001 |
| 58 | Pavona | clavus | Branching | 2M EG or 1M DMSO | 53 ± 8.9 | 20 and 10 | 30 | 15 | F12,26 = 18.187, p < 0.001 |
| 59 | Pavona | venosa | Columnar | 1M DMSO | 17 ± 18.5 | 10 | 30 | 31 | F12,26 = 39.809, p < 0.001 |
| 60 | Platygyra | daedalea | Massive | 2M DMSO | 9 ± 1.1 | 10 | 45 | 15 | F12,26 = 9.891, p < 0.001 |
| 61 | Platygyra | pini | Massive | 1M MeOH | 82 ± 11.5 | 10 | 30 | 15 | F12,26 = 39.283, p < 0.001 |
| 62 | Platygyra | ryukyuensis | Massive | 1M MeOH | 29 ± 9.3 | 10 | 30 | 45 | F12,26 = 1.866, p < 0.001 |
| 63 | Plesiastrea | versipora | Massive | 1M DMSO | 11 ± 1.7 | 10 | 30 | 30 | F12,26 = 396.433, p < 0.001 |
| 64 | Pocillopora | acuta | Branching | 1M DMSO | 9 ± 0.5 | 10 | 30 | 15 | F12,26 = 6.448, p < 0.001 |
| 65 | Pocillopora | damicornis | Branching | 1M DMSO | 21 ± 2.9 | 10 | 40 | 31 | F12,26 = 410.839, p < 0.001 |
| 66 | Pocillopora | eydouxi | Branching | 1M DMSO | 3 ± 1.1 | 20 | 50 | 16 | F12,26 = 18.488, p < 0.001 |
| 67 | Pocillopora | meandrina | Branching | 1M DMSO | 6 ± 1.1 | 10 | 50 | 16 | F12,26 = 65.020, p < 0.001 |
| 68 | Pocillopora | verrucosa | Branching | 1M DMSO | 16 ± 4.3 | 10 | 30 | 16 | F12,26 = 49.115, p < 0.001 |
| 69 | Porites | lobata | Massive | 2M DMSO | 17 ± 1.4 | 10 | 30 | 16 | F12,26 = 373.436, p < 0.001 |
| 70 | Porites | lutea | Massive | 1M MeOH | 25 ± 4.1 | 20 | 30 | 16 | F12,26 = 84.609, p < 0.001 |
| 71 | Porites | murrayensis | Massive | 1M DMSO | 10 ± 3.2 | 20 | 30 | 16 | F12,26 = 20.322, p < 0.001 |
| 72 | Pseudocriihipathes | mapia | Columnar | 2M DMSO | 23 ± 4.1 | 10 | 40 | 16 | F12,26 = 157.210, p < 0.001 |
| 73 | Seriatopora | caliendrum | Branching | 1M DMSO | 23 ± 7.0 | 20 | 30 | 16 | F12,26 = 66.962, p < 0.001 |
| 74 | Seriatopora | hystrix | Branching | 1 M MeOH | 63 ± 0.1 | 10 | 30 | 16 | F12,26 = 8.843, p < 0.001 |
| 75 | Stylophora | pistillata | Branching | 1M EG | 17 ± 5.5 | 10 | 75 | 16 | F12,26 = 58.211, p < 0.001 |
| 76 | Symphyllia | agaricia | Massive | 2M MeOH | 8 ± 1.5 | 10 | 35 | 16 | F12,26 = 71.498, p < 0.001 |
| 77 | Symphyllia | radians | Massive | 2M DMSO | 71 ± 15.3 | 10 | 30 | 30 | F12,26 = 2.322, p < 0.05 |
| 78 | Tubastraea | aurea | Massive | 1M DMSO | 12 ± 1.5 | 10 | 30 | 16 | F12,26 = 126.703, p < 0.001 |
| 79 | Turbinaria | mesenterina | Foliaceous | 1M EG | 12 ± 2.6 | 10 | 30 | 15 | F12,26 = 58.834, p < 0.001 |
| 80 | Turbinaria | peltata | Foliaceous | 2M EG | 29 ± 4.8 | 10 | 30 | 32 | F12,26 = 194.098, p < 0.001 |
| 81 | Turbinaria | stellulata | Foliaceous | 1M DMSO | 77 ± 43 | 20 | 30 | 15 | F12,26 = 4.270, p = 0.001 |
| (b) | |||||||||
| No | Genus | Species | Shape | Suitable CPA | Viability rate (%) | Equilibrium time (min) | Extraction duration (min) | No of straws | ANOVA |
| 1 | Anacropora | forbesi | Columnar | 2 M EG | 39 ± 8.5 | 10 | 30 | 15 | F12,26 = 2.905, p < 0.05 |
| 2 | Caulastrea | furcata | Massive | 1M MeOH | 22 ± 1.3 | 10 | 30 | 47 | F12,26 = 832.591, p < 0.001 |
| 3 | Echinophyllia | aspera | Laminar | 1M EG | 41 ± 12.3 | 10 | 30 | 30 | F12,26 = 28.475, p < 0.001 |
| 4 | Echinophyllia | echinata | Massive | 1M DMSO | 39 ± 5.6 | 10 | 30 | 31 | F12,26 = 211.678, p < 0.001 |
| 5 | Euphyllia | glabrescens | Branching | 1 M EG | 3 ± 0.6 | 10 | 30 | 15 | F12,26 = 2405.531, p < 0.001 |
| 6 | Euphyllia | paraancora | Branching | 1M EG | 23 ± 0.7 | 10 | 30 | 29 | F12,26 = 244.752, p < 0.001 |
| 7 | Favites | complanata | Columnar | 1M DMSO | 12 ± 1.6 | 10 | 30 | 32 | F12,26 = 623.795, p < 0.001 |
| 8 | Lobophyllia | corymbosa | Massive | 1M DMSO | 28 ± 7.8 | 10 | 30 | 32 | F12,26 = 45.394, p < 0.001 |
| 9 | Montipora | verrucosa | Foliaceous | 1M MeOH | 20 ± 5.0 | 20 | 30 | 16 | F12,26 = 73.686, p < 0.001 |
| 10 | Pavona | decussata | Foliaceous | 1M DMSO | 36 ± 12 | 10 | 30 | 15 | F12,26 = 103.974, p < 0.001 |
| 11 | Physogyra | lichtensteini | Massive | 1M MeOH | 19 ± 6.4 | 10 | 30 | 15 | F12,26 = 179.699, p < 0.001 |
| 12 | Porites | lichen | Foliaceous | 2M EG | 21 ± 1.6 | 10 | 30 | 32 | F12,26 = 165.086, p < 0.001 |
| 13 | Seriatopora | guttatus | Branching | 1M EG | 17 ± 3.5 | 10 | 30 | 16 | F12,26 = 298.055, p < 0.001 |
| 14 | Sinularia | compressa | Branching | 1M EG | 17 ± 2.4 | 10 | 30 | 16 | F12,26 = 481.321, p < 0.001 |
| 15 | Sinularia | flexibilis | Branching | 1M EG | 13 ± 1.6 | 10 | 30 | 30 | F12,26 = 1652.858, p < 0.001 |
| 16 | Sinularia | sadensis | Branching | 2M EG | 24 ± 3.5 | 10 | 30 | 15 | F12,26 = 434.807, p < 0.001 |
| 17 | Symphyllia | recta | Massive | 1M DMSO | 18 ± 4.4 | 20 | 30 | 16 | F12,26 = 19.750, p < 0.001 |
| 18 | Turbinaria | frondens | Branching | 1M EG | 8 ± 2.5 | 20 | 30 | 32 | F12,26 = 82.402, p < 0.001 |
| 19 | Turbinaria | reniformis | Foliaceous | 2M MeOH | 25 ± 3.4 | 20 | 30 | 16 | F12,26 = 41.255, p < 0.001 |
| 20 | Turbinaria | sp 1 | Foliaceous | 1M MeOH | 27 ± 2.9 | 10 | 35 | 31 | F12,26 = 244.116, p < 0.001 |
| (a) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Types Identification | ||||||||||
| No | Genus | Species | Gland Cell | Supporting Cell | Epidermal Nematocyst | Symbiodiniaceae | Symbiotic Endoderm Cell (SEC) | Spindle Cell | Cluster Cell | Gastrodermal Nematocyst |
| 1 | Acanthastrea | echinata | ● | ● | ● | ● | ● | |||
| 2 | Acanthastrea | hemprichii | ● | ● | ● | ● | ● | ● | ||
| 3 | Acropora | austera | ● | ● | ● | ● | ● | |||
| 4 | Acropora | azurea | ● | ● | ● | ● | ● | ● | ● | ● |
| 5 | Acropora | digitifera | ● | ● | ● | |||||
| 6 | Acropora | gemnifera | ● | ● | ● | ● | ● | ● | ||
| 7 | Acropora | glauca | ● | ● | ● | ● | ● | ● | ||
| 8 | Acropora | humilis | ● | ● | ● | ● | ● | ● | ● | |
| 9 | Acropora | muricata | ● | ● | ● | ● | ● | |||
| 10 | Acropora | nana | ● | ● | ● | ● | ● | |||
| 11 | Acropora | pulchra | ● | ● | ● | ● | ● | |||
| 12 | Acropora | secale | ● | ● | ● | ● | ● | |||
| 13 | Acropora | subulata | ● | ● | ● | ● | ● | ● | ||
| 14 | Acropora | tenuis | ● | ● | ● | ● | ● | ● | ● | ● |
| 15 | Alveopora | minuta | ● | ● | ● | ● | ● | ● | ● | |
| 16 | Cirrhipathes | sp 1 | ● | ● | ● | ● | ● | ● | ||
| 17 | Coeloseries | mayeri | ● | ● | ● | ● | ● | |||
| 18 | Cyphastrea | micropthalma | ● | ● | ● | ● | ● | |||
| 19 | Cyphastrea | Ocellina | ● | ● | ● | ● | ● | |||
| 20 | Cyphastrea | serailia | ● | ● | ● | ● | ● | ● | ||
| 21 | Echinopora | gemmacea | ● | ● | ● | ● | ● | ● | ● | |
| 22 | Echinopora | lamellosa | ● | ● | ● | ● | ● | |||
| 23 | Favia | favus | ● | ● | ● | ● | ● | ● | ||
| 24 | Favia | lizardensis | ● | ● | ● | ● | ● | ● | ● | |
| 25 | Favia | maritima | ● | ● | ● | ● | ● | ● | ● | ● |
| 26 | Favia | maxima | ● | ● | ● | ● | ● | |||
| 27 | Favia | pallida | ● | ● | ● | ● | ||||
| 28 | Favia | stelligera | ● | ● | ● | ● | ● | ● | ||
| 29 | Favites | abdita | ● | ● | ● | ● | ||||
| 30 | Favites | flexousa | ● | ● | ● | ● | ● | ● | ||
| 31 | Favites | halicora | ● | ● | ● | ● | ● | ● | ● | |
| 32 | Fungia | seruposa | ● | ● | ● | ● | ● | |||
| 33 | Galaxea | fascicularis | ● | ● | ● | ● | ● | ● | ● | |
| 34 | Goniopora | djiboutiensis | ● | ● | ● | ● | ● | ● | ||
| 35 | Goniastrea | edwardsi | ● | ● | ● | ● | ● | ● | ● | |
| 36 | Heliopora | coerulea | ● | ● | ● | ● | ● | ● | ● | ● |
| 37 | Hydnophora | exesa | ● | ● | ● | ● | ● | ● | ||
| 38 | Hydnophora | microconos | ● | ● | ● | ● | ● | ● | ||
| 39 | Hydnophora | rigida | ● | ● | ● | ● | ● | ● | ● | |
| 40 | Isopora | palifera | ● | ● | ● | |||||
| 41 | Junceella | fragilis | ● | ● | ● | ● | ● | |||
| 42 | Leptoria | phrygia | ● | ● | ● | ● | ● | ● | ||
| 43 | Leptoseries | foliosa | ● | ● | ● | ● | ● | |||
| 44 | Lithophyllon | undulatum | ● | ● | ● | ● | ● | ● | ||
| 45 | Lobophyllia | hemprichii | ● | ● | ● | ● | ● | ● | ||
| 46 | Merulina | ampliata | ● | ● | ● | ● | ● | ● | ||
| 47 | Millepora | tenera | ● | ● | ● | ● | ● | ● | ||
| 48 | Montastrea | colemani | ● | ● | ● | ● | ● | |||
| 49 | Montipora | grisea | ● | ● | ● | ● | ● | |||
| 50 | Montipora | informis | ● | ● | ● | ● | ||||
| 51 | Montipora | millepora | ● | ● | ● | ● | ● | |||
| 52 | Montipora | mollis | ● | ● | ● | ● | ||||
| 53 | Montipora | stellata | ● | ● | ● | ● | ● | ● | ● | |
| 54 | Montastrea | valenciennesi | ● | ● | ● | ● | ● | ● | ||
| 55 | Oxypora | glabra | ● | ● | ● | ● | ● | ● | ● | |
| 56 | Pachyseris | speciosa | ● | ● | ● | ● | ● | ● | ● | |
| 57 | Pavona | cactus | ● | ● | ● | ● | ● | ● | ||
| 58 | Pavona | clavus | ● | ● | ● | ● | ● | ● | ● | ● |
| 59 | Pavona | venosa | ● | ● | ● | ● | ● | ● | ||
| 60 | Platygyra | daedalea | ● | ● | ● | ● | ● | ● | ||
| 61 | Platygyra | pini | ● | ● | ● | ● | ||||
| 62 | Platygyra | ryukyuensis | ● | ● | ● | ● | ● | ● | ● | ● |
| 63 | Plesiastrea | versipora | ● | ● | ● | ● | ● | ● | ● | |
| 64 | Pocillopora | acuta | ● | ● | ● | ● | ● | ● | ● | ● |
| 65 | Pocillopora | damicornis | ● | ● | ● | ● | ● | ● | ||
| 66 | Pocillopora | eydouxi | ● | ● | ● | ● | ● | ● | ● | |
| 67 | Pocillopora | meandrina | ● | ● | ● | ● | ● | ● | ● | |
| 68 | Pocillopora | verrucosa | ● | ● | ● | ● | ● | ● | ● | |
| 69 | Porites | lobata | ● | ● | ● | ● | ● | ● | ● | |
| 70 | Porites | lutea | ● | ● | ● | ● | ● | ● | ● | |
| 71 | Porites | murrayensis | ● | ● | ● | ● | ● | ● | ||
| 72 | Pseudocriihipathes | mapia | ● | ● | ● | ● | ● | ● | ● | |
| 73 | Seriatopora | caliendrum | ● | ● | ● | ● | ● | |||
| 74 | Seriatopora | hystrix | ● | ● | ● | ● | ● | ● | ||
| 75 | Stylophora | pistillata | ● | ● | ● | ● | ||||
| 76 | Symphyllia | agaricia | ● | ● | ● | ● | ● | ● | ||
| 77 | Symphyllia | radians | ● | ● | ● | ● | ||||
| 78 | Tubastraea | aurea | ● | ● | ● | ● | ● | ● | ● | ● |
| 79 | Turbinaria | mesenterina | ● | ● | ● | ● | ● | ● | ||
| 80 | Turbinaria | peltata | ● | ● | ● | ● | ● | ● | ||
| 81 | Turbinaria | stellulata | ● | ● | ● | ● | ● | ● | ● | |
| (b) | ||||||||||
| Cell Types Identification | ||||||||||
| No | Genus | Species | Gland Cell | Supporting Cell | Epidermal Nematocyst | Symbiodiniaceae | Symbiotic Endoderm Cell (SEC) | Spindle Cell | Cluster Cell | Gastrodermal Nematocyst |
| 1 | Anacropora | forbesi | ● | ● | ● | ● | ||||
| 2 | Caulastrea | furcata | ● | ● | ● | |||||
| 3 | Echinophyllia | aspera | ● | ● | ● | |||||
| 4 | Echinophyllia | echinata | ● | ● | ● | |||||
| 5 | Euphyllia | glabrescens | ● | ● | ● | ● | ● | |||
| 6 | Euphyllia | paraancora | ● | ● | ● | ● | ● | |||
| 7 | Favites | complanata | ● | ● | ● | ● | ||||
| 8 | Lobophyllia | corymbosa | ● | ● | ● | ● | ||||
| 9 | Montipora | verrucosa | ● | ● | ● | ● | ||||
| 10 | Pavona | decussata | ● | ● | ● | ● | ||||
| 11 | Physogyra | lichtensteini | ● | ● | ● | ● | ● | ● | ||
| 12 | Porites | lichen | ● | ● | ● | |||||
| 13 | Seriatopora | guttatus | ● | ● | ● | ● | ● | |||
| 14 | *Sinularia | compressa | ||||||||
| 15 | Sinularia | flexibilis | ● | ● | ● | ● | ||||
| 16 | Sinularia | sadensis | ● | ● | ● | ● | ||||
| 17 | Symphyllia | recta | ● | ● | ● | |||||
| 18 | Turbinaria | frondens | ● | ● | ● | ● | ||||
| 19 | Turbinaria | reniformis | ● | ● | ● | ● | ● | ● | ||
| 20 | Turbinaria | sp 1 | ● | ● | ● | ● | ● | ● | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toh, E.-C.; Liu, K.-L.; Tsai, S.; Lin, C. Cryopreservation and Cryobanking of Cells from 100 Coral Species. Cells 2022, 11, 2668. https://doi.org/10.3390/cells11172668
Toh E-C, Liu K-L, Tsai S, Lin C. Cryopreservation and Cryobanking of Cells from 100 Coral Species. Cells. 2022; 11(17):2668. https://doi.org/10.3390/cells11172668
Chicago/Turabian StyleToh, En-Chun, Kuan-Lin Liu, Sujune Tsai, and Chiahsin Lin. 2022. "Cryopreservation and Cryobanking of Cells from 100 Coral Species" Cells 11, no. 17: 2668. https://doi.org/10.3390/cells11172668
APA StyleToh, E.-C., Liu, K.-L., Tsai, S., & Lin, C. (2022). Cryopreservation and Cryobanking of Cells from 100 Coral Species. Cells, 11(17), 2668. https://doi.org/10.3390/cells11172668


.png)




