Hmo1 Protein Affects the Nucleosome Structure and Supports the Nucleosome Reorganization Activity of Yeast FACT
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Protein Expression
- Primers: pTF1608 5′-CCGGCATATGACTACAGATCCTTCTGTCAAATTGAAG
- 5′-GGCCGGATCCGTAATAGTAACGAGTTTGTCCGTCC
2.3. Nucleosomal DNA Templates
- 5′-CCGGGATCCAG
- ATCCCGAAAATTTATCAAAAAGAGTATTGACTTAAAGTCTAACCTATAGGATACTTACAGCCATCGAGAGGGACACGGCGAAAAGCCAACCCAAGCGACACCGGCACTGGGCCCGGTTCGCGCTCCCGCCTTCCGTGTGTTGTCGTCTCTCGGGCGTCTAAGTACGCTTAGCGCACGGTAGAGCGCAATCCAAGGCTAACCACCGTGCATCGATGTTGAAAGAGGCCCTCCGTCCTTATTACTTCAAGTCCCTGGGGT-3′.
- 5′-CCCGGTTCGCGC[Cy3-dT]CCCGCCTTCCGTGTGTTGTCGTCTCTCGG-3′ and
- 5′-ACCCCAGGGACTTGAAGTAATAAGGACGGAGGGCCTCTTTCAACATCGATGCACGG[Cy5-dT]GGTTAG-3′, respectively.
- 5′-CCCGGTTCGCGCTCCCGCCTTCCGTGTGTTGTCG[Cy5-dT]CTCTCGG-3′ and
- 5′-ACCCCAGGGACTTGAAGTAATAAGGACGGAGGGCC[Cy3-dT]CTTTCAACATCGAT-3′, respectively.
- 5′-CCCGGTTCGCGCTCCCGCCTTCCGTGTGTTGTCGTCTCTCGGGCGTCTAAGTACGC[Cy3-dT]TAGGC-3′ and
- 5′-ACCCCAGGGACT[Cy5-dT]GAAGTAATAAGGACGGAGGGCCTCTTTC-3′, respectively.
- 5′-CACCGGCACGAGGGCCCGGTTC-3′ (forward primer) and
- 5′-ACTTTCTGGCAAGAAAATGAGCT-3′ (reverse primer).
- 5′-TAAGGCGAATTCACAACTTTTTGGC[Cy5-dT]AGAAAATGAGCT-3′ (forward primer) and
- 5′-ACACGGCGCACTGCCAACCCAAACGACACC[Cy3-dT]GCACGAG-3′ (reverse primer).
2.4. Nucleosome Assembly and Purification
2.5. EMSA Analysis
2.6. spFRET Experiments
2.7. Genetic Analysis
3. Results
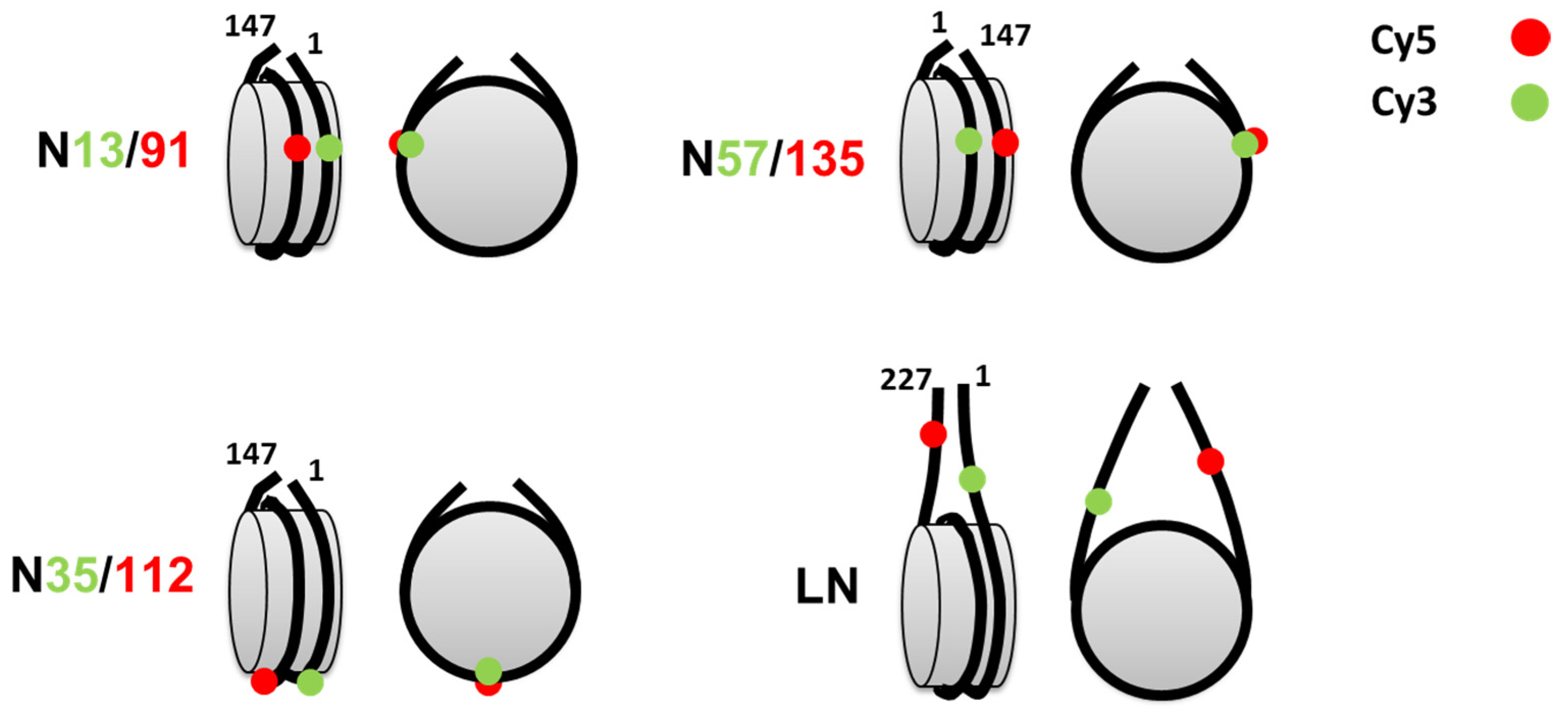
3.1. Studying Mononucleosomes by spFRET
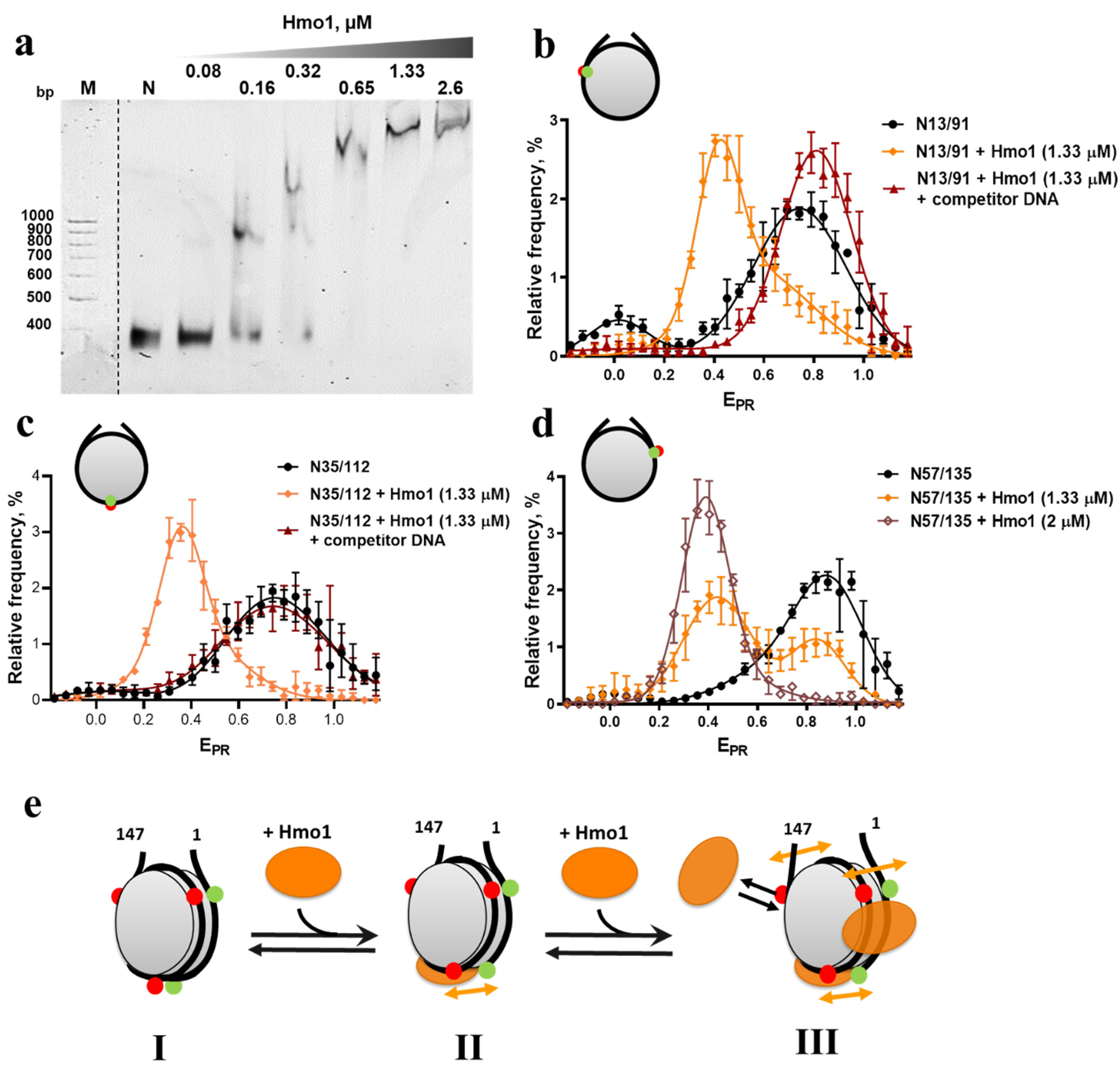
3.2. Hmo1 Affected Nucleosome Structure
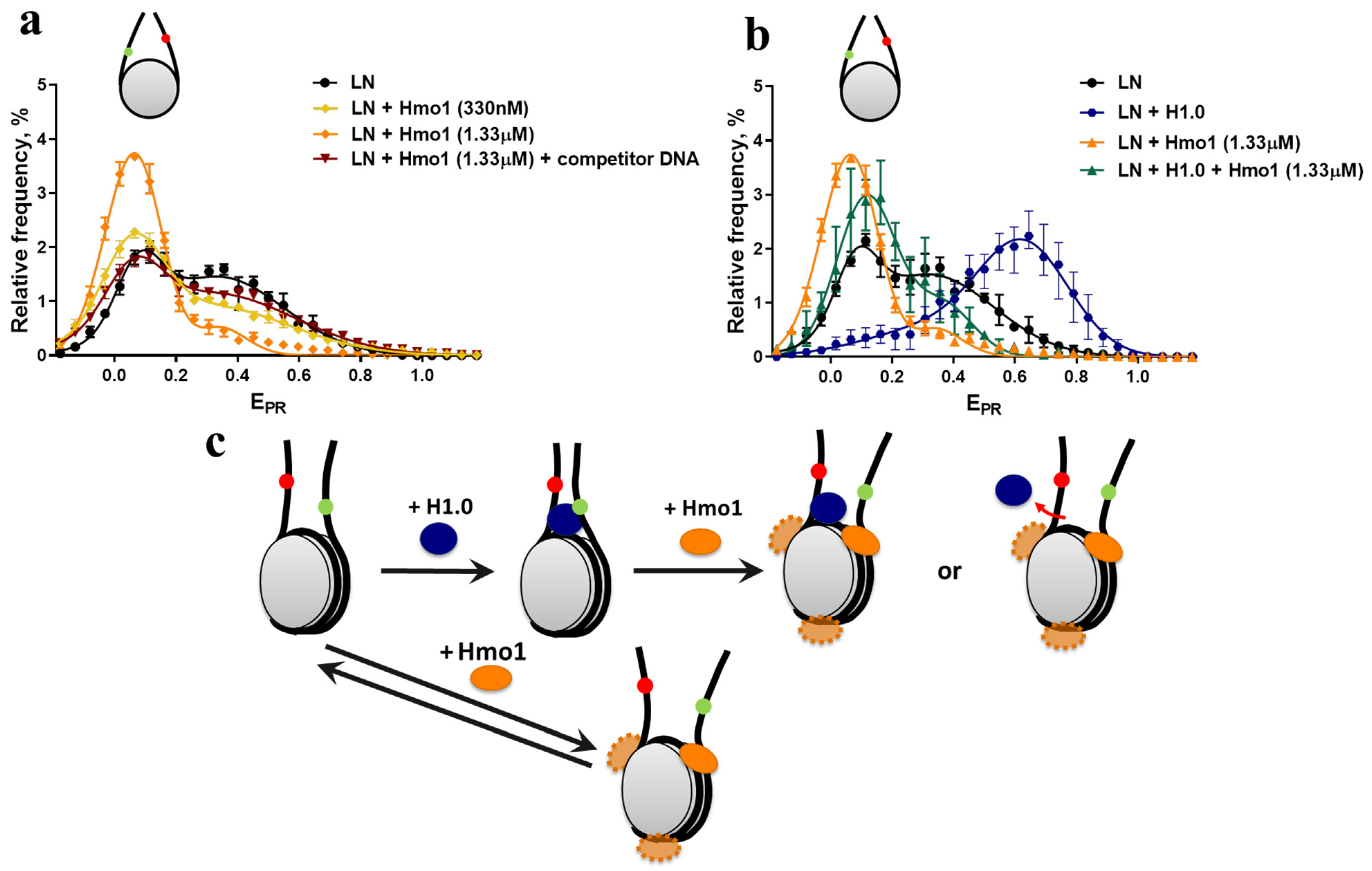
3.3. Effect of Hmo1 on Linker DNA in Nucleosomes and Chromatosomes
3.4. Hmo1 Facilitates Unwrapping of Nucleosomal DNA by FACT
3.5. Hmo1 Supports FACT Function In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Bustin, M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef]
- Oberbeckmann, E.; Wolff, M.; Krietenstein, N.; Heron, M.; Ellins, J.L.; Schmid, A.; Krebs, S.; Blum, H.; Gerland, U.; Korber, P. Absolute nucleosome occupancy map for the Saccharomyces cerevisiae genome. Genome Res. 2019, 29, 1996–2009. [Google Scholar] [CrossRef]
- Ho, B.; Baryshnikova, A.; Brown, G.W. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst. 2018, 6, 192–205.e3. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, A.L.; Valieva, M.E.; Maluchenko, N.V.; Studitsky, V.M. HMGB Proteins as DNA Chaperones That Modulate Chromatin Activity. Mol. Biol. (Mosk). 2018, 52, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Tsujita, R.; Tsubota, M.; Saeki, H.; Sekiguchi, F.; Honda, G.; Kawabata, A. Human soluble thrombomodulin-induced blockade of peripheral HMGB1-dependent allodynia in mice requires both the lectin-like and EGF-like domains. Biochem. Biophys. Res. Commun. 2018, 495, 634–638. [Google Scholar] [CrossRef]
- Kolodrubetz, D.; Burgum, A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem. 1990, 265, 3234–3239. [Google Scholar] [CrossRef]
- Kassavetis, G.A.; Steiner, D.F. Nhp6 is a transcriptional initiation fidelity factor for RNA polymerase III transcription in vitro and in vivo. J. Biol. Chem. 2006, 281, 7445–7451. [Google Scholar] [CrossRef]
- Paull, T.T.; Carey, M.; Johnson, R.C. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996, 10, 2769–2781. [Google Scholar] [CrossRef]
- Lopez, S.; Livingstone-Zatchej, M.; Jourdain, S.; Thoma, F.; Sentenac, A.; Marsolier, M.C. High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol. 2001, 21, 3096–3104. [Google Scholar] [CrossRef][Green Version]
- Hepp, M.I.; Smolle, M.; Gidi, C.; Amigo, R.; Valenzuela, N.; Arriagada, A.; Maureira, A.; Gogol, M.M.; Torrejón, M.; Workman, J.L.; et al. Role of Nhp6 and Hmo1 in SWI/SNF occupancy and nucleosome landscape at gene regulatory regions. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2017, 1860, 316–326. [Google Scholar] [CrossRef]
- Szerlong, H.; Saha, A.; Cairns, B.R. The nuclear actin-related proteins Arp7 and Arp9: A dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003, 22, 3175–3187. [Google Scholar] [CrossRef] [PubMed]
- Maluchenko, N.V.; Chang, H.W.; Kozinova, M.T.; Valieva, M.E.; Gerasimova, N.S.; Kitashov, A.V.; Kirpichnikov, M.P.; Georgiev, P.G.; Studitsky, V.M. Inhibiting the pro-tumor and transcription factor FACT: Mechanisms. Mol. Biol. (Mosk). 2016, 50, 532–541. [Google Scholar] [CrossRef]
- Kemble, D.J.; McCullough, L.L.; Whitby, F.G.; Formosa, T.; Hill, C.P. FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs. Mol. Cell 2015, 60, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Sivkina, A.L.; Karlova, M.G.; Valieva, M.E.; McCullough, L.L.; Formosa, T.; Shaytan, A.K.; Feofanov, A.V.; Kirpichnikov, M.P.; Sokolova, O.S.; Studitsky, V.M. Electron microscopy analysis of ATP-independent nucleosome unfolding by FACT. Commun. Biol. 2022, 5, 2. [Google Scholar] [CrossRef]
- Wittmeyer, J.; Formosa, T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 1997, 17, 4178–4190. [Google Scholar] [CrossRef]
- Malone, E.A.; Clark, C.D.; Chiang, A.; Winston, F. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 5710–5717. [Google Scholar]
- Cheung, V.; Chua, G.; Batada, N.N.; Landry, C.R.; Michnick, S.W.; Hughes, T.R.; Winston, F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008, 6, e277. [Google Scholar] [CrossRef]
- Jeronimo, C.; Poitras, C.; Robert, F. Histone Recycling by FACT and Spt6 during Transcription Prevents the Scrambling of Histone Modifications. Cell Rep. 2019, 28, 1206–1218 e8. [Google Scholar] [CrossRef]
- Mason, P.B.; Struhl, K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 2003, 23, 8323–8333. [Google Scholar] [CrossRef]
- Petrenko, N.; Jin, Y.; Dong, L.; Wong, K.H.; Struhl, K. Requirements for RNA polymerase II preinitiation complex formation in vivo. Elife 2019, 8, e43654. [Google Scholar] [CrossRef]
- Formosa, T.; Eriksson, P.; Wittmeyer, J.; Ginn, J.; Yu, Y.; Stillman, D.J. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001, 20, 3506–3517. [Google Scholar] [CrossRef]
- Brewster, N.K.; Johnston, G.C.; Singer, R.A. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 2001, 21, 3491–3502. [Google Scholar] [CrossRef]
- Brewster, N.K.; Johnston, G.C.; Singer, R.A. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 1998, 273, 21972–21979. [Google Scholar] [CrossRef] [PubMed]
- Orphanides, G.; Wu, W.H.; Lane, W.S.; Hampsey, M.; Reinberg, D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 1999, 400, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Stillman, D.J. Nhp6: A small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2010, 1799, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Albert, B.; Colleran, C.; Léger-Silvestre, I.; Berger, A.B.; Dez, C.; Normand, C.; Perez-Fernandez, J.; McStay, B.; Gadal, O. Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-Box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Res. 2013, 41, 10135–10149. [Google Scholar] [CrossRef]
- Amigo, R.; Farkas, C.; Gidi, C.; Hepp, M.I.; Cartes, N.; Tarifeño, E.; Workman, J.L.; Gutiérrez, J.L. The linker histone Hho1 modulates the activity of ATP-dependent chromatin remodeling complexes. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2022, 1865, 194781. [Google Scholar] [CrossRef]
- Hall, D.B.; Wade, J.T.; Struhl, K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006, 26, 3672–3679. [Google Scholar] [CrossRef]
- Schlichter, A.; Kasten, M.M.; Parnell, T.J.; Cairns, B.R. Specialization of the chromatin remodeler RSC to mobilize partially-unwrapped nucleosomes. Elife 2020, 9, e58130. [Google Scholar] [CrossRef]
- Lu, J.; Kobayashi, R.; Brill, S.J. Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem. 1996, 271, 33678–33685. [Google Scholar] [CrossRef] [PubMed]
- Kamau, E.; Bauerle, K.T.; Grove, A. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J. Biol. Chem. 2004, 279, 55234–55240. [Google Scholar] [CrossRef] [PubMed]
- Bauerle, K.T.; Kamau, E.; Grove, A. Interactions between N- and C-terminal domains of the Saccharomyces cerevisiae high-mobility group protein HMO1 are required for DNA bending. Biochemistry 2006, 45, 3635–3645. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Williams, A.M.; Grove, A. The C-terminal domain of yeast high mobility group protein HMO1 mediates lateral protein accretion and in-phase DNA bending. Biochemistry 2010, 49, 4051–4059. [Google Scholar] [CrossRef] [PubMed]
- Masse, J.E.; Wong, B.; Yen, Y.M.; Allain, F.H.; Johnson, R.C.; Feigon, J. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002, 323, 263–284. [Google Scholar] [CrossRef]
- Hepp, M.I.; Alarcon, V.; Dutta, A.; Workman, J.L.; Gutiérrez, J.L. Nucleosome remodeling by the SWI/SNF complex is enhanced by yeast high mobility group box (HMGB) proteins. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2014, 1839, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Panday, A.; Grove, A. Yeast HMO1: Linker Histone Reinvented. Microbiol. Mol. Biol. Rev. 2017, 81, e00037-16. [Google Scholar] [CrossRef]
- Panday, A.; Grove, A. The high mobility group protein HMO1 functions as a linker histone in yeast. Epigenetics Chromatin 2016, 9, 13. [Google Scholar] [CrossRef]
- Paull, T.T.; Johnson, R.C. DNA Looping by Saccharomyces-Cerevisiae High-Mobility Group Proteins Nhp6a/B—Consequences for Nucleoprotein Complex Assembly and Chromatin Condensation. J. Biol. Chem. 1995, 270, 8744–8754. [Google Scholar] [CrossRef]
- Ruone, S.; Rhoades, A.R.; Formosa, T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 2003, 278, 45288–45295. [Google Scholar] [CrossRef]
- Biswas, D.; Yu, Y.; Prall, M.; Formosa, T.; Stillman, D.J. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 2005, 25, 5812–5822. [Google Scholar] [CrossRef]
- Wittmeyer, J.; Joss, L.; Formosa, T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 1999, 38, 8961–8971. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.R.; Feng, H.; Ghirlando, R.; Li, S.; Schwieters, C.D.; Bai, Y. A Small Number of Residues Can Determine if Linker Histones Are Bound on or off Dyad in the Chromatosome. J. Mol. Biol. 2016, 428, 3948–3959. [Google Scholar] [CrossRef] [PubMed]
- Kulaeva, O.I.; Gaykalova, D.A.; Pestov, N.A.; Golovastov, V.V.; Vassylyev, D.G.; Artsimovitch, I.; Studitsky, V.M. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 2009, 16, 1272–1278. [Google Scholar] [CrossRef]
- Lyubitelev, A.V.; Kudryashova, K.S.; Mikhaylova, M.S.; Malyuchenko, N.V.; Chertkov, O.V.; Studitsky, V.M.; Feofanov, A.V.; Kirpichnikov, M.P. Change in linker DNA conformation upon histone H1.5 binding to nucleosome: Fluorescent microscopy of single complexes. Mosc. Univ. Biol. Sci. Bull. 2016, 71, 108–113. [Google Scholar] [CrossRef]
- Thåström, A.; Lowary, P.T.; Widlund, H.R.; Cao, H.; Kubista, M.; Widom, J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 1999, 288, 213–229. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar] [PubMed]
- Kudryashova, K.S.; Chertkov, O.V.; Nikitin, D.V.; Pestov, N.A.; Kulaeva, O.I.; Efremenko, A.V.; Solonin, A.S.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Preparation of mononucleosomal templates for analysis of transcription with RNA polymerase using spFRET. Methods Mol. Biol. 2015, 1288, 395–412. [Google Scholar]
- Joo, C.; Ha, T. Single-molecule FRET with total internal reflection microscopy. Cold Spring Harb. Protoc 2012, pdb.top072058. [Google Scholar] [CrossRef]
- Thastrom, A.; Bingham, L.M.; Widom, J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 2004, 338, 695–709. [Google Scholar] [CrossRef]
- Valieva, M.E.; Armeev, G.A.; Kudryashova, K.S.; Gerasimova, N.S.; Shaytan, A.K.; Kulaeva, O.I.; McCullough, L.L.; Formosa, T.; Georgiev, P.G.; Kirpichnikov, M.P.; et al. Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 2016, 23, 1111–1116. [Google Scholar] [CrossRef]
- Kasahara, K.; Higashino, A.; Unzai, S.; Yoshikawa, H.; Kokubo, T. Oligomerization of Hmo1 mediated by box A is essential for DNA binding in vitro and in vivo. Genes Cells 2016, 21, 1333–1352. [Google Scholar] [CrossRef] [PubMed]
- Rajakumara, E.; Satish, M.; Abhishek, S. In vitro studies on non-canonical DNA binding specificities of KAP6 and HMO1 and mechanistic insights into DNA bound and unbinding dynamics of KAP6. Int. J. Biol. Macromol. 2020, 160, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Levitus, M.; Bustamante, C.; Widom, J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005, 12, 46–53. [Google Scholar] [CrossRef] [PubMed]
- VanDemark, A.P.; Xin, H.; McCullough, L.; Rawlins, R.; Bentley, S.; Heroux, A.; Stillman, D.J.; Hill, C.P.; Formosa, T. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J. Biol. Chem. 2008, 283, 5058–5068. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, M.B.; Formosa, T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 2000, 155, 1593–1606. [Google Scholar] [CrossRef]
- Murugesapillai, D.; McCauley, M.J.; Huo, R.; Nelson Holte, M.H.; Stepanyants, A.; Maher, L.J., 3rd; Israeloff, N.E.; Williams, M.C. DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin. Nucleic Acids Res. 2014, 42, 8996–9004. [Google Scholar] [CrossRef]
- Zlatanova, J.; Seebart, C.; Tomschik, M. The linker-protein network: Control of nucleosomal DNA accessibility. Trends Biochem. Sci. 2008, 33, 247–253. [Google Scholar] [CrossRef]
- Cato, L.; Stott, K.; Watson, M.; Thomas, J.O. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J. Mol. Biol. 2008, 384, 1262–1272. [Google Scholar] [CrossRef]
- Polanska, E.; Pospisilova, S.; Stros, M. Binding of histone H1 to DNA is differentially modulated by redox state of HMGB1. PLoS ONE 2014, 9, e89070. [Google Scholar] [CrossRef]
- Takahata, S.; Yu, Y.; Stillman, D.J. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 2009, 34, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Voth, W.P.; Takahata, S.; Nishikawa, J.L.; Metcalfe, B.M.; Näär, A.M.; Stillman, D.J. A role for FACT in repopulation of nucleosomes at inducible genes. PLoS ONE 2014, 9, e84092. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Takahata, S.; Blanksma, M.; McCullough, L.; Stillman, D.J.; Formosa, T. yFACT Induces Global Accessibility of Nucleosomal DNA without H2A-H2B Displacement. Mol. Cell 2009, 35, 365–376. [Google Scholar] [CrossRef] [PubMed]
- McCullough, L.; Rawlins, R.; Olsen, A.; Xin, H.; Stillman, D.; Formosa, T. Insight Into the Mechanism of Nucleosome Reorganization From Histone Mutants That Suppress Defects in the FACT Histone Chaperone. Genetics 2011, 188, 835–846. [Google Scholar] [CrossRef]
- McCullough, L.; Poe, B.; Connell, Z.; Xin, H.; Formosa, T. The FACT Histone Chaperone Guides Histone H4 Into Its Nucleosomal Conformation in Saccharomyces cerevisiae. Genetics 2013, 195, 101–113. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malinina, D.K.; Sivkina, A.L.; Korovina, A.N.; McCullough, L.L.; Formosa, T.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Hmo1 Protein Affects the Nucleosome Structure and Supports the Nucleosome Reorganization Activity of Yeast FACT. Cells 2022, 11, 2931. https://doi.org/10.3390/cells11192931
Malinina DK, Sivkina AL, Korovina AN, McCullough LL, Formosa T, Kirpichnikov MP, Studitsky VM, Feofanov AV. Hmo1 Protein Affects the Nucleosome Structure and Supports the Nucleosome Reorganization Activity of Yeast FACT. Cells. 2022; 11(19):2931. https://doi.org/10.3390/cells11192931
Chicago/Turabian StyleMalinina, Daria K., Anastasiia L. Sivkina, Anna N. Korovina, Laura L. McCullough, Tim Formosa, Mikhail P. Kirpichnikov, Vasily M. Studitsky, and Alexey V. Feofanov. 2022. "Hmo1 Protein Affects the Nucleosome Structure and Supports the Nucleosome Reorganization Activity of Yeast FACT" Cells 11, no. 19: 2931. https://doi.org/10.3390/cells11192931
APA StyleMalinina, D. K., Sivkina, A. L., Korovina, A. N., McCullough, L. L., Formosa, T., Kirpichnikov, M. P., Studitsky, V. M., & Feofanov, A. V. (2022). Hmo1 Protein Affects the Nucleosome Structure and Supports the Nucleosome Reorganization Activity of Yeast FACT. Cells, 11(19), 2931. https://doi.org/10.3390/cells11192931






