Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics
2.2. Construction of Plasmids
2.3. Cell Culture, Transfection, and Imaging of Transfected Cells
2.4. Quantification of Nuclear Import
3. Results
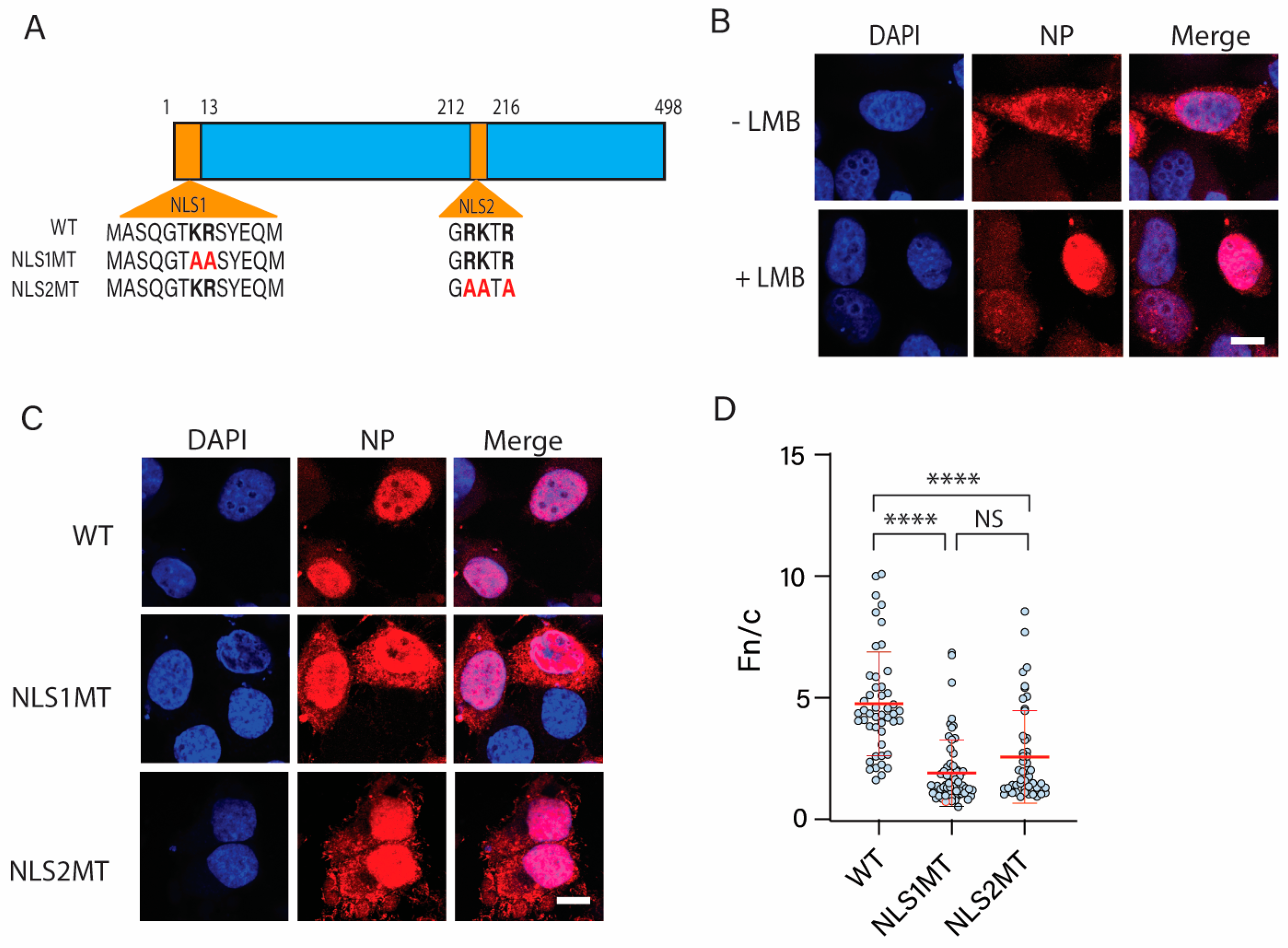
3.1. NLS2 Contributes to the Nuclear Import of NP to the Same Extent as NLS1
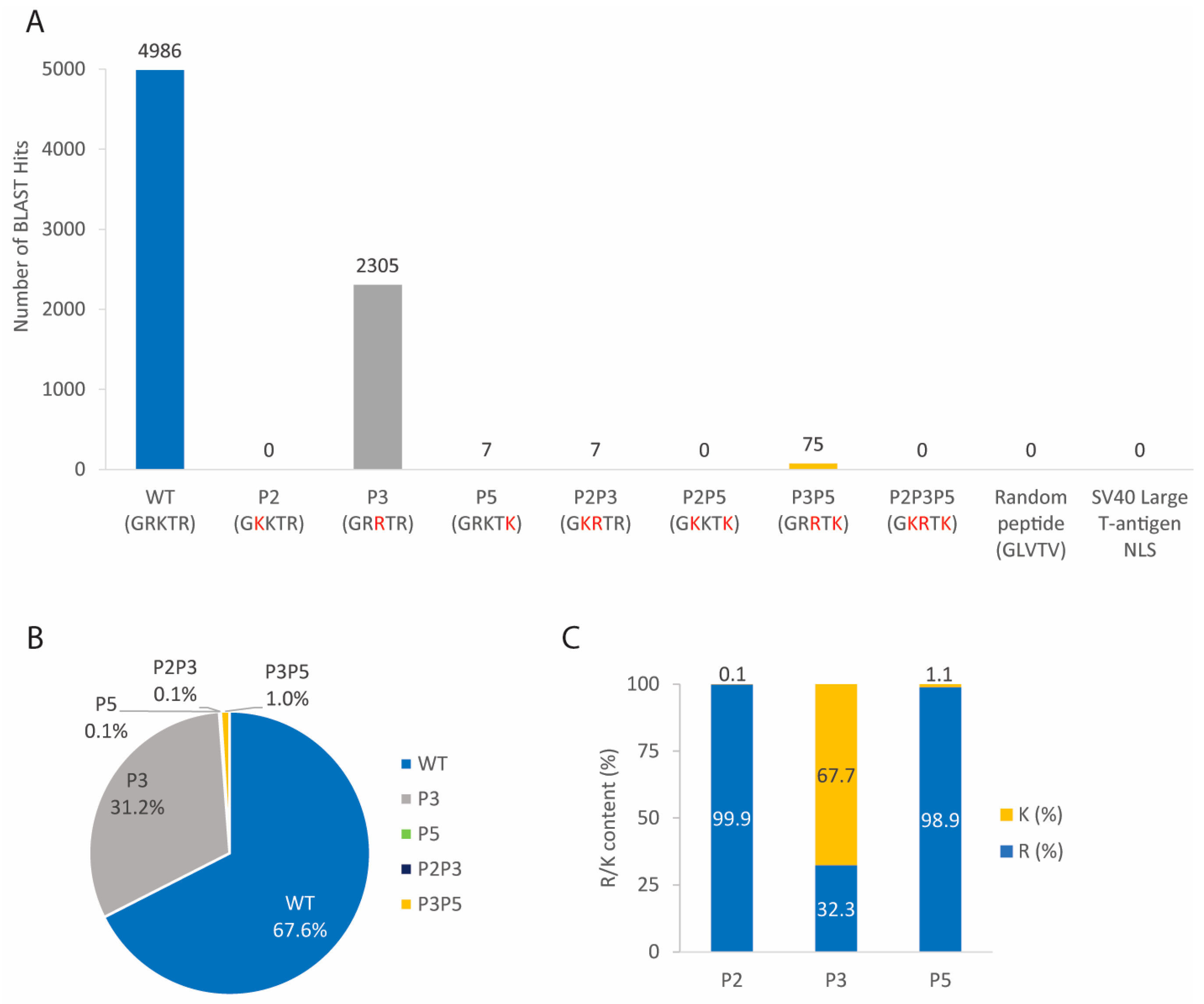
3.2. Five NLS2 Variants Are Present in NP of Different Influenza A Virus Strains, but Most Strains Contain the Wild-Type Sequence
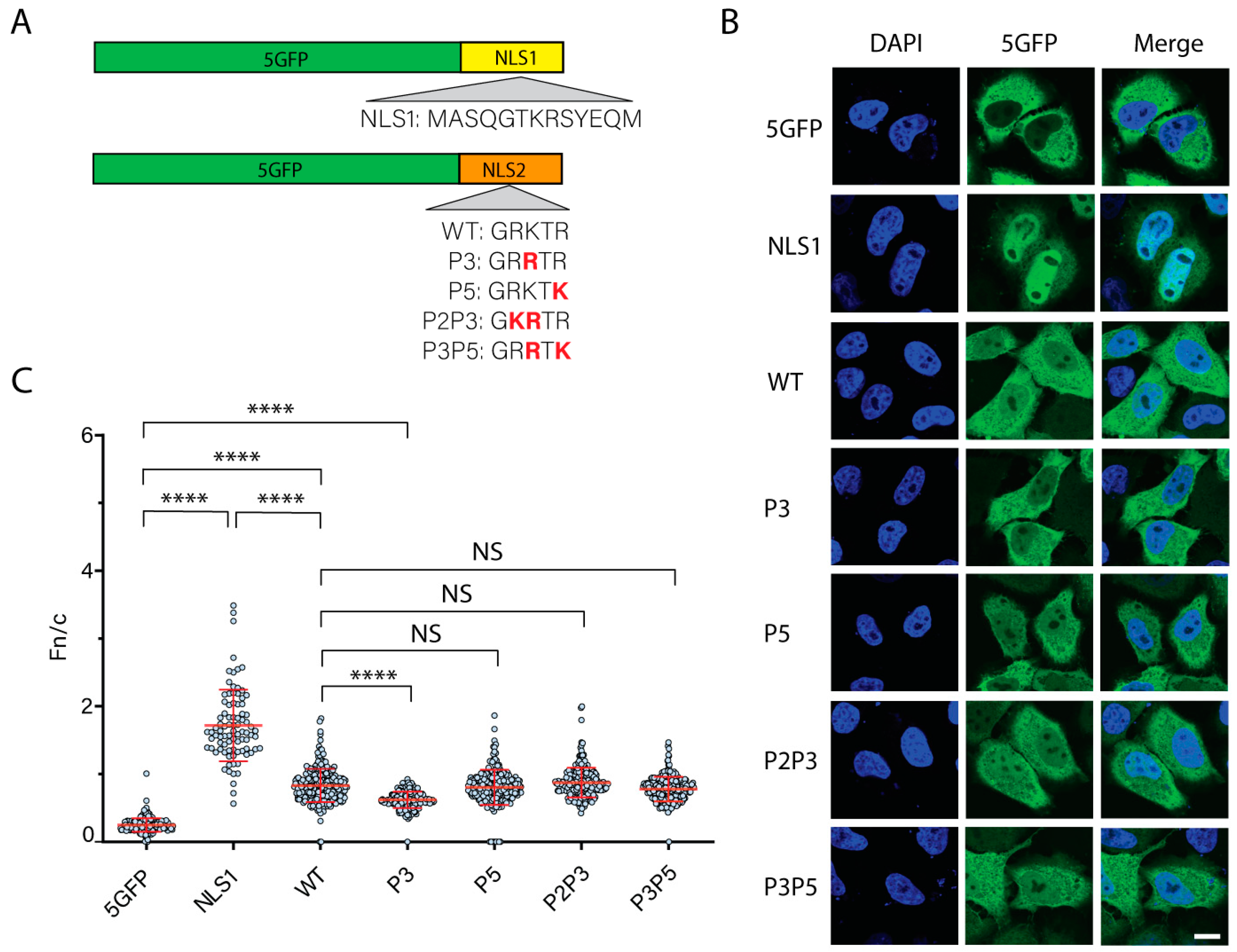
3.3. The Five NLS2 Variants Present in NP of Influenza A Virus Mediate Nuclear Import of Chimeric Proteins
3.4. From All Viral Protein Sequences from DNA Viruses in the Database, NLS2 and Its Variants Are Present Only in a Few Viral Proteins
3.5. The Sequence GRKTR and Its Variants Are Present in a Very Low Proportion of Nuclear Proteins
3.6. Identification of a Novel NLS in Nucleolar Protein 14
3.7. NLS2 Plays a Role in the Nucleolar Localization of Nucleolar Protein 14
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohr, D.; Frey, S.; Fischer, T.; Güttler, T.; Görlich, D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009, 28, 2541–2553. [Google Scholar] [CrossRef] [PubMed]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed]
- Roggero, V.R.; Zhang, J.; Parente, L.E.; Doshi, Y.; Dziedzic, R.C.; McGregor, E.L.; Allison, L.A. Nuclear import of the thyroid hormone receptor alpha1 is mediated by importin 7, importin beta1, and adaptor importin alpha1. Mol. Cell Endocrinol. 2016, 419, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Bunn, C.F.; Neidig, J.A.; Freidinger, K.E.; Stankiewicz, T.A.; Weaver, B.S.; McGrew, J.; Allison, L.A. Nucleocytoplasmic shuttling of the thyroid hormone receptor alpha. Mol. Endocrinol. 2001, 15, 512–533. [Google Scholar]
- Adachi, M.; Fukuda, M.; Nishida, E. Two co-existing mechanisms for nuclear import of MAP kinase: Passive diffusion of a monomer and active transport of a dimer. EMBO J. 1999, 18, 5347–5358. [Google Scholar] [CrossRef]
- Chook, Y.M.; Suel, K.E. Nuclear import by karyopherin-betas: Recognition and inhibition. Biochim. Biophys. Acta 2011, 1813, 1593–1606. [Google Scholar] [CrossRef]
- Wing, C.E.; Fung, H.Y.J.; Chook, Y.M. Karyopherin-mediated nucleocytoplasmic transport. Nat. Rev. Mol. Cell Biol. 2022, 23, 307–328. [Google Scholar] [CrossRef]
- Conti, E.; Uy, M.; Leighton, L.; Blobel, G.; Kuriyan, J. Crystallographic Analysis of the Recognition of a Nuclear Localization Signal by the Nuclear Import Factor Karyopherin α. Cell 1998, 94, 193–204. [Google Scholar] [CrossRef]
- Leung, S.W.; Harreman, M.T.; Hodel, M.R.; Hodel, A.E.; Corbett, A.H. Dissection of the karyopherin alpha nuclear localization signal (NLS)-binding groove: Functional requirements for NLS binding. J. Biol. Chem. 2003, 278, 41947–41953. [Google Scholar] [CrossRef]
- Oka, M.; Yoneda, Y. Importin α: Functions as a nuclear transport factor and beyond. Proc. Jpn. Acad. Ser. B 2018, 94, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.J.; Cansizoglu, A.E.; Suel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef]
- Truant, R.; Cullen, B.R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell Biol. 1999, 19, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Wu, Y.; Chen, Q.; Zhang, Z.; Chen, X.; Zhang, Y. The Arginine/Lysine-Rich Element within the DNA-Binding Domain Is Essential for Nuclear Localization and Function of the Intracellular Pathogen Resistance 1. PLoS ONE 2016, 11, e0162832. [Google Scholar] [CrossRef]
- Lam, M.H.; Hu, W.; Xiao, C.Y.; Gillespie, M.T.; Jans, D.A. Molecular dissection of the importin beta1-recognized nuclear targeting signal of parathyroid hormone-related protein. Biochem. Biophys. Res. Commun. 2001, 282, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Vogel, F.; Mills, A.D.; Hartmann, E.; Laskey, R.A. Distinct functions for the two importin subunits in nuclear protein import. Nature 1995, 377, 246–248. [Google Scholar] [CrossRef]
- Chi, N.C.; Adam, E.J.; Adam, S. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol. 1995, 130, 265–274. [Google Scholar] [CrossRef]
- Imamoto, N.; Shimamoto, T.; Kose, S.; Takao, T.; Tachibana, T.; Matsubae, M.; Sekimoto, T.; Shimonishi, Y.; Yoneda, Y. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995, 368, 415–419. [Google Scholar] [CrossRef]
- Kobe, B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Genet. 1999, 6, 388–397. [Google Scholar] [CrossRef]
- Fauquet, C.; Fargette, D. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol. J. 2005, 2, 64. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef]
- Arranz, R.; Coloma, R.; Chichón, F.J.; Conesa, J.J.; Carrascosa, J.L.; Valpuesta, J.M.; Ortín, J.; Martín-Benito, J. The Structure of Native Influenza Virion Ribonucleoproteins. Science 2012, 338, 1634–1637. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.R.; Torian, U.; McCraw, D.M.; Harris, A.K. Structural studies of influenza virus RNPs by electron microscopy indicate molecular contortions within NP supra-structures. J. Struct. Biol. 2017, 197, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Digard, P. The influenza virus nucleoprotein: A multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002, 83, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Compans, R.W.; Content, J.; Duesberg, P.H. Structure of the Ribonucleoprotein of Influenza Virus. J. Virol. 1972, 10, 795–800. [Google Scholar] [CrossRef]
- Cros, J.F.; García-Sastre, A.; Palese, P. An Unconventional NLS is Critical for the Nuclear Import of the Influenza A Virus Nucleoprotein and Ribonucleoprotein. Traffic 2005, 6, 205–213. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.E.; Jaskunas, R.; Blobel, G.; Palese, P.; Moroianu, J. Nuclear Import of Influenza Virus RNA Can Be Mediated by Viral Nucleoprotein and Transport Factors Required for Protein Import. J. Biol. Chem. 1995, 270, 22701–22704. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sankhala, R.S.; Florio, T.J.; Zhou, L.; Nguyen, N.L.T.; Lokareddy, R.K.; Cingolani, G.; Panté, N. Synergy of two low-affinity NLSs determines the high avidity of influenza A virus nucleoprotein NP for human importin α isoforms. Sci. Rep. 2017, 7, 11381. [Google Scholar] [CrossRef]
- Vreede, F.T.; Jung, T.E.; Brownlee, G.G. Model Suggesting that Replication of Influenza Virus Is Regulated by Stabilization of Replicative Intermediates. J. Virol. 2004, 78, 9568–9572. [Google Scholar] [CrossRef]
- Ye, Q.; Krug, R.M.; Tao, Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 2006, 444, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Chenavas, S.; Estrozi, L.F.; Slama-Schwok, A.; Delmas, B.; Di Primo, C.; Baudin, F.; Li, X.; Crépin, T.; Ruigrok, R.W.H. Monomeric Nucleoprotein of Influenza A Virus. PLOS Pathog. 2013, 9, e1003275. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-S.; Xu, S.; Chen, Y.-W.; Wang, J.-H.; Shaw, P.-C. Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction. Nucleic Acids Res. 2021, 49, 4144–4154. [Google Scholar] [CrossRef] [PubMed]
- Knight, M.L.; Fan, H.; Bauer, D.L.V.; Grimes, J.M.; Fodor, E.; Keown, J.R. Structure of an H3N2 influenza virus nucleoprotein. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2021, 77, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Nakada, R.; Hirano, H.; Matsuura, Y. Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci. Rep. 2015, 5, 15055. [Google Scholar] [CrossRef] [PubMed]
- Pumroy, R.A.; Nardozzi, J.D.; Hart, D.J.; Root, M.J.; Cingolani, G. Nucleoporin Nup50 Stabilizes Closed Conformation of Armadillo repeat 10 in Importin α5. J. Biol. Chem. 2012, 287, 2022–2031. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Couñago, R.L.M.; Williams, S.J.; Bodén, M.; Kobe, B. Crystal Structure of Rice Importin-α and Structural Basis of Its Interaction with Plant-Specific Nuclear Localization Signals. Plant Cell 2012, 24, 5074–5088. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y.; Lange, A.; Harreman, M.T.; Corbett, A.H.; Stewart, M. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 2003, 22, 5358–5369. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-H.; Ben-Efraim, I.; Mitrousis, G.; Walker-Kopp, N.; Sims, P.J.; Cingolani, G. Phospholipid Scramblase 1 Contains a Nonclassical Nuclear Localization Signal with Unique Binding Site in Importin α. J. Biol. Chem. 2005, 280, 10599–10606. [Google Scholar] [CrossRef]
- Lott, K.; Bhardwaj, A.; Sims, P.J.; Cingolani, G. A Minimal Nuclear Localization Signal (NLS) in Human Phospholipid Scramblase 4 That Binds Only the Minor NLS-binding Site of Importin α1. J. Biol. Chem. 2011, 286, 28160–28169. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Kochs, G.; Gruber, S.; Haller, O. A Classical Bipartite Nuclear Localization Signal on Thogoto and Influenza A Virus Nucleoproteins. Virology 1998, 250, 9–18. [Google Scholar] [CrossRef]
- Takeda, A.A.; de Barros, A.C.; Chang, C.-W.; Kobe, B.; Fontes, M.R. Structural Basis of Importin-α-Mediated Nuclear Transport for Ku70 and Ku80. J. Mol. Biol. 2011, 412, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.W.; Sun, Y.-H.B.; Panté, N. Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol. J. 2007, 4, 49. [Google Scholar] [CrossRef]
- Miyamoto, S.; Nakano, M.; Morikawa, T.; Hirabayashi, A.; Tamura, R.; Fujita-Fujiharu, Y.; Hirose, N.; Muramoto, Y.; Noda, T. Migration of Influenza Virus Nucleoprotein into the Nucleolus Is Essential for Ribonucleoprotein Complex Formation. mBio 2022, 13, e03315-21. [Google Scholar] [CrossRef]
- Ozawa, M.; Fujii, K.; Muramoto, Y.; Yamada, S.; Yamayoshi, S.; Takada, A.; Goto, H.; Horimoto, T.; Kawaoka, Y. Contributions of Two Nuclear Localization Signals of Influenza A Virus Nucleoprotein to Viral Replication. J. Virol. 2007, 81, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Nguyen, B.A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Faleiro, L.; Lazebnik, Y. Caspases Disrupt the Nuclear-Cytoplasmic Barrier. J. Cell Biol. 2000, 151, 951–960. [Google Scholar] [CrossRef]
- Wang, R.; Brattain, M.G. The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett. 2007, 581, 3164–3170. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Castrucci, M.R.; Kawaoka, Y. Nuclear import and export of influenza virus nucleoprotein. J. Virol. 1997, 71, 9690–9700. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Roy, A.-M.M.; Whittaker, G.R. Nuclear Export of Influenza Virus Ribonucleoproteins: Identification of an Export Intermediate at the Nuclear Periphery. Virology 2001, 282, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Nishi, K.; Yoshida, M.; Fujiwara, D.; Nishikawa, M.; Horinouchi, S.; Beppu, T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 1994, 269, 6320–6324. [Google Scholar] [CrossRef]
- Catimel, B.; Teh, T.; Fontes, M.; Jennings, I.; Jans, D.; Howlett, G.J.; Nice, E.C.; Kobe, B. Biophysical Characterization of Interactions Involving Importin-α during Nuclear Import. J. Biol. Chem. 2001, 276, 34189–34198. [Google Scholar] [CrossRef]
- Marfori, M.; Lonhienne, T.G.; Forwood, J.K.; Kobe, B. Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-α. Traffic 2012, 13, 532–548. [Google Scholar] [CrossRef]
- Ugai, H.; Dobbins, G.C.; Wang, M.; Le, L.P.; Matthews, D.; Curiel, D.T. Adenoviral protein V promotes a process of viral assembly through nucleophosmin 1. Virology 2012, 432, 283–295. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, X.; Tikoo, S.K. Nuclear and Nucleolar Localization of Bovine Adenovirus-3 Protein, V. Front. Microbiol. 2021, 11, 579593. [Google Scholar] [CrossRef]
- Mysiak, M.E.; Holthuizen, P.E.; Van Der Vliet, P.C. The adenovirus priming protein pTP contributes to the kinetics of initiation of DNA replication. Nucleic Acids Res. 2004, 32, 3913–3920. [Google Scholar] [CrossRef]
- Webster, A.; Leith, I.R.; Nicholson, J.; Hounsell, J.; Hay, R.T. Role of preterminal protein processing in adenovirus replication. J. Virol. 1997, 71, 6381–6389. [Google Scholar] [CrossRef]
- Wu, K.; Guimet, D.; Hearing, P. The Adenovirus L4-33K Protein Regulates both Late Gene Expression Patterns and Viral DNA Packaging. J. Virol. 2013, 87, 6739–6747. [Google Scholar] [CrossRef]
- Kulshreshtha, V.; Ayalew, L.E.; Islam, A.; Tikoo, S.K. Conserved Arginines of Bovine Adenovirus-3 33K Protein Are Important for Transportin-3 Mediated Transport and Virus Replication. PLoS ONE 2014, 9, e101216. [Google Scholar] [CrossRef][Green Version]
- Nemerow, G.R.; Stewart, P.L.; Reddy, V.S. Structure of human adenovirus. Curr. Opin. Virol. 2012, 2, 115–121. [Google Scholar] [CrossRef]
- Lee, T.W.R.; Lawrence, F.J.; Dauksaite, V.; Akusjarvi, G.; Blair, G.E.; Matthews, D.A. Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol. 2004, 85, 185–196. [Google Scholar] [CrossRef]
- Gustin, K.E.; Imperiale, M.J. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 1998, 72, 7860–7870. [Google Scholar] [CrossRef] [PubMed]
- Hasson, T.B.; A Ornelles, D.; Shenk, T. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J. Virol. 1992, 66, 6133–6142. [Google Scholar] [CrossRef]
- Vellinga, J.; Van Der Heijdt, S.; Hoeben, R. The adenovirus capsid: Major progress in minor proteins. J. Gen. Virol. 2005, 86, 1581–1588. [Google Scholar] [CrossRef]
- Ayalew, L.E.; Gaba, A.; Kumar, P.; Tikoo, S.K. Conserved regions of bovine adenovirus-3 pVIII contain functional domains involved in nuclear localization and packaging in mature infectious virions. J. Gen. Virol. 2014, 95, 1743–1754. [Google Scholar] [CrossRef]
- Russell, W.C. Adenoviruses: Update on structure and function. J. Gen. Virol. 2009, 90, 1–20. [Google Scholar] [CrossRef]
- Gottlieb, J.; I Marcy, A.; Coen, D.M.; Challberg, M.D. The herpes simplex virus type 1 UL42 gene product: A subunit of DNA polymerase that functions to increase processivity. J. Virol. 1990, 64, 5976–5987. [Google Scholar] [CrossRef]
- AuCoin, D.P.; Smith, G.B.; Meiering, C.D.; Mocarski, E.S. Betaherpesvirus-Conserved Cytomegalovirus Tegument Protein ppUL32 (pp150) Controls Cytoplasmic Events during Virion Maturation. J. Virol. 2006, 80, 8199–8210. [Google Scholar] [CrossRef] [PubMed]
- Hensel, G.; Meyer, H.; Gärtner, S.; Brand, G.; Kern, H.F. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 1995, 76, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Mbong, E.F.; Woodley, L.; Dunkerley, E.; Schrimpf, J.E.; Morrison, L.A.; Duffy, C. Deletion of the herpes simplex virus 1 UL49 gene results in mRNA and protein translation defects that are complemented by secondary mutations in UL41. J. Virol. 2012, 86, 12351–12361. [Google Scholar] [CrossRef]
- Plafker, S.M.; Gibson, W. Cytomegalovirus assembly protein precursor and proteinase precursor contain two nuclear localization signals that mediate their own nuclear translocation and that of the major capsid protein. J. Virol. 1998, 72, 7722–7732. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Liao, Z.; Chen, T.; Wang, P.; Zou, X.; Wang, Y.; Xu, Z.; Jiang, S.; Huang, J.; Chen, D.; et al. Characterization of the subcellular localization of Epstein-Barr virus encoded proteins in live cells. Oncotarget 2017, 8, 70006–70034. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heath, L.; Williamson, A.-L.; Rybicki, E. The Capsid Protein of Beak and Feather Disease Virus Binds to the Viral DNA and Is Responsible for Transporting the Replication-Associated Protein into the Nucleus. J. Virol. 2006, 80, 7219–7225. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Tikoo, S.K.; Babiuk, L.A. Nuclear Localization of the ORF2 Protein Encoded by Porcine Circovirus Type 2. Virology 2001, 285, 91–99. [Google Scholar] [CrossRef]
- Bouchard, M.J.; Schneider, R.J. The Enigmatic X Gene of Hepatitis B Virus. J. Virol. 2004, 78, 12725–12734. [Google Scholar] [CrossRef]
- VanderWeele, T.J.; Ding, P. Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann. Intern. Med. 2017, 167, 268–274. [Google Scholar] [CrossRef]
- Haneuse, S.; VanderWeele, T.J.; Arterburn, D. Using the E-Value to Assess the Potential Effect of Unmeasured Confounding in Observational Studies. JAMA 2019, 321, 602–603. [Google Scholar] [CrossRef]
- Liu, P.C.C.; Thiele, D.J. Novel Stress-responsive Genes EMG1 and NOP14 Encode Conserved, Interacting Proteins Required for 40S Ribosome Biogenesis. Mol. Biol. Cell 2001, 12, 3644–3657. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Lambie, E.J.; Snyder, M. NuMA: An unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. 1992, 116, 1303–1317. [Google Scholar] [CrossRef] [PubMed]
- Fournier, G.; Chiang, C.; Munier, S.; Tomoiu, A.; Demeret, C.; Vidalain, P.-O.; Jacob, Y.; Naffakh, N. Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA Splicing. PLOS Pathog. 2014, 10, e1004164. [Google Scholar] [CrossRef]
- Brandt, D.T.; Baarlink, C.; Kitzing, T.M.; Kremmer, E.; Ivaska, J.; Nollau, P.; Grosse, R. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of β1-integrin. Nat. Cell Biol. 2009, 11, 557–568. [Google Scholar] [CrossRef]
- Xia, S.; Zhu, Z.; Hao, L.; Chen, J.-G.; Xiao, L.; Zhang, Y.; Li, X. Negative Regulation of Systemic Acquired Resistance by Replication Factor C Subunit3 in Arabidopsis. Plant Physiol. 2009, 150, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, J.; Azumano, M.; Takeda, T. Nuclear localization of transcription factor Sp1. Nucleic Acids Symp. Ser. 1999, 42, 244–293. [Google Scholar] [CrossRef]
- Dittmer, J.; Gerloff, A.; Dittmer, A.; Oerlecke, I.; Holzhausen, H.-J.; Dittmer, J. Protein expression of the Ets transcription factor Elf-1 in breast cancer cells is negatively correlated with histological grading, but not with clinical outcome. Oncol. Rep. 2011, 26, 1121–1125. [Google Scholar] [CrossRef][Green Version]
- Yeung, M.-L.; Tam, T.S.M.; Tsang, A.C.C.; Yao, K.-M. Proteolytic cleavage of PDZD2 generates a secreted peptide containing two PDZ domains. EMBO Rep. 2003, 4, 412–418. [Google Scholar] [CrossRef]
- Taha, M.S.; Nouri, K.; Milroy, L.G.; Moll, J.M.; Herrmann, C.; Brunsveld, L.; Piekorz, R.P.; Ahmadian, M.R. Subcellular Fractionation and Localization Studies Reveal a Direct Interaction of the Fragile X Mental Retardation Protein (FMRP) with Nucleolin. PLoS ONE 2014, 9, e91465. [Google Scholar] [CrossRef] [PubMed]
- Tummala, H.; Walne, A.J.; Williams, M.; Bockett, N.; Collopy, L.; Cardoso, S.; Ellison, A.; Wynn, R.; Leblanc, T.; Fitzgibbon, J.; et al. DNAJC21 Mutations Link a Cancer-Prone Bone Marrow Failure Syndrome to Corruption in 60S Ribosome Subunit Maturation. Am. J. Hum. Genet. 2016, 99, 115–124. [Google Scholar] [CrossRef]
- Lutz, T.; Stöger, R.; Nieto, A. CHD6 is a DNA-dependent ATPase and localizes at nuclear sites of mRNA synthesis. FEBS Lett. 2006, 580, 5851–5857. [Google Scholar] [CrossRef]
- Alfonso, R.; Lutz, T.; Rodriguez, A.; Chavez, J.P.; Rodriguez, P.; Gutierrez, S.; Nieto, A. CHD6 chromatin remodeler is a negative modulator of influenza virus replication that relocates to inactive chromatin upon infection. Cell. Microbiol. 2011, 13, 1894–1906. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-M.; Kohn, M.J.; Bruinsma, M.W.; Vech, C.; Intine, R.V.; Fuhrmann, S.; Grinberg, A.; Mukherjee, I.; Love, P.E.; Ko, M.S.; et al. The Multifunctional RNA-Binding Protein La Is Required for Mouse Development and for the Establishment of EmbryonicStem Cells. Mol. Cell. Biol. 2006, 26, 1445–1451. [Google Scholar] [CrossRef]
- Wang, W.; Xue, Y.; Zhou, S.; Kuo, A.; Cairns, B.R.; Crabtree, G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996, 10, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Li, J.; Song, T.; Lu, M.; Kan, P.-Y.; Lee, M.G.; Sha, B.; Shi, X. Recognition of Histone H3K4 Trimethylation by the Plant Homeodomain of PHF2 Modulates Histone Demethylation. J. Biol. Chem. 2010, 285, 9322–9326. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.C.; Willadsen, K.; Buske, F.A.; Cao, K.-A.L.; Bailey, T.L.; Dellaire, G.; Bodén, M. Sorting the nuclear proteome. Bioinformatics 2011, 27, i7–i14. [Google Scholar] [CrossRef]
- Bonnet, J.; Lindeboom, R.G.; Pokrovsky, D.; Stricker, G.; Çelik, M.H.; Rupp, R.A.; Gagneur, J.; Vermeulen, M.; Imhof, A.; Müller, J. Quantification of Proteins and Histone Marks in Drosophila Embryos Reveals Stoichiometric Relationships Impacting Chromatin Regulation. Dev. Cell 2019, 51, 632–644.e6. [Google Scholar] [CrossRef]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Tajrishi, M.M.; Tuteja, R.; Tuteja, N. Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun. Integr. Biol. 2011, 4, 267–275. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Brameier, M.; Krings, A.; Maccallum, R.M. NucPred Predicting nuclear localization of proteins. Bioinformatics 2007, 23, 1159–1160. [Google Scholar] [CrossRef]
- Gorman, O.T.; Bean, W.J.; Kawaoka, Y.; Donatelli, I.; Guo, Y.J.; Webster, R.G. Evolution of influenza A virus nucleoprotein genes: Implications for the origins of H1N1 human and classical swine viruses. J. Virol. 1991, 65, 3704–3714. [Google Scholar] [CrossRef]
- Shu, L.L.; Bean, W.J.; Webster, R.G. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J. Virol. 1993, 67, 2723–2729. [Google Scholar] [CrossRef]
- Dong, G.; Peng, C.; Luo, J.; Wang, C.; Han, L.; Wu, B.; Ji, G.; He, H. Adamantane-Resistant Influenza A Viruses in the World (1902–2013): Frequency and Distribution of M2 Gene Mutations. PLoS ONE 2015, 10, e0119115. [Google Scholar] [CrossRef]
- Thyagarajan, B.; Bloom, J.D. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife 2014, 3, e03300. [Google Scholar] [CrossRef]
- Marfori, M.; Mynott, A.; Ellis, J.J.; Mehdi, A.M.; Saunders, N.F.; Curmi, P.M.; Forwood, J.K.; Bodén, M.; Kobe, B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 2011, 1813, 1562–1577. [Google Scholar] [CrossRef] [PubMed]
- Kalderon, D.; Richardson, W.D.; Markham, A.F.; Smith, A.E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 1984, 311, 33–38. [Google Scholar] [CrossRef]
- Colledge, W.H.; Richardson, W.D.; Edge, M.D.; E Smith, A. Extensive mutagenesis of the nuclear location signal of simian virus 40 large-T antigen. Mol. Cell. Biol. 1986, 6, 4136–4139. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.I.; Dombrovski, A.K.; Swarbrick, C.M.; Raidal, S.R.; Forwood, J.K. Structural determination of importin alpha in complex with beak and feather disease virus capsid nuclear localization signal. Biochem. Biophys. Res. Commun. 2013, 438, 680–685. [Google Scholar] [CrossRef]
- Wolff, T.; Unterstab, G.; Heins, G.; Richt, J.A.; Kann, M. Characterization of an Unusual Importin α Binding Motif in the Borna Disease Virus p10 Protein That Directs Nuclear Import. J. Biol. Chem. 2002, 277, 12151–12157. [Google Scholar] [CrossRef]
- Lombardo, E.; Ramírez, J.C.; García-Pérez, J.; Almendral, J.M. Complementary Roles of Multiple Nuclear Targeting Signals in the Capsid Proteins of the Parvovirus Minute Virus of Mice during Assembly and Onset of Infection. J. Virol. 2002, 76, 7049–7059. [Google Scholar] [CrossRef]
- Yu, H.T.; Chan, W.W.; Chai, K.H.; Lee, C.W.; Chang, R.C.; Yu, M.S. Transcriptional regulation of human FE65, a ligand of Alzheimer’s disease amyloid precursor protein, by Sp1. J Cell Biochem. 2010, 109, 782–793. [Google Scholar] [PubMed]





| Construct Name | Primer Sequence (F Indicates Forward; R Indicates Reverse) |
|---|---|
| NLS1 MT | F′5′-GATCCAATGGCGTCTCAAGGCACCAAACGATCATATGAACAATGCCG-3′ R′5′-GATCCGGCATTTGTTCATACGATCGTTTGGTGCCTTGAGACGCCATTG-3′ |
| NLS2 MT | F′5′-GAGGGGTGAAAATGGAGCAAAGACAGCGCCGAATC-3′ R′5′-GATCCGGCGCTGTCTTTGCTCCATTTTCACCCCTC-3′ |
| NOP14-mutNLS2 | F′5′-ACGGCCCACGACGTGGGACTGCCC-3′ R′3′-CGCCGCGCCCAGGATCTGGAACTTCTG-5′ |
| NOP14-mutSeq3 and NOP14-mutNLS2/Seq3 | F′5′-TCTGGCGGCGGCAGCGTTCAAAAAAACGCG-3′ R′3′-GCCTTCCATTCGCCTTCC-5′ |
| P2 | F′5′-TGAAAATGGAAAAAAGACAAGGCCG-3′ R′3′-CCCCTCCAGAAATTTCGG-5′ |
| P3 | F′5′-AAATGGACGAAGGACAAGGCCGG-3′ R′3′-TCACCCCTCCAGAAATTTCG-5′ |
| P5 | F′5′-ACGAAAGACAAAGCCGGATCCAC-3′ R′3′-CCATTTTCACCCCTCCAGAAATTTC-5′ |
| P2P3 | F′5′-TGAAAATGGAAAAAGGACAAGGCCG-3′ R′3′-CCCCTCCAGAAATTTCGG-5′ |
| P2P5 | F′5′-TGAAAATGGAAAAAGGACAAGGCCG-3′ R′3′-CCCCTCCAGAAATTTCGG-5′ |
| P3P5 | F′5′-ACGAAGGACAAAGCCGGATCCAC-3′ R′3′-CCATTTTCACCCCTCCAGAAATTTC-5′ |
| P2P3P5 | F′5′-AAATGGAAAAAGGACAAAGCCGG-3′ R′3′-TCACCCCTCCAGAAATTTC-5′ |
| NLS2 Variant | Position at the Importin-α Major Binding Site 1 P1 P2 P3 P4 P5 |
|---|---|
| WT | 212G R K T R216 |
| P2: R→ K at 213 | 212G K K T R216 |
| P3: K→ R at 214 | 212G R R T R216 |
| P5: R→ K at 216 | 212G R K T K216 |
| P2P3: R→ K at 213 and K→ R at 214 | 212G K R T R216 |
| P2P5: R→ K at 213 and R→ K at 216 | 212G K K T K216 |
| P3P5: K→ R at 214 and R→ K at 216 | 212G R R T K216 |
| P2P3P5: R→ K at 213 and K→ R at 214 and R→K at 216 | 212G K R T K216 |
| Protein 1 | Protein Function 2 [References] | Virus/Host | Putative NLS2 Variant | Predicted NLS 3 |
|---|---|---|---|---|
| Adenoviridae family | ||||
| Minor core protein pV | Participates in capsid assembly in the nucleus [57,58] | Harbour porpoise adenovirus 1 | P2: 130GKKTR134 | 21RKRKTPKREPKTEIKIERVKTEDVKPFKKGKRRKH55 |
| Human mastadenovirus B and several human adenoviruses (16, 3 + 7, 68, 66, 7d2) | P2P3: 117GKRTR121 | 297YKPPKRQYRKRKTRRVRQGRR317 | ||
| Precursor terminal protein pTP | Participates in viral replication [59,60] | Titi monkey adenovirus ECC-2011 | P3: 350GRRTR354 | 339GARPGLRRRPTAGRR353 |
| Squirrel monkey adenovirus | P3: 351GRRTR355 | 389RLPIRRRRRRAPP401 | ||
| Viral RNA splicing factor L4-33 kDa | Required for genome packaging and capsid assembly in the nucleus [61,62] | Deer mastadenovirus B and murine adenovirus 3 | P2: 74GKKTR78 108GKKTR112 | 126RGRRR130 |
| Late L2 mu core protein pX | Condenses the viral pro-chromatin for encapsidation (reviewed in [63,64]) | Murine adenovirus 2 and canine adenovirus 1 | P3: 26GRRTR30 31GRRTR35 | 15RSRRLRRRLGGGGCSSGRRTRRRSYRRRRGLR46 |
| Encapsidation protein L1-52/55 kDa | Involved in genome packaging in the nucleus [65,66] | Duck adenovirus 4 | P3: 23GRRTR27 | No NLS predicted |
| Hexon-associated structural protein pVIII precursor | Capsid assembly in the nucleus by connecting the major structural units with each other and with the viral core (reviewed in [67,68,69]) | Bovine adenovirus 1 | P3P5: 111GRRTK115 | No NLS predicted |
| Viral transcription factor L4-22 kDa | Required for genome packaging and capsid assembly in the nucleus [61,62] | Murine adenovirus 3 | P2: 84GKKTR88 | No NLS predicted |
| Herpesviridae family | ||||
| DNA polymerase processivity subunit | Involved in viral DNA replication [70] | Wood mouse herpesvirus, murid gammaherpesvirus 4, and 68 | P2: 210GKKTR214 | 373KRPPPKKEKEPTPKRPK389 |
| Tegument protein UL32 | Associates with nuclear capsids prior to DNA encapsidation and preserves the integrity of capsids through secondary envelopment [71,72] | Cynomolgus macaque cytomegalovirus strain Ottawa | P5: 312GRKTK316 | 546PKAKRRLILKPKTKKNVPKPKP567 |
| Tegument protein VP22 | Regulates the activity of the viral endonuclease vhs [73] | Pteropus lylei-associated alpha herpesvirus | P3: 148GRRTR152 | 89RRGRGAARPAAARAPTARRAPASGGAASARGTRGAAAS126 144ASASGRRTRRP154 |
| Assembly protein M80 | Coordinates capsid assembly in the nucleus [74,75] | Murine betaherpesvirus 1 | P2P3: 504GKRTR508 505GKRTR509 506GKRTR510 507GKRTR511 508GKRTR512 | 507GGKRTRQRGSADSGRKRRRRG527 |
| Circoviridae family | ||||
| Capsid protein | Binds and transports the viral genome through the NPC [76,77] | Capybara-associated cyclovirus 1 | P3: 9GRRTR13 | 5RRFKGRRTRLPWRRSRFVRRRRGRFSRRTRRNYRR39 |
| Hepadnaviridae family | ||||
| X protein | Regulates transcription through direct interaction with different transcription factors [78] | Human hepatitis B virus | P2P5: 124GKKTK128 | No NLS predicted |
| Protein 1 [Localization References] | Organism | Putative NLS2 Variant | Predicted NLS 2 |
|---|---|---|---|
| Nucleolar protein 14 (NOP14) [81] | H. sapiens M. musculus | WT: 45GRKTR49 | 3KAKKVGARRKASGAPAGARGGPAKA27 784KEEQERKRLIHKHKREFKGAVRE807 848KALKRKKFKK857 |
| Nuclear mitotic apparatus protein 1 isoform X1 [82] | H. sapiens M. musculus | WT: 1819GRKTR1823 1805GRKTR1809 1801GRKTR1805 1797GRKTR1801 1787GRKTR1791 1783GRKTR1787 1748GRKTR1752 1734GRKTR1738 1705GRKTR1709 | 2083RRGASKKALSKASP2096 2127AKGKAKH2133 |
| Spliceosomal factor RED [83] | H. sapiens M. musculus D. rerio A. thaliana | WT: 532GRKTR536 513GRKTR517 504GRKTR508 | 73RRRKKKS79, 541KRK543 294RNKKLKKKDKGKLEEKKP311 334RDKERERYRERERDRERDRDRDRERERERDRERERERDREREEEKKRH381 |
| Serum response factor [84] | H. sapiens M. musculus D. rerio | P2: 137GKKTR141 133GKKTR137 116GKKTR120 | 135KPGKKTRGRVKIK146 |
| Replication factor C subunit 3 [85] | H. sapiens M. musculus D. rerio D. melanogaster C. elegans S. cerevisiae D. discoideum | P2: 49GKKTR53 48GKKTR52 47GKKTR51 46GKKTR50 | No NLS predicted |
| Transcription factor Sp1 [86] | H. sapiens M. musculus | P3: 596GRRTR600 594GRRTR598 589 GRRTR593 587 GRRTR591 548 GRRTR552 | No NLS predicted |
| ETS-related transcription factor Elf-1 [87] | H. sapiens M. musculus | P5: 177 GRKTK181 153 GRKTK157 140 GRKTK144 118 GRKTK122 | 171QRKRKKGRKTKPPRP185 |
| PDZ domain-containing protein 2 [88] | H. sapiens M. musculus D. rerio | P2P3: 230GKRTR234 213GKRTR217 48GKRTR52 39GKRTR43 | 99KRRGGKKRK107 210AKKGKRTRKFGVISR224 |
| Nucleolin [89] | H. sapiens M. musculus D. rerio | P2P5: 739GKKTK743 735GKKTK739 704GKKTK708 701GKKTK705 685GKKTK689 | 277AAPGKRKKEMTKQKEAPEAKK297 382KPKGRDSKKVR392 644PKGEGGFGGRGGGRGGFGGRGGGRGGRGGFGGRGRGGFGGRGGFRGGRGGGGDFKPQGKKTK705 |
| DnaJ homolog subfamily C member 21 (DNJC21) [90] | H. sapiens M. musculus | P2P5: 604GKKTK608 546GKKTK550 517GKKTK521 504GKKTK508 459GKKTK463 472GKKTK476 334GKKTK338 | 181KRAMEKENKKIRDRARKEKNELVRQLVAFIRKRDKRVQAHRKLV224 230EKARKAE236 380QKLSKKQKKKKQKS393 452KSVPKSKGKKTKDVKKSVK470 523NKKEKRRSR531 |
| Chromo-domain helicase DNA-binding protein 6 (CHD6) [91,92] | H. sapiens M. musculus | P2P5: 1187GKKTK1191 1186GKKTK1190 1185GKKTK1189 1184GKKTK1188 1164GKKTK1168 1163GKKTK1167 1139GKKTK1143 852GKKTK 856 509GKKTK513 | 175GSRTKSKKASREQGPTPVERKKKGKRK201 236RSGRQVKR243 1181RGRKGKK1187 2284RRRRGRRK2291 2437GPRRRGRRPR2446 2652KRKKKKTK2659 |
| Lupus La protein [93] | H. sapiens D. rerio | P2P5: 359GKKTK363 357GKKTK361 | 328KWKSKGRRFKGKGKGNKAAQPGSGKGKV355 |
| Brefeldin A-inhibited guanine nucleotide-exchange protein 1 [89] | H. sapiens M. musculus D. rerio | P2P5: 4GKKTK8 | No NLS predicted |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 [94] | H. sapiens M. musculus | P2P5: 69GKKTK73 | No NLS predicted |
| Lysine-specific demethylase PHF2 [95] | H. sapiens M. musculus | P2P3P5: 1069GKRTK1073 1068GKRTK1072 1034GKRTK1038 | 63KKKR66, 888KKR890, 942KNRKKKNTKRKP953 487KVSKKKTSKTVKMPKPSKIPKPPKSPKPPKTLKLKDGSKKKGKK530 827RKIGGGNKGTGKRLLKR843 1066AKGKRTKKGMATAKQRLGKILKIHRN1091 |
| Thymocyte nuclear protein 1 [46] | H. sapiens M. musculus | P2P3P5: 44GKRTK48 21GKRTK25 20GKRTK24 | 26RPRKRQTGTAGPDRKKLSGKR46 |
| NLS Type | Minor Binding Site P1′ P2′ P3′ P4′ P5′ | Linker | Major Binding Site P1 P2 P3 P4 P5 | PDB id |
|---|---|---|---|---|
| SV40 large T-antigen | K K R K | K K K R K | 1EJL/1BK6 1 | |
| hPLSCR1-NLS | G K I S K | 1Y2A | ||
| hPLSCR4-NLS | I R KW N | 3Q5U | ||
| Guα -NLS | K R S F | 3ZIN | ||
| A89-NLS | K R K Y W | 4B8P 2 | ||
| B54-NLS | K R K R H | 2YNS 2 | ||
| TPX2 | K R K H | V K M I K | 3KND | |
| C-Myc | K R V K L | A K R V K | 1EE4 1 | |
| Nucleoplasmin | K R P A A | TKKAG | K K K K L | 1EJY/1EE5 1 |
| Kap60-IBB | R R R R D | TQQVELRKAKRDEA | A K R R N | 1WA5 1 |
| h1NLS | K R K D P | DSDDWSES | S K E N K | 4XZR 1 |
| h2NLS | K R K R E | QISTDNEAKMQIQEEKS | K K K R K | 4PVZ 1 |
| hRCC1 | K R R S | PPADAIP | S K K V K | 5TBK |
| yRCC1 | K R T V A | TNGDASGAH | K K M S K | 5T94 1 |
| BFDV Cap NLS | Y R R R R R Y | 4HTV | ||
| Influenza A NP-NLS1 | K R S Y E | 4ZDU | ||
| WT NLS2 | R K T R | G R K T R | 5V5O | |
| NLS2 P3 variant | R R T R | G R R T R | 5V5P |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.L.T.; Panté, N. Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein. Cells 2022, 11, 2957. https://doi.org/10.3390/cells11192957
Nguyen NLT, Panté N. Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein. Cells. 2022; 11(19):2957. https://doi.org/10.3390/cells11192957
Chicago/Turabian StyleNguyen, Nhan L. T., and Nelly Panté. 2022. "Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein" Cells 11, no. 19: 2957. https://doi.org/10.3390/cells11192957
APA StyleNguyen, N. L. T., & Panté, N. (2022). Bioinformatics and Functional Analysis of a New Nuclear Localization Sequence of the Influenza A Virus Nucleoprotein. Cells, 11(19), 2957. https://doi.org/10.3390/cells11192957






