Micro-RNA193a-3p Inhibits Breast Cancer Cell Driven Growth of Vascular Endothelial Cells by Altering Secretome and Inhibiting Mitogenesis: Transcriptomic and Functional Evidence
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Conditioned Media (CM) Formation
2.3. Spheroid Formation
2.4. Transfection with miRNA
2.5. Cell Proliferation Assay
2.6. Migration Studies
2.7. Microvessel Formation Assay
2.8. Western Blot
2.9. Microarray
2.10. Quantitative RT-PCR
2.11. Angiogenesis Proteome Array
2.12. Immunohistochemistry
2.13. Statistical Analysis
3. Results
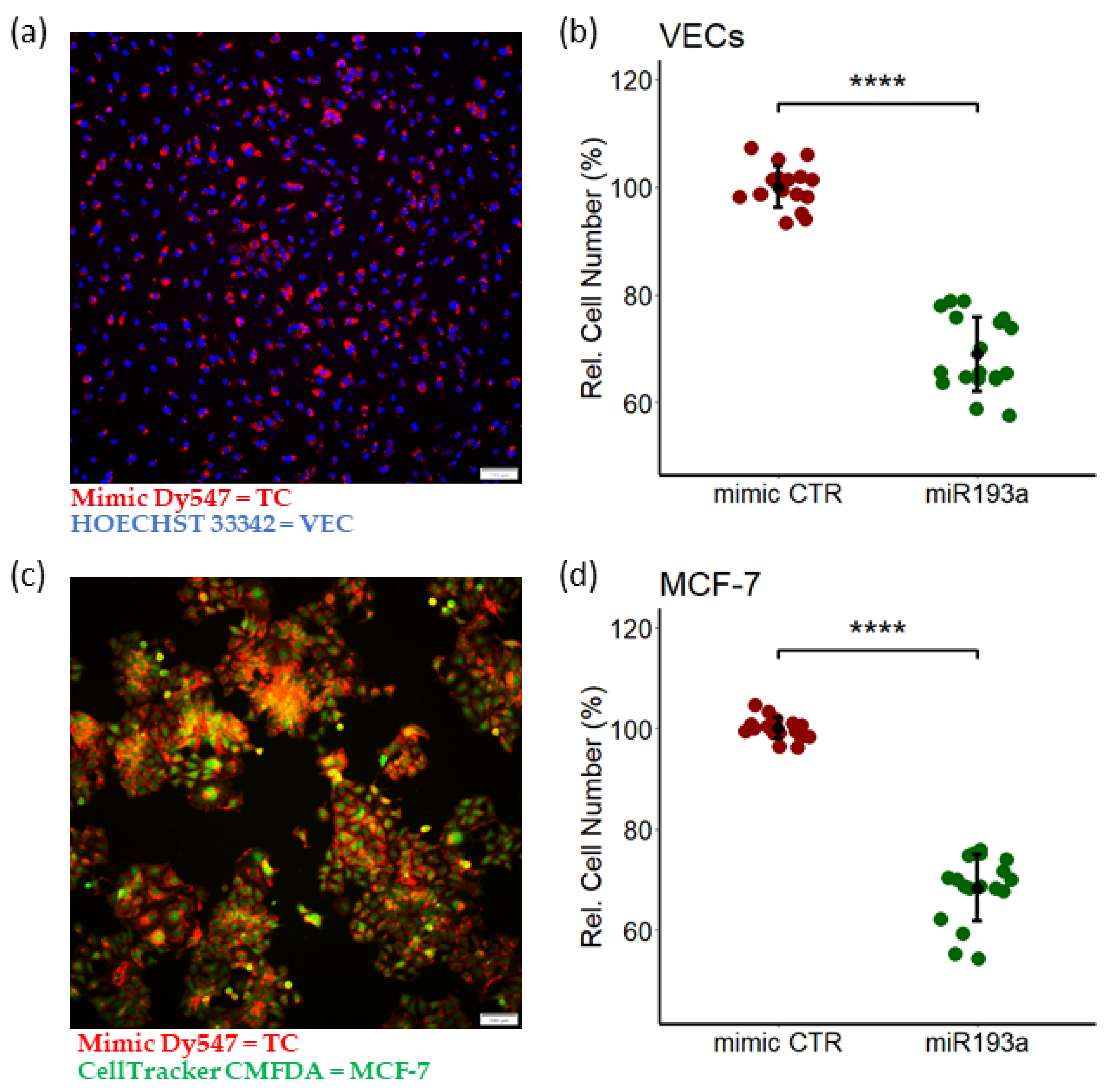
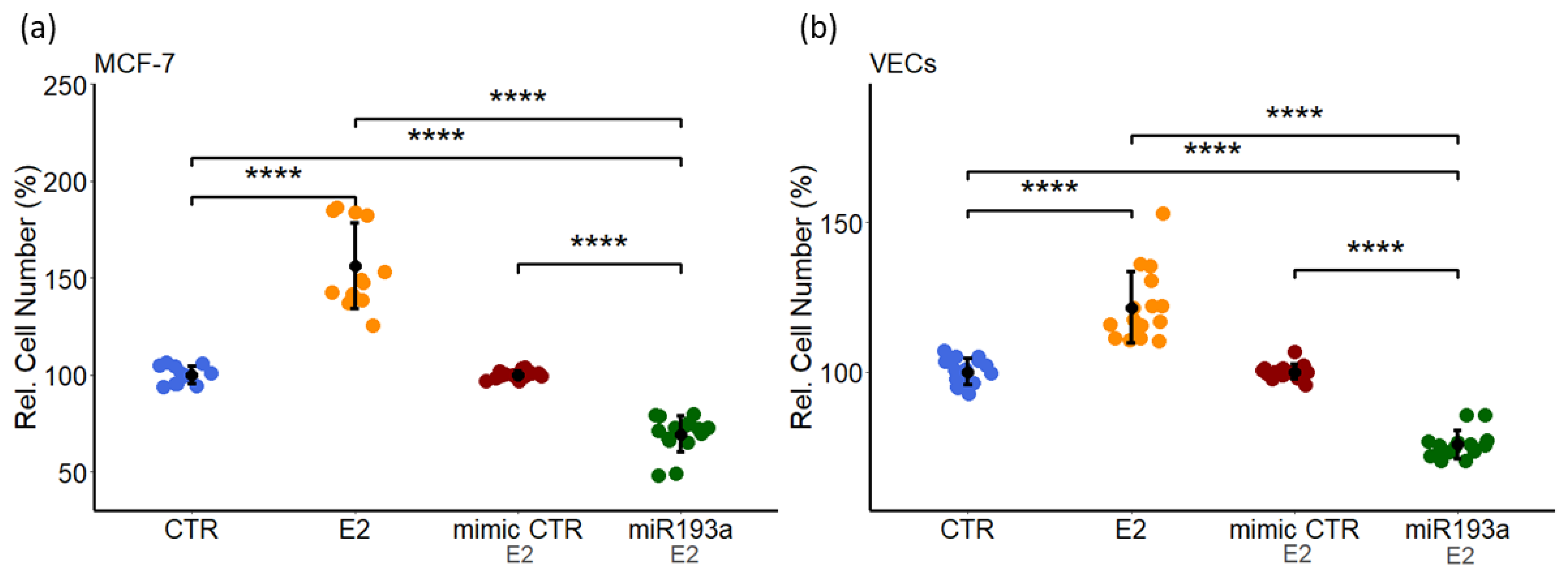
3.1. miR193a-3p Inhibits Serum-Induced Growth of MCF-7 Cells and VECs
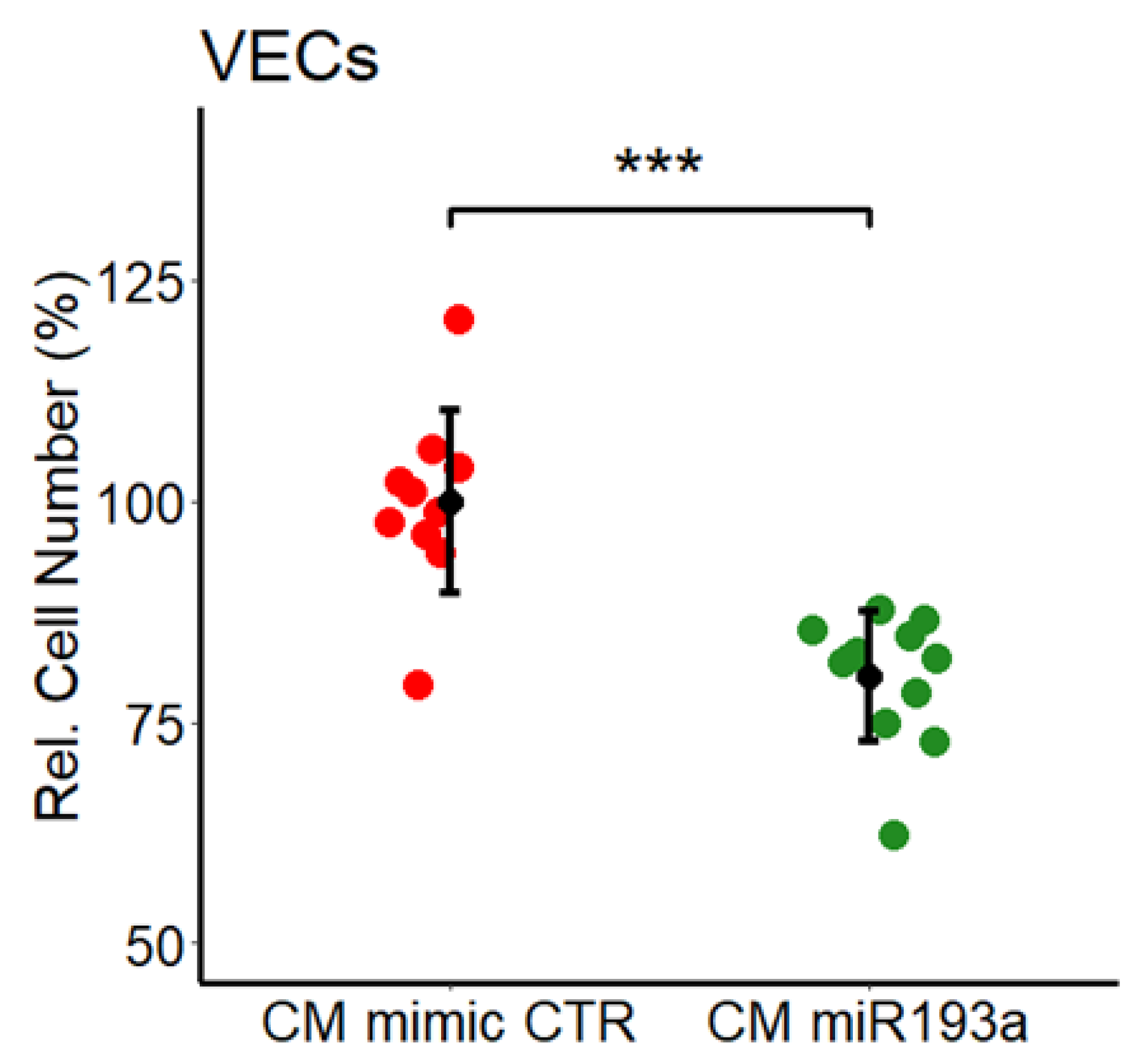
3.2. Effect of miR193a-3p Conditioned Media on VECs
3.3. Effect of miR193a-3p Conditioned Media on cell Migration/Wound Closure
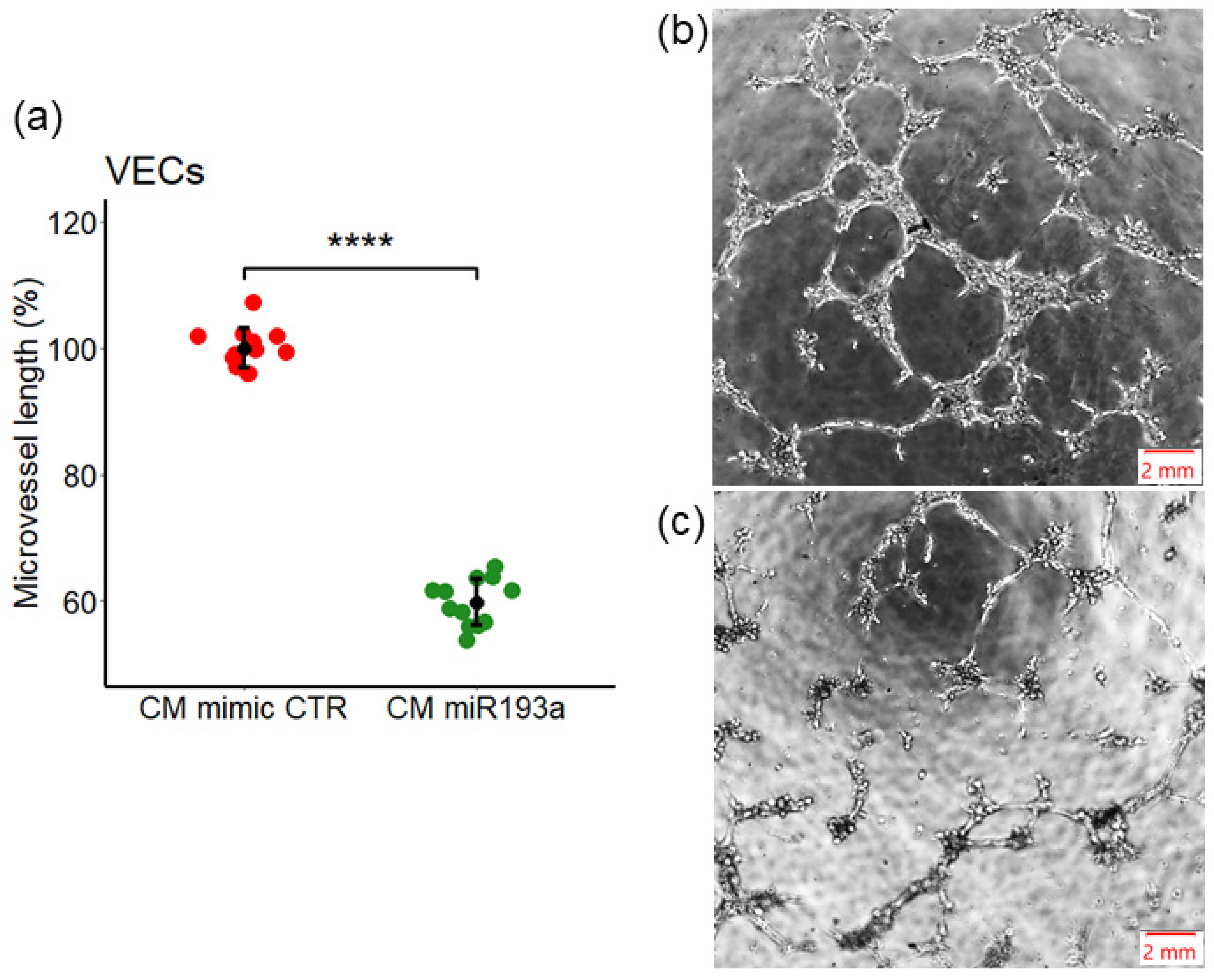
3.4. Effect of miR193a-3p Conditioned Media on Tube/Capillary Formation by VECs
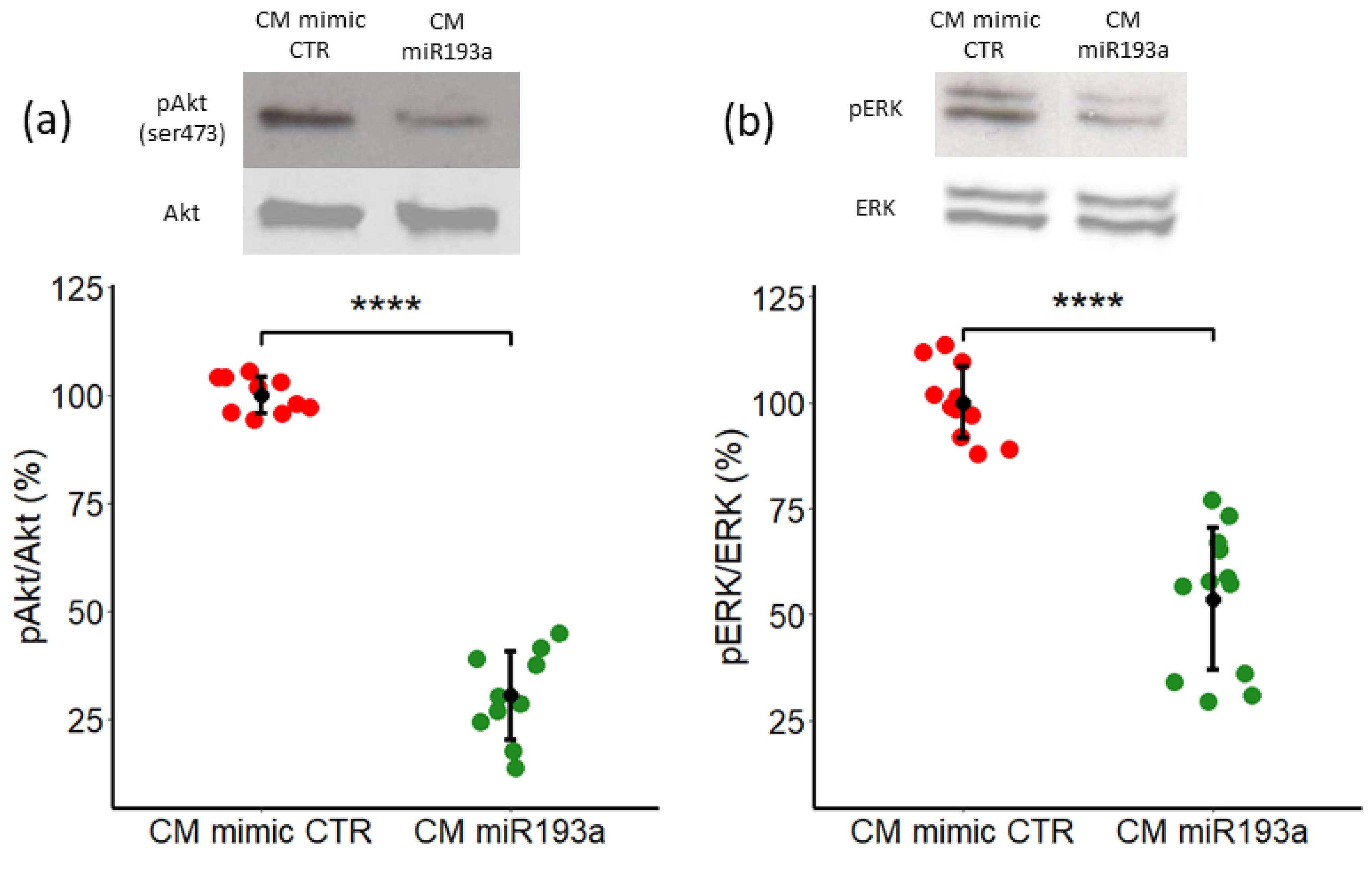
3.5. Secretome/CM from miRNA193a-3p Transfected MCF-7 Inhibits VEC Growth by Downregulating ERK1/2 and PI3K-Akt Pathways
3.6. Microarray Analysis
3.6.1. Differentially Regulated Genes
3.6.2. Validation of Differentially Regulated Genes by Quantitative Real Time PCR
3.6.3. Pathway Enrichment Analysis of DRGs
3.6.4. Gene’s Identification in Biological Pathways
3.7. Angiogenesis Proteome Profiler in VECs
3.8. miR193a-3p Abrogates E2 Induced Growth in VECs and MCF-7 Cells
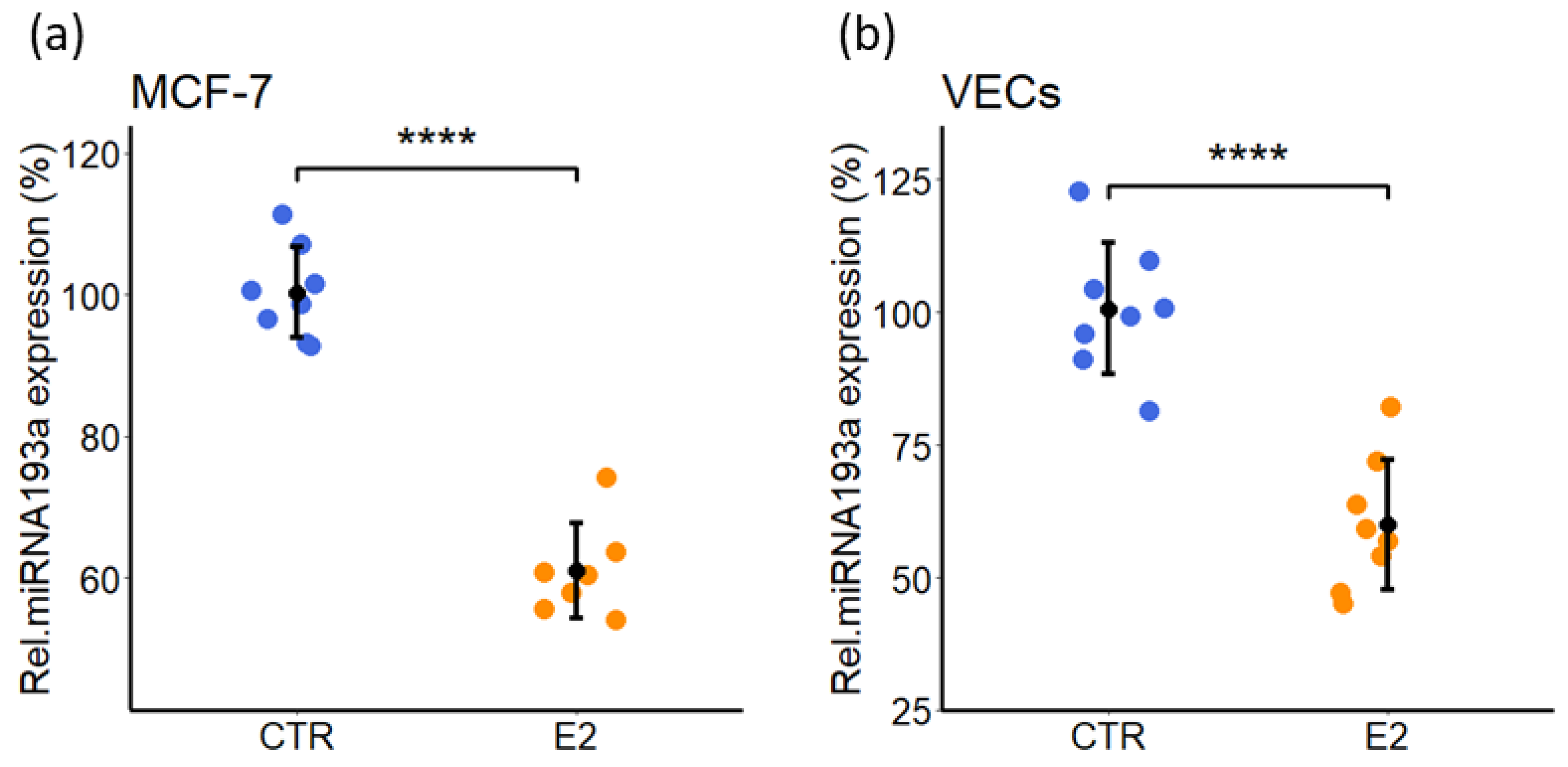
3.9. Estradiol (E2) Decreases miRNA193a-3p Expression in MCF-7 and VECs
3.10. Growth Effects of E2 on MCF-7 and MCF-7 Plus VEC Spheroids
3.11. Inhibitory Effects of miR193a-3p on E2 and FCS Induced Growth of Spheroids
3.12. Immunohistochemistry
3.13. MDA-MB-231 Secretome Induced VEC Proliferation are Lost in Secretome from miR193a-3p Transfected Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Comşa, Ş.; Cîmpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar] [PubMed]
- Libson, S.; Lippman, M. A review of clinical aspects of breast cancer. Int. Rev. Psychiatry 2014, 26, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, L.; Jordāo, M.J.C.; Joyce, J.A. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov. 2021, 11, 933–959. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Tsang, J.Y.; Tse, G.M. Tumor Microenvironment in Breast Cancer-Updates on Therapeutic Implications and Pathologic Assessment. Cancers 2021, 13, 4233. [Google Scholar] [CrossRef]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef]
- Lee, E.; Pandey, N.B.; Popel, A.S. Crosstalk between cancer cells and blood endothelial and lymphatic endothelial cells in tumour and organ microenvironment. Expert Rev. Mol. Med. 2015, 17, e3. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal 2020, 18, 59. [Google Scholar] [CrossRef]
- Madu, C.O.; Wang, S.; Madu, C.O.; Lu, Y. Angiogenesis in Breast Cancer Progression, Diagnosis, and Treatment. J. Cancer 2020, 11, 4474–4494. [Google Scholar] [CrossRef]
- Ayoub, N.M.; Jaradat, S.K.; Al-Shami, K.M.; Alkhalifa, A.E. Targeting Angiogenesis in Breast Cancer: Current Evidence and Future Perspectives of Novel Anti-Angiogenic Approaches. Front. Pharmacol. 2022, 13, 838133. [Google Scholar] [CrossRef]
- Aalders, K.C.; Tryfonidis, K.; Senkus, E.; Cardoso, F. Anti-angiogenic treatment in breast cancer: Facts, successes, failures and future perspectives. Cancer Treat. Rev. 2017, 53, 98–110. [Google Scholar] [CrossRef]
- Azzarito, G.; Visentin, M.; Leeners, B.; Dubey, R.K. Transcriptomic and Functional Evidence for Differential Effects of MCF-7 Breast Cancer Cell-Secretome on Vascular and Lymphatic Endothelial Cell Growth. Int. J. Mol. Sci. 2022, 23, 7192. [Google Scholar] [CrossRef]
- Klinge, C.M. miRNAs regulated by estrogens, tamoxifen, and endocrine disruptors and their downstream gene targets. Mol. Cell Endocrinol. 2015, 418 Pt 3, 273–297. [Google Scholar] [CrossRef]
- Hydbring, P.; Wang, Y.; Fassl, A.; Li, X.; Matia, V.; Otto, T.; Choi, Y.J.; Sweeney, K.E.; Suski, J.M.; Yin, H.; et al. Cell-Cycle-Targeting MicroRNAs as Therapeutic Tools against Refractory Cancers. Cancer Cell 2017, 31, 576–590. [Google Scholar] [CrossRef]
- Kayani, M.; Kayani, M.A.; Malik, F.A.; Faryal, R. Role of miRNAs in breast cancer. Asian Pac. J. Cancer Prev. 2011, 12, 3175–3180. [Google Scholar]
- Farazi, T.A.; Spitzer, J.I.; Morozov, P.; Tuschl, T. miRNAs in human cancer. J. Pathol. 2011, 223, 102–115. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef]
- Fridrichova, I.; Zmetakova, I. MicroRNAs Contribute to Breast Cancer Invasiveness. Cells 2019, 8, 1361. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Evangelista, A.F.; Oliveira, R.J.; VA, O.S.; RA, D.C.V.; Reis, R.M.; MM, C.M. Integrated analysis of mRNA and miRNA profiles revealed the role of miR-193 and miR-210 as potential regulatory biomarkers in different molecular subtypes of breast cancer. BMC Cancer 2021, 21, 76. [Google Scholar] [CrossRef]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Ejifcc 2019, 30, 114–127. [Google Scholar] [PubMed]
- Khordadmehr, M.; Shahbazi, R.; Ezzati, H.; Jigari-Asl, F.; Sadreddini, S.; Baradaran, B. Key microRNAs in the biology of breast cancer; emerging evidence in the last decade. J. Cell. Physiol. 2019, 234, 8316–8326. [Google Scholar] [CrossRef] [PubMed]
- Travis, R.C.; Key, T.J. Oestrogen exposure and breast cancer risk. Breast Cancer Res. 2003, 5, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Type and timing of menopausal hormone therapy and breast cancer risk: Individual participant meta-analysis of the worldwide epidemiological evidence. Lancet 2019, 394, 1159–1168. [CrossRef]
- Guo, L.; Xu, J.; Qi, J.; Zhang, L.; Wang, J.; Liang, J.; Qian, N.; Zhou, H.; Wei, L.; Deng, L. MicroRNA-17-92a upregulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. J. Cell Sci. 2013, 126, 978–988. [Google Scholar] [CrossRef]
- Lv, H.; Sun, Y.; Zhang, Y. MiR-133 is Involved in Estrogen Deficiency-Induced Osteoporosis through Modulating Osteogenic Differentiation of Mesenchymal Stem Cells. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2015, 21, 1527–1534. [Google Scholar] [CrossRef][Green Version]
- Fu, J.; Hao, L.; Tian, Y.; Liu, Y.; Gu, Y.; Wu, J. miR-199a-3p is involved in estrogen-mediated autophagy through the IGF-1/mTOR pathway in osteocyte-like MLO-Y4 cells. J. Cell. Physiol. 2018, 233, 2292–2303. [Google Scholar] [CrossRef]
- Vrtačnik, P.; Ostanek, B.; Mencej-Bedrač, S.; Marc, J. The many faces of estrogen signaling. Biochem. Med. 2014, 24, 329–342. [Google Scholar] [CrossRef]
- Bhat-Nakshatri, P.; Wang, G.; Collins, N.R.; Thomson, M.J.; Geistlinger, T.R.; Carroll, J.S.; Brown, M.; Hammond, S.; Srour, E.F.; Liu, Y.; et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009, 37, 4850–4861. [Google Scholar] [CrossRef]
- Wickramasinghe, N.S.; Manavalan, T.T.; Dougherty, S.M.; Riggs, K.A.; Li, Y.; Klinge, C.M. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009, 37, 2584–2595. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, S.; Wang, M.; Liu, D.; Liu, Y.; Zhang, Y.; Zhang, Q. Epigenetically altered miR-193a-3p promotes HER2 positive breast cancer aggressiveness by targeting GRB7. Int. J. Mol. Med. 2019, 43, 2352–2360. [Google Scholar] [CrossRef]
- Xie, F.; Hosany, S.; Zhong, S.; Jiang, Y.; Zhang, F.; Lin, L.; Wang, X.; Gao, S.; Hu, X. MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS ONE 2017, 12, e0185565. [Google Scholar] [CrossRef]
- Zhuo, Y.; Li, X.; Zheng, Q.; Fan, X.; Ma, W.; Chen, J.; Zhao, X.; Zhao, P.; Liu, X.; Tang, F.; et al. Estrogen enhances tumor growth and angiogenesis indirectly via mediation of bone marrow-derived cells as well as directly through stimulation of tumor and endothelial cells. Oncol. Rep. 2018, 40, 2147–2156. [Google Scholar] [CrossRef]
- Azzarito, G.; Szutkowska, M.E.; Saltari, A.; Jackson, E.K.; Leeners, B.; Rosselli, M.; Dubey, R.K. Mammary Epithelial and Endothelial Cell Spheroids as a Potential Functional In vitro Model for Breast Cancer Research. J. Vis. Exp. 2021, 173, e62940. [Google Scholar] [CrossRef]
- Tahiri, A.; Leivonen, S.K.; Lüders, T.; Steinfeld, I.; Ragle Aure, M.; Geisler, J.; Mäkelä, R.; Nord, S.; Riis, M.L.; Yakhini, Z.; et al. Deregulation of cancer-related miRNAs is a common event in both benign and malignant human breast tumors. Carcinogenesis 2014, 35, 76–85. [Google Scholar] [CrossRef]
- Yu, M.; Liu, Z.; Liu, Y.; Zhou, X.; Sun, F.; Liu, Y.; Li, L.; Hua, S.; Zhao, Y.; Gao, H.; et al. PTP1B markedly promotes breast cancer progression and is regulated by miR-193a-3p. FEBS J. 2019, 286, 1136–1153. [Google Scholar] [CrossRef]
- Khordadmehr, M.; Shahbazi, R.; Sadreddini, S.; Baradaran, B. miR-193: A new weapon against cancer. J. Cell. Physiol. 2019, 234, 16861–16872. [Google Scholar] [CrossRef]
- Sheng, S.; Qiao, M.; Pardee, A.B. Metastasis and AKT activation. J. Cell. Physiol. 2009, 218, 451–454. [Google Scholar] [CrossRef]
- Li, Q.T.; Feng, Y.M.; Ke, Z.H.; Qiu, M.J.; He, X.X.; Wang, M.M.; Li, Y.N.; Xu, J.; Shi, L.L.; Xiong, Z.F. KCNN4 promotes invasion and metastasis through the MAPK/ERK pathway in hepatocellular carcinoma. J. Investig. Med. 2020, 68, 68–74. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef]
- Jung, H.Y.; Fattet, L.; Yang, J. Molecular pathways: Linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Pandey, N.B.; Popel, A.S. Lymphatic endothelial cells support tumor growth in breast cancer. Sci. Rep. 2014, 4, 5853. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Liao, Q.; Su, M.; Huang, K.; Jin, J.; Cao, D. AKT and ERK dual inhibitors: The way forward? Cancer Lett. 2019, 459, 30–40. [Google Scholar] [CrossRef]
- Minn, A.J. Interferons and the Immunogenic Effects of Cancer Therapy. Trends Immunol. 2015, 36, 725–737. [Google Scholar] [CrossRef]
- Cheon, H.; Borden, E.C.; Stark, G.R. Interferons and their stimulated genes in the tumor microenvironment. Semin. Oncol. 2014, 41, 156–173. [Google Scholar] [CrossRef]
- Saleiro, D.; Platanias, L.C. Interferon signaling in cancer. Non-canonical pathways and control of intracellular immune checkpoints. Semin. Immunol. 2019, 43, 101299. [Google Scholar] [CrossRef]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Zhang, C.; Peng, L.; Zhang, Y.; Liu, Z.; Li, W.; Chen, S.; Li, G. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med. Oncol. 2017, 34, 101. [Google Scholar] [CrossRef]
- Chen, W.; Qin, Y.; Liu, S. CCL20 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1231, 53–65. [Google Scholar] [CrossRef]
- Forey, P.; Cros-Perrial, E.; Dumontet, C.; Jordheim, L.P. Targeting the Nucleotide Metabolism Proteins of the NUDIX Family and SAMHD1 in Cancer. Curr. Med. Chem. 2021, 28, 4088–4116. [Google Scholar] [CrossRef]
- Hatakeyama, S. TRIM proteins and cancer. Nat. Rev. Cancer 2011, 11, 792–804. [Google Scholar] [CrossRef]
- Huang, P.; Liao, R.; Chen, X.; Wu, X.; Li, X.; Wang, Y.; Cao, Q.; Dong, C. Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer. Theranostics 2020, 10, 4644–4658. [Google Scholar] [CrossRef]
- Pidugu, V.K.; Pidugu, H.B.; Wu, M.M.; Liu, C.J.; Lee, T.C. Emerging Functions of Human IFIT Proteins in Cancer. Front. Mol. Biosci. 2019, 6, 148. [Google Scholar] [CrossRef]
- Martín-Martín, N.; Piva, M.; Urosevic, J.; Aldaz, P.; Sutherland, J.D.; Fernández-Ruiz, S.; Arreal, L.; Torrano, V.; Cortazar, A.R.; Planet, E.; et al. Stratification and therapeutic potential of PML in metastatic breast cancer. Nat. Commun. 2016, 7, 12595. [Google Scholar] [CrossRef]
- Chang, Z.; Liu, X.; Zhao, W.; Xu, Y. Identification and Characterization of the Copy Number Dosage-Sensitive Genes in Colorectal Cancer. Mol. Ther. Methods Clin. Dev. 2020, 18, 501–510. [Google Scholar] [CrossRef]
- Henle, A.M.; Nassar, A.; Puglisi-Knutson, D.; Youssef, B.; Knutson, K.L. Downregulation of TAP1 and TAP2 in early stage breast cancer. PLoS ONE 2017, 12, e0187323. [Google Scholar] [CrossRef]
- Chou, F.C.; Chen, H.Y.; Kuo, C.C.; Sytwu, H.K. Role of Galectins in Tumors and in Clinical Immunotherapy. Int. J. Mol. Sci. 2018, 19, 430. [Google Scholar] [CrossRef]
- Aljohani, A.I.; Joseph, C.; Kurozumi, S.; Mohammed, O.J.; Miligy, I.M.; Green, A.R.; Rakha, E.A. Myxovirus resistance 1 (MX1) is an independent predictor of poor outcome in invasive breast cancer. Breast Cancer Res. Treat. 2020, 181, 541–551. [Google Scholar] [CrossRef]
- Hu, M.M.; Yang, Q.; Zhang, J.; Liu, S.M.; Zhang, Y.; Lin, H.; Huang, Z.F.; Wang, Y.Y.; Zhang, X.D.; Zhong, B.; et al. TRIM38 inhibits TNFα- and IL-1β-triggered NF-κB activation by mediating lysosome-dependent degradation of TAB2/3. Proc. Natl. Acad. Sci. USA 2014, 111, 1509–1514. [Google Scholar] [CrossRef]
- Juraleviciute, M.; Pozniak, J.; Nsengimana, J.; Harland, M.; Randerson-Moor, J.; Wernhoff, P.; Bassarova, A.; Øy, G.F.; Trøen, G.; Flørenes, V.A.; et al. MX 2 is a novel regulator of cell cycle in melanoma cells. Pigment. Cell Melanoma Res. 2020, 33, 446–457. [Google Scholar] [CrossRef]
- Li, F.; Yu, L.; Zhu, J. LncRNA PSMA3-AS1 Promotes Lung Cancer Growth and Invasion via Sponging MiR-4504. Cancer Manag. Res. 2020, 12, 5277–5283. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, C.; He, Z.; Chen, S.; Li, L.; Sun, C. LncRNA PSMB8-AS1 contributes to pancreatic cancer progression via modulating miR-382-3p/STAT1/PD-L1 axis. J. Exp. Clin. Cancer Res. 2020, 39, 179. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Ao, L.; Miller, Z.; Kim, K.; Wu, Y.; Jang, E.R.; Lee, E.Y.; Kim, K.B.; Lee, W. PSMB9 codon 60 polymorphisms have no impact on the activity of the immunoproteasome catalytic subunit B1i expressed in multiple types of solid cancer. PLoS ONE 2013, 8, e73732. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Guo, S.; Stiles, J.K. The emerging role of CXCL10 in cancer (Review). Oncol. Lett. 2011, 2, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qiu, X.; Lin, S.; Chen, X.; Wang, T.; Liao, T. Knockdown of IFI27 inhibits cell proliferation and invasion in oral squamous cell carcinoma. World J. Surg. Oncol. 2018, 16, 64. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Tang, T.H.; Ng, S.K. Interferon Regulatory Factor 9 Structure and Regulation. Front. Immunol. 2018, 9, 1831. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, C. Prognostic characterization of OAS1/OAS2/OAS3/OASL in breast cancer. BMC Cancer 2020, 20, 575. [Google Scholar] [CrossRef]
- Alteber, Z.; Sharbi-Yunger, A.; Pevsner-Fischer, M.; Blat, D.; Roitman, L.; Tzehoval, E.; Elinav, E.; Eisenbach, L. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol. Cell Biol. 2018, 96, 284–297. [Google Scholar] [CrossRef]
- Li, L.; Bai, J.; Fan, H.; Yan, J.; Li, S.; Jiang, P. E2 ubiquitin-conjugating enzyme UBE2L6 promotes Senecavirus A proliferation by stabilizing the viral RNA polymerase. PLoS Pathog. 2020, 16, e1008970. [Google Scholar] [CrossRef]
- Tan, Y.; Zhou, G.; Wang, X.; Chen, W.; Gao, H. USP18 promotes breast cancer growth by upregulating EGFR and activating the AKT/Skp2 pathway. Int. J. Oncol. 2018, 53, 371–383. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z. STAT1 in cancer: Friend or foe? Discov. Med. 2017, 24, 19–29. [Google Scholar]
- Makise, M.; Nakamura, H.; Kuniyasu, A. The role of vimentin in the tumor marker Nup88-dependent multinucleated phenotype. BMC Cancer 2018, 18, 519. [Google Scholar] [CrossRef]
- Tang, J.; Yang, Q.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Ren, J.; Gong, Y.; Wu, G. Weighted gene correlation network analysis identifies RSAD2, HERC5, and CCL8 as prognostic candidates for breast cancer. J. Cell. Physiol. 2020, 235, 394–407. [Google Scholar] [CrossRef]
- Qi, Y.; Li, Y.; Zhang, Y.; Zhang, L.; Wang, Z.; Zhang, X.; Gui, L.; Huang, J. IFI6 Inhibits Apoptosis via Mitochondrial-Dependent Pathway in Dengue Virus 2 Infected Vascular Endothelial Cells. PLoS ONE 2015, 10, e0132743. [Google Scholar] [CrossRef]
- Hanak, L.; Slaby, O.; Lauerova, L.; Kren, L.; Nenutil, R.; Michalek, J. Expression pattern of HLA class I antigens in renal cell carcinoma and primary cell line cultures: Methodological implications for immunotherapy. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2009, 15, Cr638–Cr643. [Google Scholar]
- Jeong, S.I.; Kim, J.W.; Ko, K.P.; Ryu, B.K.; Lee, M.G.; Kim, H.J.; Chi, S.G. XAF1 forms a positive feedback loop with IRF-1 to drive apoptotic stress response and suppress tumorigenesis. Cell Death Dis. 2018, 9, 806. [Google Scholar] [CrossRef]
- Herbert, A. ADAR and Immune Silencing in Cancer. Trends Cancer 2019, 5, 272–282. [Google Scholar] [CrossRef]
- Erdreich-Epstein, A.; Robison, N.; Ren, X.; Zhou, H.; Xu, J.; Davidson, T.B.; Schur, M.; Gilles, F.H.; Ji, L.; Malvar, J.; et al. PID1 (NYGGF4), a new growth-inhibitory gene in embryonal brain tumors and gliomas. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 827–836. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Zhang, Y.; Bai, X.; Peng, Y.; He, P. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 5747–5752. [Google Scholar] [CrossRef]
- Meng, J.; Wang, L.; Hou, J.; Yang, X.; Lin, K.; Nan, H.; Li, M.; Wu, X.; Chen, X. CCL23 suppresses liver cancer progression through the CCR1/AKT/ESR1 feedback loop. Cancer Sci. 2021, 112, 3099–3110. [Google Scholar] [CrossRef]
- Wu, S.; Lu, X.; Zhang, Z.L.; Lei, P.; Hu, P.; Wang, M.; Huang, B.; Xing, W.; Jiang, X.T.; Liu, H.J.; et al. CC chemokine ligand 21 enhances the immunogenicity of the breast cancer cell line MCF-7 upon assistance of TLR2. Carcinogenesis 2011, 32, 296–304. [Google Scholar] [CrossRef]
- Kim, T.H.; Cho, S.G. Kisspeptin inhibits cancer growth and metastasis via activation of EIF2AK2. Mol. Med. Rep. 2017, 16, 7585–7590. [Google Scholar] [CrossRef]
- Luo, J.; Wang, H.; Wang, L.; Wang, G.; Yao, Y.; Xie, K.; Li, X.; Xu, L.; Shen, Y.; Ren, B. lncRNA GAS6-AS1 inhibits progression and glucose metabolism reprogramming in LUAD via repressing E2F1-mediated transcription of GLUT1. Mol. Ther. Nucleic Acids 2021, 25, 11–24. [Google Scholar] [CrossRef]
- Feng, Q.; Sekula, D.; Guo, Y.; Liu, X.; Black, C.C.; Galimberti, F.; Shah, S.J.; Sempere, L.F.; Memoli, V.; Andersen, J.B.; et al. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol. Cancer Ther. 2008, 7, 3780–3788. [Google Scholar] [CrossRef]
- Vyas, S.; Chesarone-Cataldo, M.; Todorova, T.; Huang, Y.H.; Chang, P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013, 4, 2240. [Google Scholar] [CrossRef]
- Pidugu, V.K.; Wu, M.M.; Yen, A.H.; Pidugu, H.B.; Chang, K.W.; Liu, C.J.; Lee, T.C. IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene 2019, 38, 3232–3247. [Google Scholar] [CrossRef]
- Ogony, J.; Choi, H.J.; Lui, A.; Cristofanilli, M.; Lewis-Wambi, J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res. 2016, 18, 25. [Google Scholar] [CrossRef]
- Walter, K.R.; Goodman, M.L.; Singhal, H.; Hall, J.A.; Li, T.; Holloran, S.M.; Trinca, G.M.; Gibson, K.A.; Jin, V.X.; Greene, G.L.; et al. Interferon-Stimulated Genes Are Transcriptionally Repressed by PR in Breast Cancer. Mol. Cancer Res. 2017, 15, 1331–1340. [Google Scholar] [CrossRef]
- Popson, S.A.; Hughes, C.C.W. A role for IFITM proteins in angiogenesis. FASEB J. 2010, 24, 750.1. [Google Scholar] [CrossRef]
- Popson, S.A.; Ziegler, M.E.; Chen, X.; Holderfield, M.T.; Shaaban, C.I.; Fong, A.H.; Welch-Reardon, K.M.; Papkoff, J.; Hughes, C.C. Interferon-induced transmembrane protein 1 regulates endothelial lumen formation during angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.S.; Zhao, X.; Cheng, X.; Guan, D.; Mahabeleshwar, G.H.; Liu, Y.; Borden, E.; Jain, M.K.; Kao, H.Y. Dual regulation of Stat1 and Stat3 by the tumor suppressor protein PML contributes to interferon α-mediated inhibition of angiogenesis. J. Biol. Chem. 2017, 292, 10048–10060. [Google Scholar] [CrossRef] [PubMed]
- Ciccarese, F.; Grassi, A.; Pasqualini, L.; Rosano, S.; Noghero, A.; Montenegro, F.; Bussolino, F.; Di Camillo, B.; Finesso, L.; Toffolo, G.M.; et al. Genetic perturbation of IFN-α transcriptional modulators in human endothelial cells uncovers pivotal regulators of angiogenesis. Comput. Struct. Biotechnol. J. 2020, 18, 3977–3986. [Google Scholar] [CrossRef]
- Burkart, C.; Arimoto, K.; Tang, T.; Cong, X.; Xiao, N.; Liu, Y.C.; Kotenko, S.V.; Ellies, L.G.; Zhang, D.E. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-λ and elevated secretion of Cxcl10. EMBO Mol. Med. 2013, 5, 1035–1050. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.J.; Shu, Q.; Stinson, W.A.; Tsou, P.S.; Ruth, J.H.; Isozaki, T.; Campbell, P.L.; Ohara, R.A.; Koch, A.E.; Fox, D.A.; et al. A unique role for galectin-9 in angiogenesis and inflammatory arthritis. Arthritis Res. Ther. 2018, 20, 31. [Google Scholar] [CrossRef] [PubMed]
- Griffioen, A.W.; Thijssen, V.L. Galectins in tumor angiogenesis. Ann. Transl. Med. 2014, 2, 90. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, H.; Long, Y.; Hua, H.; Jiang, Y.; Jing, J. PARP9 is overexpressed in human breast cancer and promotes cancer cell migration. Oncol. Lett. 2018, 16, 4073–4077. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, M.; Zhang, M.; Zhang, J.; Du, Z.; Zhang, H.; Hua, W.; Mao, Y. DDX60 Is Associated With Glioma Malignancy and Serves as a Potential Immunotherapy Biomarker. Front. Oncol. 2021, 11, 665360. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, Y.; Zhao, W.; You, S.; Feng, M.; Xie, C.; Chi, X.; Zhang, Y.; Wang, X. As a downstream target of the AKT pathway, NPTX1 inhibits proliferation and promotes apoptosis in hepatocellular carcinoma. Biosci. Rep. 2019, 39, BSR20181662. [Google Scholar] [CrossRef]
- Vadija, R.; Mustyala, K.K.; Nambigari, N.; Dulapalli, R.; Dumpati, R.K.; Ramatenki, V.; Vellanki, S.P.; Vuruputuri, U. Homology modeling and virtual screening studies of FGF-7 protein-a structure-based approach to design new molecules against tumor angiogenesis. J. Chem. Biol. 2016, 9, 69–78. [Google Scholar] [CrossRef]
- Seo, H.R.; Jeong, H.E.; Joo, H.J.; Choi, S.C.; Park, C.Y.; Kim, J.H.; Choi, J.H.; Cui, L.H.; Hong, S.J.; Chung, S.; et al. Intrinsic FGF2 and FGF5 promotes angiogenesis of human aortic endothelial cells in 3D microfluidic angiogenesis system. Sci. Rep. 2016, 6, 28832. [Google Scholar] [CrossRef]
- Mehta, V.B.; Besner, G.E. HB-EGF promotes angiogenesis in endothelial cells via PI3-kinase and MAPK signaling pathways. Growth Factors 2007, 25, 253–263. [Google Scholar] [CrossRef]
- Cao, R.; Bråkenhielm, E.; Li, X.; Pietras, K.; Widenfalk, J.; Ostman, A.; Eriksson, U.; Cao, Y. Angiogenesis stimulated by PDGF-CC, a novel member in the PDGF family, involves activation of PDGFR-alphaalpha and -alphabeta receptors. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002, 16, 1575–1583. [Google Scholar] [CrossRef]
- Barron, G.A.; Goua, M.; Wahle, K.W.J.; Bermano, G. Circulating levels of angiogenesis-related growth factors in breast cancer: A study to profile proteins responsible for tubule formation. Oncol. Rep. 2017, 38, 1886–1894. [Google Scholar] [CrossRef]
- Schuler, L.A.; O’Leary, K.A. Prolactin: The Third Hormone in Breast Cancer. Front. Endocrinol. 2022, 13, 910978. [Google Scholar] [CrossRef]
- Reuwer, A.Q.; Nowak-Sliwinska, P.; Mans, L.A.; van der Loos, C.M.; von der Thüsen, J.H.; Twickler, M.T.; Spek, C.A.; Goffin, V.; Griffioen, A.W.; Borensztajn, K.S. Functional consequences of prolactin signalling in endothelial cells: A potential link with angiogenesis in pathophysiology? J. Cell. Mol. Med. 2012, 16, 2035–2048. [Google Scholar] [CrossRef]
- Cao, Y.; Linden, P.; Farnebo, J.; Cao, R.; Eriksson, A.; Kumar, V.; Qi, J.H.; Claesson-Welsh, L.; Alitalo, K. Vascular endothelial growth factor C induces angiogenesis in vivo. Proc. Natl. Acad. Sci. USA 1998, 95, 14389–14394. [Google Scholar] [CrossRef]
- De Falco, S. The discovery of placenta growth factor and its biological activity. Exp. Mol. Med. 2012, 44, 1–9. [Google Scholar] [CrossRef]
- Parr, C.; Watkins, G.; Boulton, M.; Cai, J.; Jiang, W.G. Placenta growth factor is over-expressed and has prognostic value in human breast cancer. Eur. J. Cancer 2005, 41, 2819–2827. [Google Scholar] [CrossRef]
- Tahergorabi, Z.; Khazaei, M. Leptin and its cardiovascular effects: Focus on angiogenesis. Adv. Biomed. Res. 2015, 4, 79. [Google Scholar] [CrossRef]
- Mira, E.; Lacalle, R.A.; Buesa, J.M.; de Buitrago, G.G.; Jiménez-Baranda, S.; Gómez-Moutón, C.; Martínez, A.C.; Mañes, S. Secreted MMP9 promotes angiogenesis more efficiently than constitutive active MMP9 bound to the tumor cell surface. J. Cell Sci. 2004, 117, 1847–1857. [Google Scholar] [CrossRef]
- Stamatovic, S.M.; Keep, R.F.; Mostarica-Stojkovic, M.; Andjelkovic, A.V. CCL2 regulates angiogenesis via activation of Ets-1 transcription factor. J. Immunol. 2006, 177, 2651–2661. [Google Scholar] [CrossRef]
- Yu, X.; Zhao, R.; Lin, S.; Bai, X.; Zhang, L.; Yuan, S.; Sun, L. CXCL16 induces angiogenesis in autocrine signaling pathway involving hypoxia-inducible factor 1α in human umbilical vein endothelial cells. Oncol. Rep. 2016, 35, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Bajdak-Rusinek, K.; Kupnicka, P.; Kapczuk, P.; Simińska, D.; Chlubek, D.; Baranowska-Bosiacka, I. The Role of CXCL16 in the Pathogenesis of Cancer and Other Diseases. Int. J. Mol. Sci. 2021, 22, 3490. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Wu, Z.S.; Qian, P.X.; Kang, J.; Liu, D.X.; Zhu, T.; Lobie, P.E. ARTEMIN promotes de novo angiogenesis in ER negative mammary carcinoma through activation of TWIST1-VEGF-A signalling. PLoS ONE 2012, 7, e50098. [Google Scholar] [CrossRef] [PubMed]
- Bluff, J.E.; Brown, N.J.; Reed, M.W.; Staton, C.A. Tissue factor, angiogenesis and tumour progression. Breast Cancer Res. 2008, 10, 204. [Google Scholar] [CrossRef] [PubMed]
- Bagnato, A.; Spinella, F. Emerging role of endothelin-1 in tumor angiogenesis. Trends Endocrinol. Metab. 2003, 14, 44–50. [Google Scholar] [CrossRef]


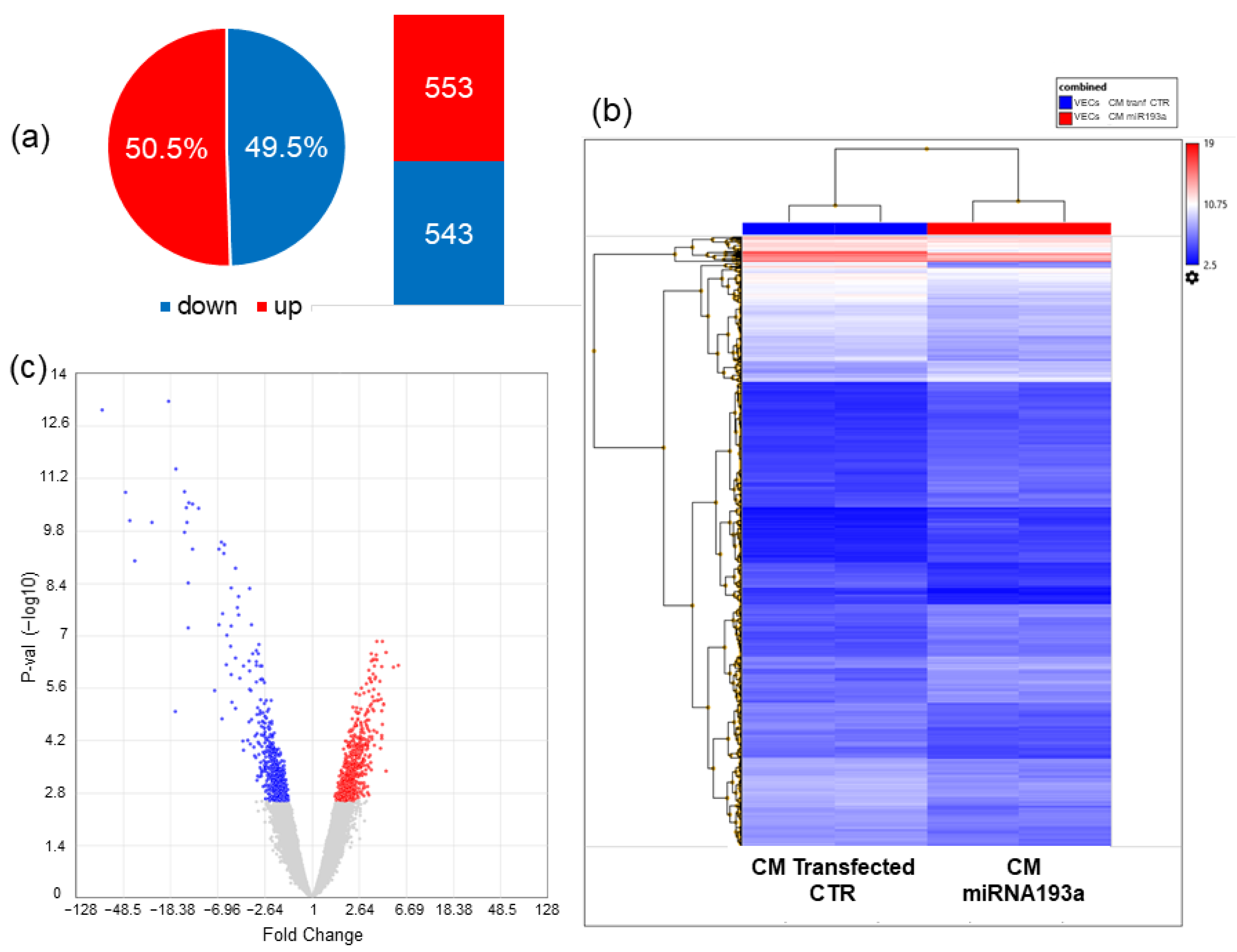
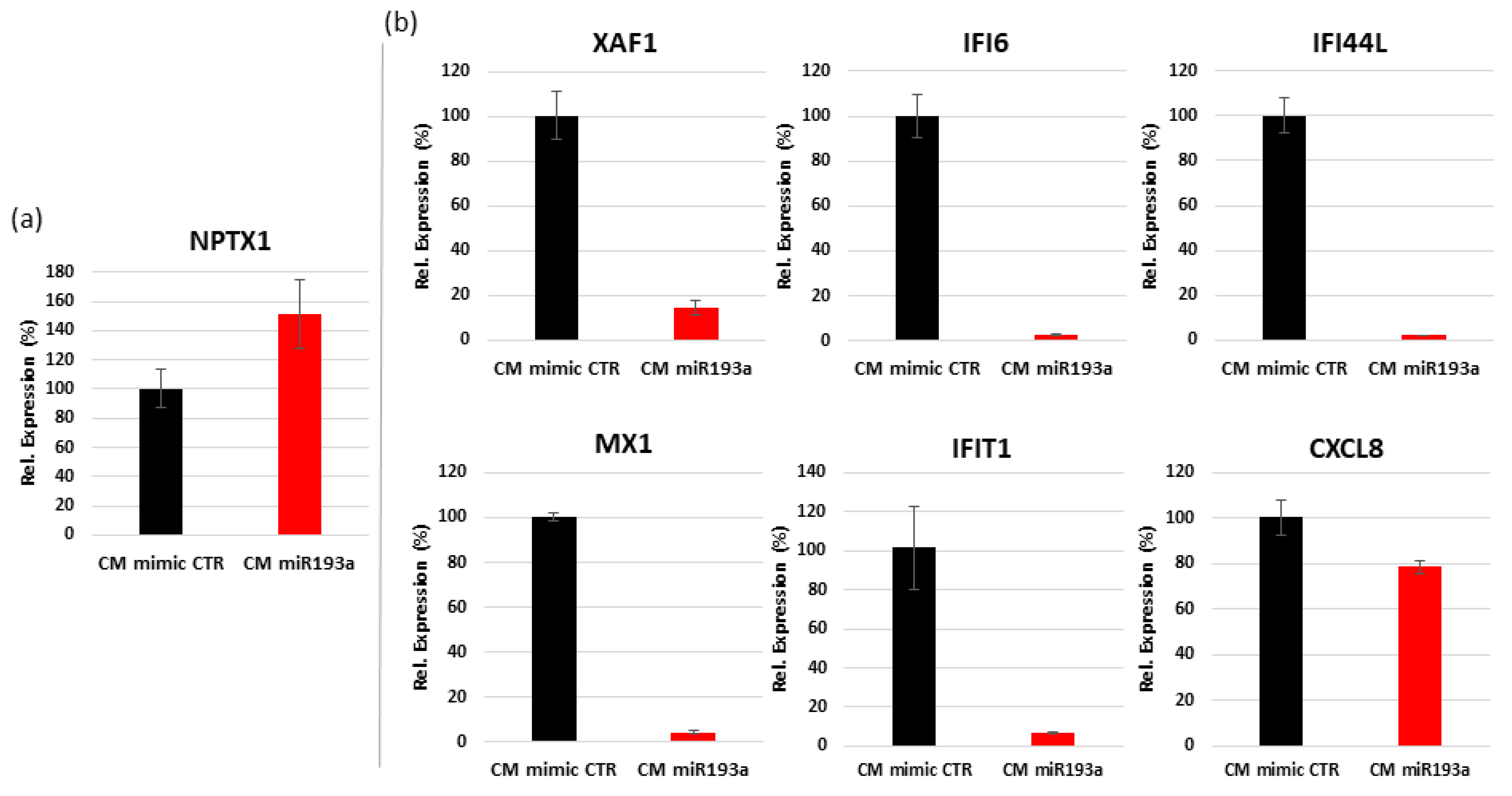
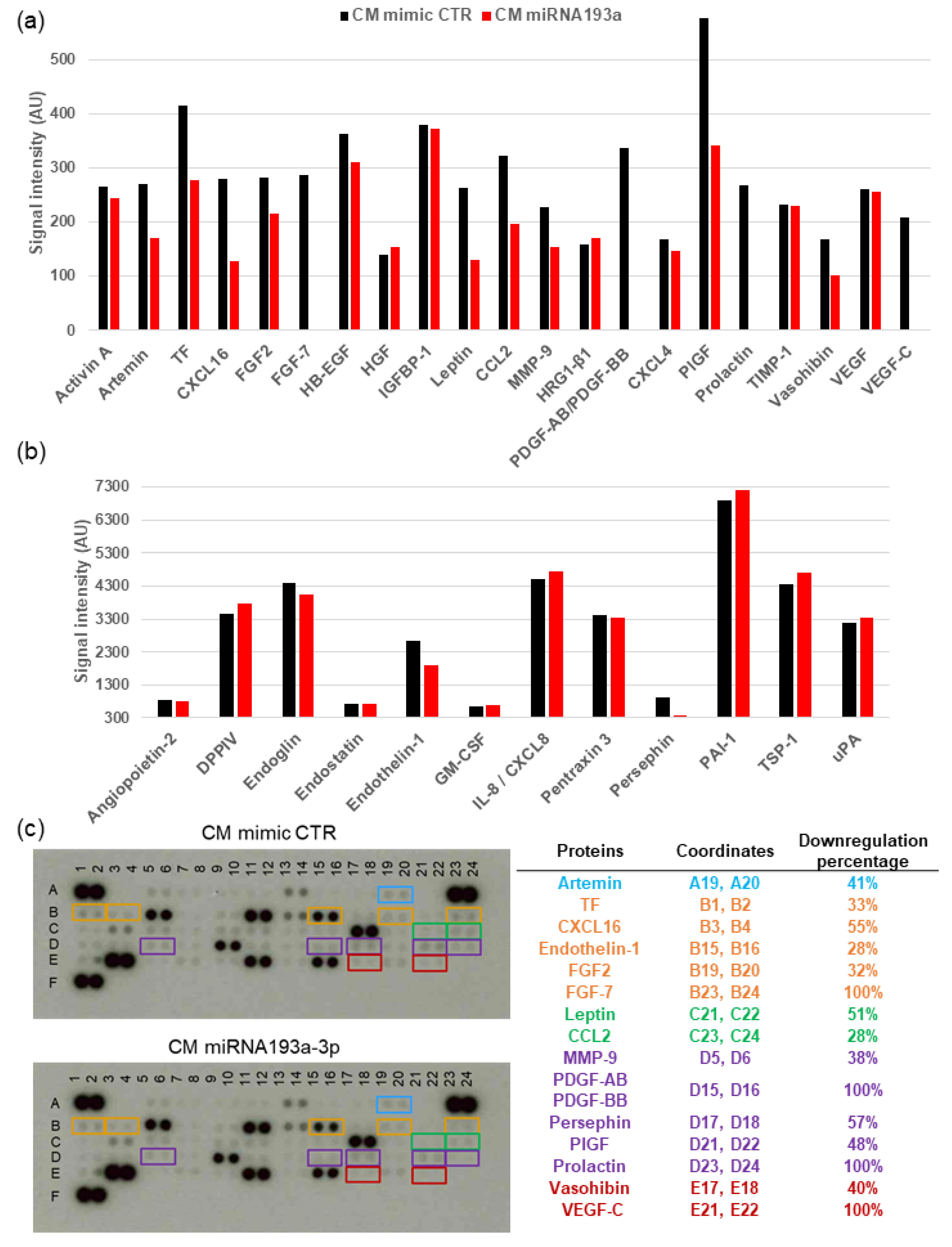



| Gene Symbol | Description | Log2 FC | FDR p-Value |
|---|---|---|---|
| PARP8 | poly(ADP-ribose) polymerase family member 8 | 2.58 | 0.0002 |
| ATP6V1G3 | ATPase H+ transporting V1 subunit G3 | 2.43 | 0.0003 |
| NANOGNB | NANOG neighbor homeobox | 2.21 | 0.018 |
| CALB1 | calbindin 1 | 2.2 | 0.0002 |
| CCDC172 | coiled-coil domain containing 172 | 2.14 | 0.0015 |
| PCP4 | Purkinje cell protein 4 | 2.12 | 0.0015 |
| TDO2 | tryptophan 2,3-dioxygenase | 2.1 | 8.91 × 10−5 |
| AK9 | adenylate kinase 9 | 2.09 | 0.0034 |
| YEATS2 | YEATS domain containing 2 | 2.08 | 0.0054 |
| WBP4 | WW domain binding protein 4 | 2.07 | 0.0002 |
| Gene Symbol | Description | Log2 FC | FDR p-Value |
|---|---|---|---|
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | −6.24 | 1 × 10−9 |
| IFITM1 | interferon induced transmembrane protein 1 | −4.27 | 1 × 10−9 |
| OAS2 | 2-5-oligoadenylate synthetase 2 | −4.04 | 2.55 × 10−8 |
| IFI44L | interferon-induced protein 44-like | −5.53 | 6.27 × 10−8 |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | −3.77 | 6.27 × 10−8 |
| LGALS9 | lectin, galactoside-binding, soluble, 9 | −3.56 | 9.37 × 10−8 |
| MX2 | MX dynamin-like GTPase 2 | −3.66 | 9.37 × 10−8 |
| DDX60 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 | −3.37 | 9.57 × 10−8 |
| USP18 | ubiquitin specific peptidase 18 | −3.73 | 9.57 × 10−8 |
| PARP9 | poly(ADP-ribose) polymerase family member 9 | −3.71 | 1.72 × 10−7 |
| Pathway | Overlap | Adj. p-Value |
|---|---|---|
| BioPlanet | ||
| Interferon alpha/beta signaling | 25/64 | 1.74 × 10−12 |
| Interferon signaling | 35/168 | 4.98 × 10−9 |
| Immune system signaling by interferons, interleukins, prolactin, and growth hormones | 45/280 | 4.47 × 10−8 |
| Type II interferon signaling (interferon-gamma) | 13/50 | 6.11 × 10−4 |
| Antigen processing: cross presentation | 15/79 | 0.005 |
| Endosomal/vacuolar pathway | 5/9 | 0.009 |
| GO Biological Process | ||
| cellular response to type I interferon (GO:0071357) | 29/65 | 1.93 × 10−16 |
| type I interferon signaling pathway (GO:0060337) | 29/65 | 1.93 × 10−16 |
| negative regulation of viral process (GO:0048525) | 23/70 | 1.95 × 10−9 |
| defense response to symbiont (GO:0140546) | 30/124 | 4.78 × 10−9 |
| defense response to virus (GO:0051607) | 31/133 | 4.85 × 10−9 |
| negative regulation of viral genome replication (GO:0045071) | 18/54 | 1.96 × 10−7 |
| regulation of viral genome replication (GO:0045069) | 19/67 | 1.21 × 10−6 |
| cellular response to interferon-gamma (GO:0071346) | 22/121 | 3.59 × 10−4 |
| response to interferon-beta (GO:0035456) | 10/28 | 5.61 × 10−4 |
| response to interferon-alpha (GO:0035455) | 8/18 | 8.38 × 10−4 |
| interferon-gamma-mediated signaling pathway (GO:0060333) | 15/68 | 0.001 |
| response to cytokine (GO:0034097) | 23/150 | 0.003 |
| antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent (GO:0002479) | 15/73 | 0.003 |
| antigen processing and presentation of exogenous peptide antigen via MHC class I (GO:0042590) | 15/78 | 0.006 |
| antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent (GO:0002480) | 5/8 | 0.006 |
| Gene Symbol | Description | Function | Angiogenesis | Log2 FC | FDR p-Value |
|---|---|---|---|---|---|
| ADAR | adenosine deaminase, RNA-specific | A to I RNA Editing | Pro | −1.15 | 0.007 |
| AIF1 | allograft inflammatory factor 1 | Inflammation | Pro | 1.52 | 0.0326 |
| B2M | beta-2-microglobulin | Immune response | Pro | −1.02 | 0.0344 |
| BST2 | bone marrow stromal cell antigen 2 | Immunomodulatory | Pro | −3.68 | 3.98 × 10−6 |
| CCL20 | chemokine (C-C motif) ligand 20 | Inflammation | Pro | −1.2 | 0.0348 |
| CCL23 | chemokine (C-C motif) ligand 23 | Immune response | Pro | 1.87 | 0.0004 |
| CCR7 | chemokine (C-C motif) receptor 7 | Immune response | Pro | 1.19 | 0.0204 |
| CD47 | CD47 molecule | Immune/Integrin | Anti | −1.12 | 0.0216 |
| CXCL10 | chemokine (C-X-C motif) ligand 10 | Immunomodulatory | Anti | −2.21 | 1.53 × 10−5 |
| CYBA | cytochrome b-245, alpha polypeptide | Antiviral/Immunomodulatory | Pro | −0.83 | 0.0297 |
| DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | Innate Immunity | Anti | −1.87 | 0.0002 |
| EIF2AK2 | eukaryotic translation initiation factor 2-alpha kinase 2 | Innate Immunity | Pro | −2.18 | 2.21 × 10−5 |
| EIF4A2 | eukaryotic translation initiation factor 4A2 | Cancer growth | NK | −0.73 | 0.0491 |
| GAS6-AS1 | GAS6 antisense RNA 1 | Inflammation/Tumor growth | NK | 1.03 | 0.0314 |
| GBP1 | guanylate binding protein 1, interferon-inducible | Autonomous Immunity | Anti | −1.45 | 0.0304 |
| GBP3 | guanylate binding protein 3, interferon-inducible | Innate Immunity | NK | 1.53 | 0.0124 |
| HLA-A | major histocompatibility complex, class I, A | Immune defense/anti-viral | Pro | −1.3 | 0.0036 |
| HLA-B | major histocompatibility complex, class I, B | Immune defense/viral immunity | Pro | −1.65 | 0.0005 |
| HLA-C | major histocompatibility complex, class I, C | Immune defense/viral immunity | Pro | −1.,63 | 0.0049 |
| HLA-F | major histocompatibility complex, class I, F | Immune defense/viral immunity | Pro | −1.48 | 0.0002 |
| IFI27 | interferon, alpha-inducible protein 27 | Innate Immunity | Pro | −2.89 | 0.0008 |
| IFI6 | interferon, alpha-inducible protein 6 | Immunomodulatory/anti-apoptosis/ | Pro | −5.41 | 1.72 × 10−7 |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | Innate Immunity | Pro | −6.24 | 1 × 10−9 |
| IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | Innate Immunity | Anti | −1.26 | 0.0041 |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | Innate Immunity | Pro | −3.77 | 6.27 × 10−8 |
| IFIT5 | interferon-induced protein with tetratricopeptide repeats 5 | Innate Immunity | Pro | −2.4 | 5.05 × 10−6 |
| IFITM1 | interferon induced transmembrane protein 1 | Immunomodulatory | Pro | −4.27 | 1 × 10−9 |
| IFITM2 | interferon induced transmembrane protein 2 | Immunomodulatory | Pro | −1.58 | 0.0001 |
| IFITM3 | interferon induced transmembrane protein 3 | Immunomodulatory | Pro | −1.86 | 5.05 × 10−6 |
| IFNB1 | interferon, beta 1, fibroblast | Innate Immunity | Anti | −1.04 | 0.0384 |
| IRF9 | interferon regulatory factor 9 | Immunity/anti-viral | Anti | −0.96 | 0.0178 |
| ISG15 | ISG15 ubiquitin-like modifier | Anti-viral/Transcription factor | Pro | −2.4 | 0.0004 |
| LEF1 | lymphoid enhancer-binding factor 1 | Transcription factor/Wnt signaling | Pro | 1.58 | 0.0023 |
| LGALS9 | lectin, galactoside-binding, soluble, 9 | Inflammation/Immune response | Pro | −3.56 | 9.37 × 10−8 |
| MAP4K3 | mitogen-activated protein kinase kinase kinase kinase 3 | Autoimmune/cell stress | Pro | −0.79 | 0.0331 |
| MME | membrane metallo-endopeptidase | Protease/Metabolism | Anti | 1.23 | 0.0217 |
| MX1 | MX dynamin-like GTPase 1 | Anti-viral/Immunity | Pro | −5.26 | 1.10 × 10−6 |
| MX2 | MX dynamin-like GTPase 2 | Innate immunity | Pro | −3.66 | 9.37 × 10−8 |
| NCF1 | neutrophil cytosolic factor 1 | Innate immunity | Pro | 1.27 | 0.0272 |
| NCF2 | neutrophil cytosolic factor 2 | Autoimmunity | Pro | 1.45 | 0.0262 |
| NUP88 | nucleoporin 88kDa | Nuclear trafficking/oncogenic | Pro | −1.22 | 0.0172 |
| OAS1 | 2-5-oligoadenylate synthetase 1 | Innate anti-viral | Pro | −4.76 | 1.72 × 10−7 |
| OAS2 | 2-5-oligoadenylate synthetase 2 | Innate anti-viral | Pro | −4.04 | 2.55 × 10−8 |
| OAS3 | 2-5-oligoadenylate synthetase 3 | Innate anti-viral | Pro | −2.54 | 6.41 × 10−5 |
| PID1 | phosphotyrosine interaction domain containing 1 | Adipocyte growth | NK | 0.79 | 0.0263 |
| PLSCR1 | phospholipid scramblase 1 | Anti-viral immunity | Pro | −2.61 | 5.21 × 10−7 |
| PML | promyelocytic leukemia | Anti-viral/tumor suppressor | Anti | −1.08 | 0.0138 |
| PNPT1 | polyribonucleotide nucleotidyltransferase 1 | RNA metabolism | NK | −1.56 | 0.0072 |
| PSMA3 | proteasome subunit alpha 3 | Proteolytic | NK | −1.1 | 0.0171 |
| PSMB8 | proteasome subunit beta 8 | Proteolytic/Immunoproteasome | Pro | −1.51 | 0.0036 |
| PSMB9 | proteasome subunit beta 9 | Immunoproteasome | Anti | −2.42 | 0.0001 |
| PSMF1 | proteasome inhibitor subunit 1 | Immunoproteasome | NK | −0.82 | 0.0298 |
| RSAD2 | radical S-adenosyl methionine domain containing 2 | Innate immunity | NK | −2.38 | 0.0013 |
| SAMHD1 | SAM domain and HD domain 1 | Anti-viral/tumor suppressor | NK | −2.7 | 4.9 × 10−7 |
| STAT1 | signal transducer and activator of transcription 1 | Growth signaling | Pro/Anti | −2.63 | 7.37 × 10−7 |
| TAP1 | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | ABC transporter/multidrug resistance | NK | −1.05 | 0.04 |
| TAP2 | transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | ABC transporter/multidrug resistance | NK | −2.4 | 3.83 × 10−5 |
| TAPBP | TAP binding protein (tapasin) | MHC I/associated glycoprotein | NK | −1.77 | 0.0002 |
| TDGF1 | teratocarcinoma-derived growth factor 1 | Growth EGF signaling | Pro | 1.36 | 0.0076 |
| TRIM22 | tripartite motif containing 22 | Innate Immunity/Ant-viral | Pro | −0.94 | 0.018 |
| TRIM31 | tripartite motif containing 31 | SRC induced growth regulator | NK | 1.29 | 0.0461 |
| TRIM38 | tripartite motif containing 38 | Innate Immunity | NK | −1.04 | 0.0218 |
| UBA7 | ubiquitin-like modifier activating enzyme 7 | Activates ubiquitin ISGylation | NK | −2.03 | 0.0088 |
| UBE2L6 | ubiquitin-conjugating enzyme E2L 6 | Ubiquitation of FLT3 | Pro | −2.13 | 0.0004 |
| USP18 | ubiquitin specific peptidase 18 | Negative regulator of interferon signaling | Pro | −3.73 | 9.57 × 10−8 |
| XAF1 | XIAP associated factor 1 | Apoptotic protein(s) inhibitor | Anti | −4.07 | 0.002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzarito, G.; Kurmann, L.; Leeners, B.; Dubey, R.K. Micro-RNA193a-3p Inhibits Breast Cancer Cell Driven Growth of Vascular Endothelial Cells by Altering Secretome and Inhibiting Mitogenesis: Transcriptomic and Functional Evidence. Cells 2022, 11, 2967. https://doi.org/10.3390/cells11192967
Azzarito G, Kurmann L, Leeners B, Dubey RK. Micro-RNA193a-3p Inhibits Breast Cancer Cell Driven Growth of Vascular Endothelial Cells by Altering Secretome and Inhibiting Mitogenesis: Transcriptomic and Functional Evidence. Cells. 2022; 11(19):2967. https://doi.org/10.3390/cells11192967
Chicago/Turabian StyleAzzarito, Giovanna, Lisa Kurmann, Brigitte Leeners, and Raghvendra K. Dubey. 2022. "Micro-RNA193a-3p Inhibits Breast Cancer Cell Driven Growth of Vascular Endothelial Cells by Altering Secretome and Inhibiting Mitogenesis: Transcriptomic and Functional Evidence" Cells 11, no. 19: 2967. https://doi.org/10.3390/cells11192967
APA StyleAzzarito, G., Kurmann, L., Leeners, B., & Dubey, R. K. (2022). Micro-RNA193a-3p Inhibits Breast Cancer Cell Driven Growth of Vascular Endothelial Cells by Altering Secretome and Inhibiting Mitogenesis: Transcriptomic and Functional Evidence. Cells, 11(19), 2967. https://doi.org/10.3390/cells11192967






