Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation and Pathological Analysis of Coronary Arteries
2.2. Cell Cultures
2.3. RNA Preparation
2.4. RNA-Seq Library Construction and Sequencing
2.5. MeRIP-Seq Library Construction and Sequencing
2.6. Bioinformatics Analysis
2.7. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.8. Western Blotting (WB)
2.9. Immunohistochemistry of Coronary Artery Tissues
2.10. Cell Counting Kit 8 (CCK-8)
2.11. Transwell Assay
2.12. Statistical Analysis
3. Results
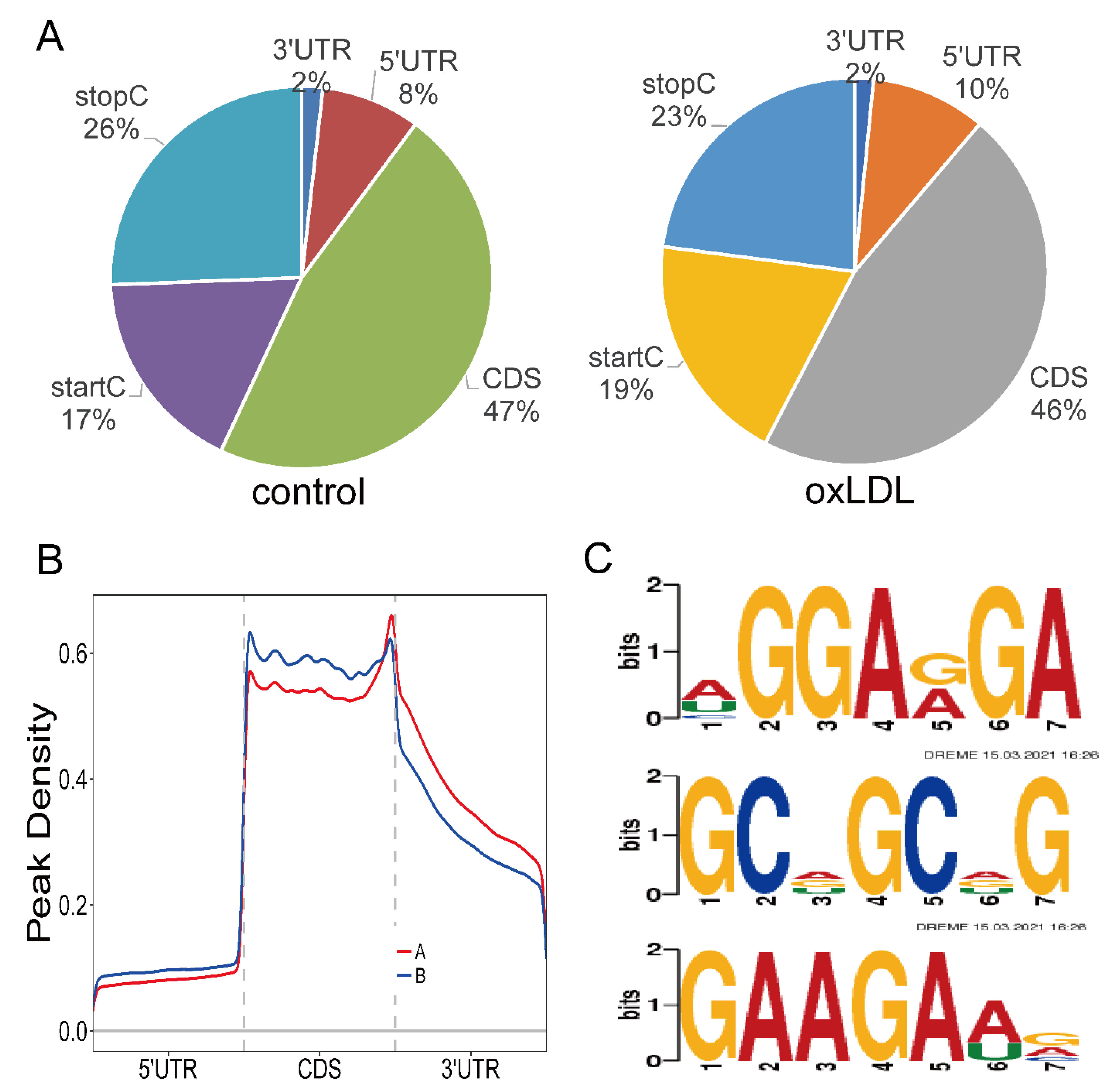
3.1. Overview of mRNA m6A Methylation in Proliferation and Migration Models of HCASMCs
3.2. Analysis of Sources of mRNA Methylation and Motif Analysis
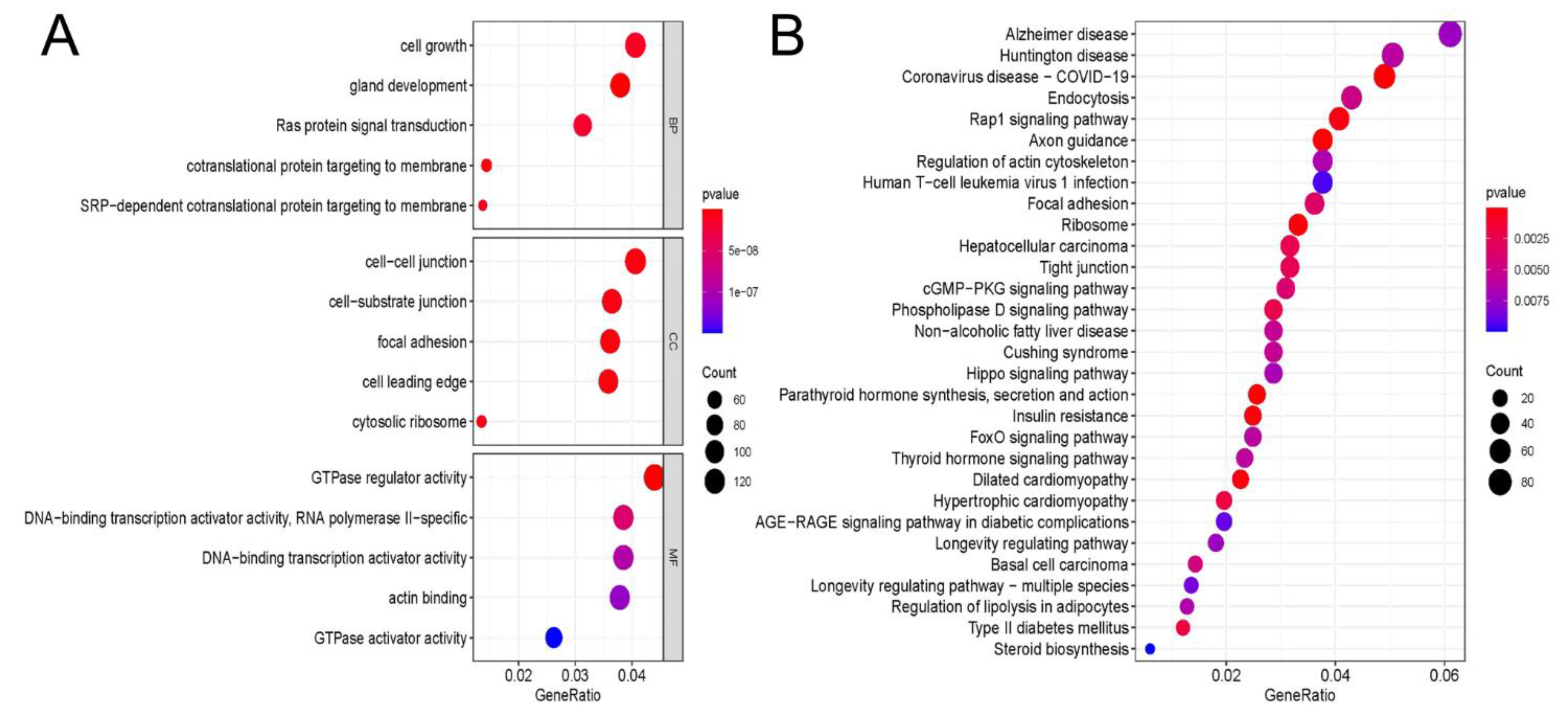
3.3. GO and KEGG Enrichment Analyses of m6A Sites
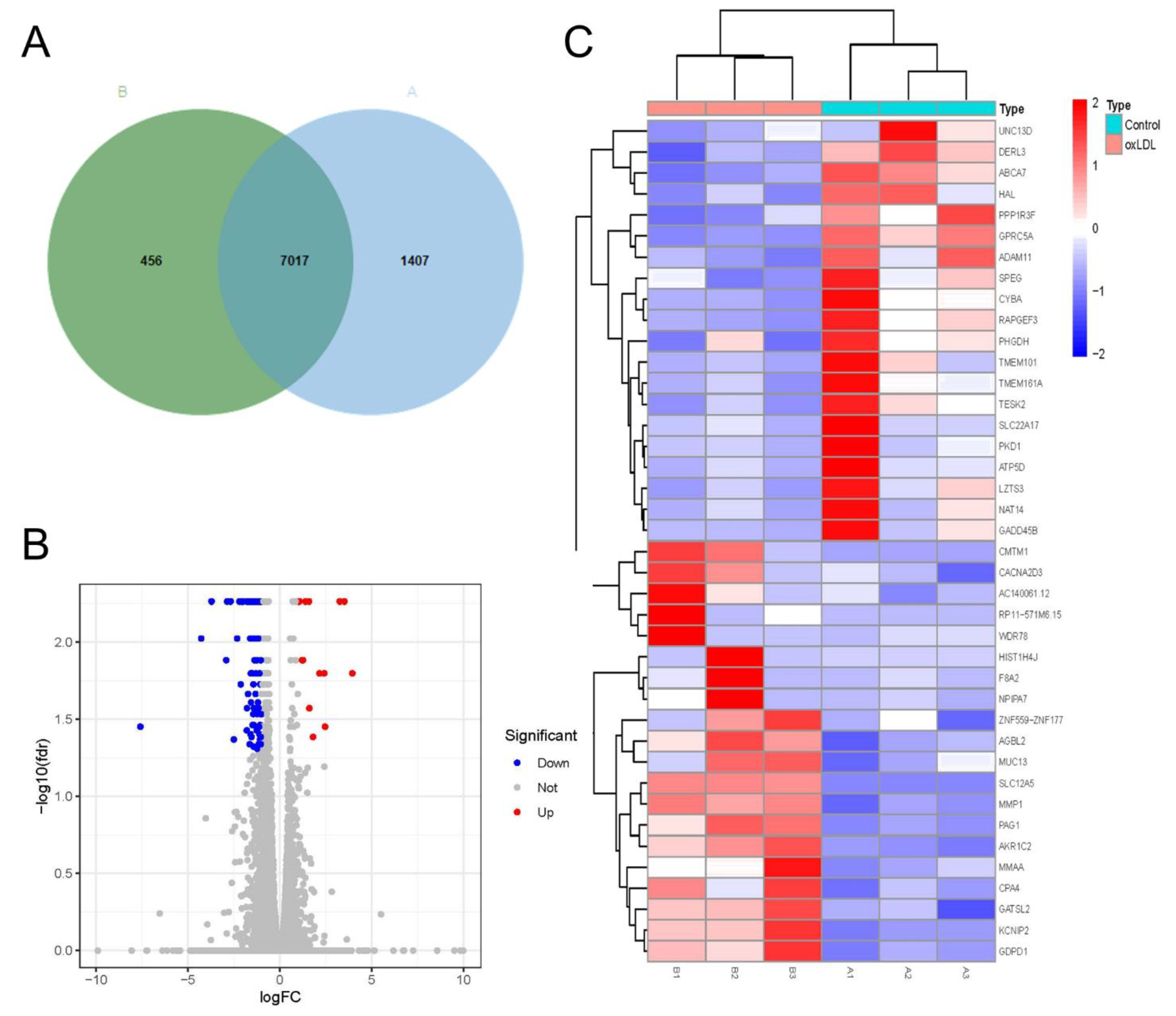
3.4. Characteristics of mRNA Expression Profiles in Pathological Models of HCASMCs
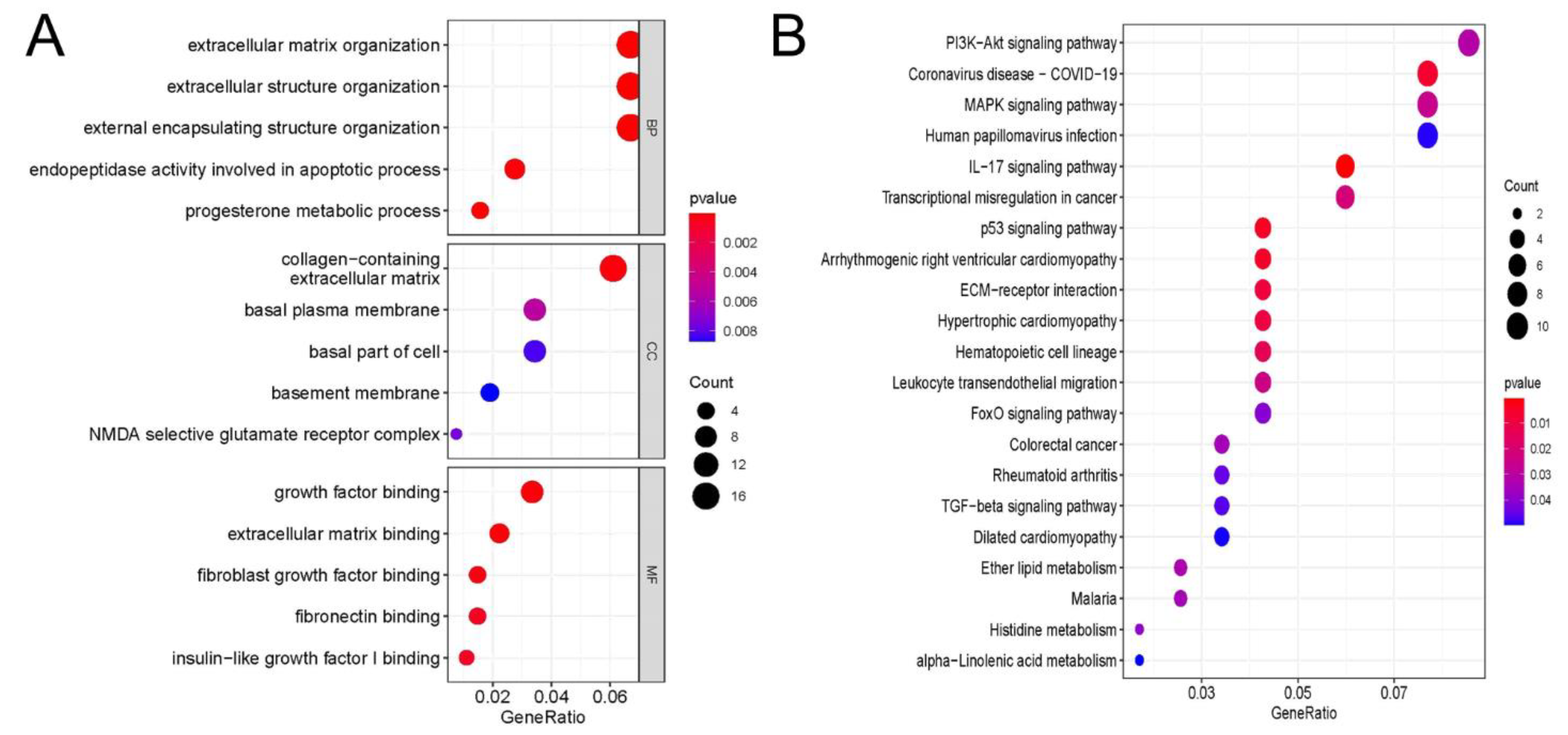
3.5. GO and KEGG Analyses of Differentially Expressed Genes
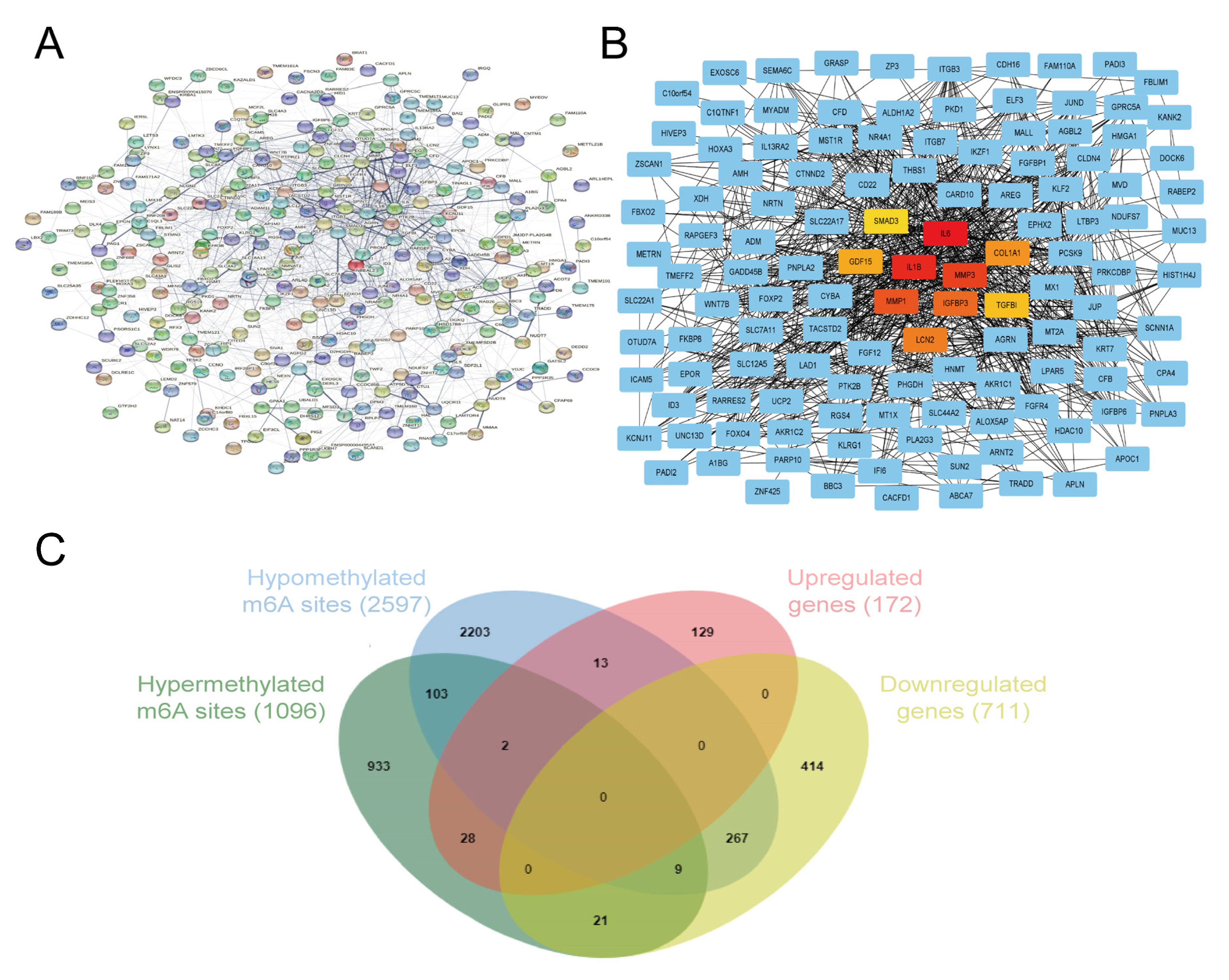
3.6. Identification of Hub Genes and Conjoint Analysis of RNA-Seq and MeRIP-Seq
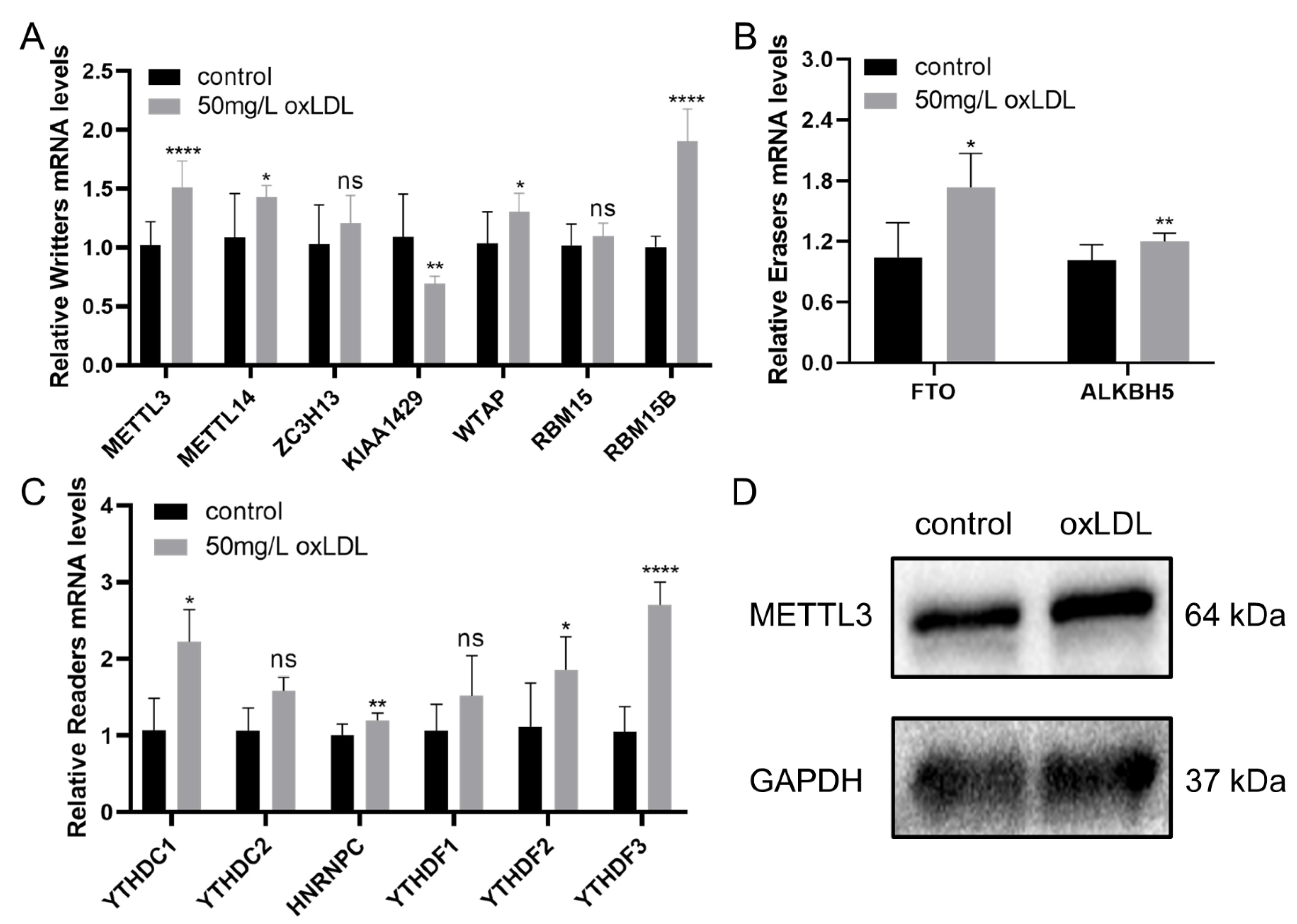
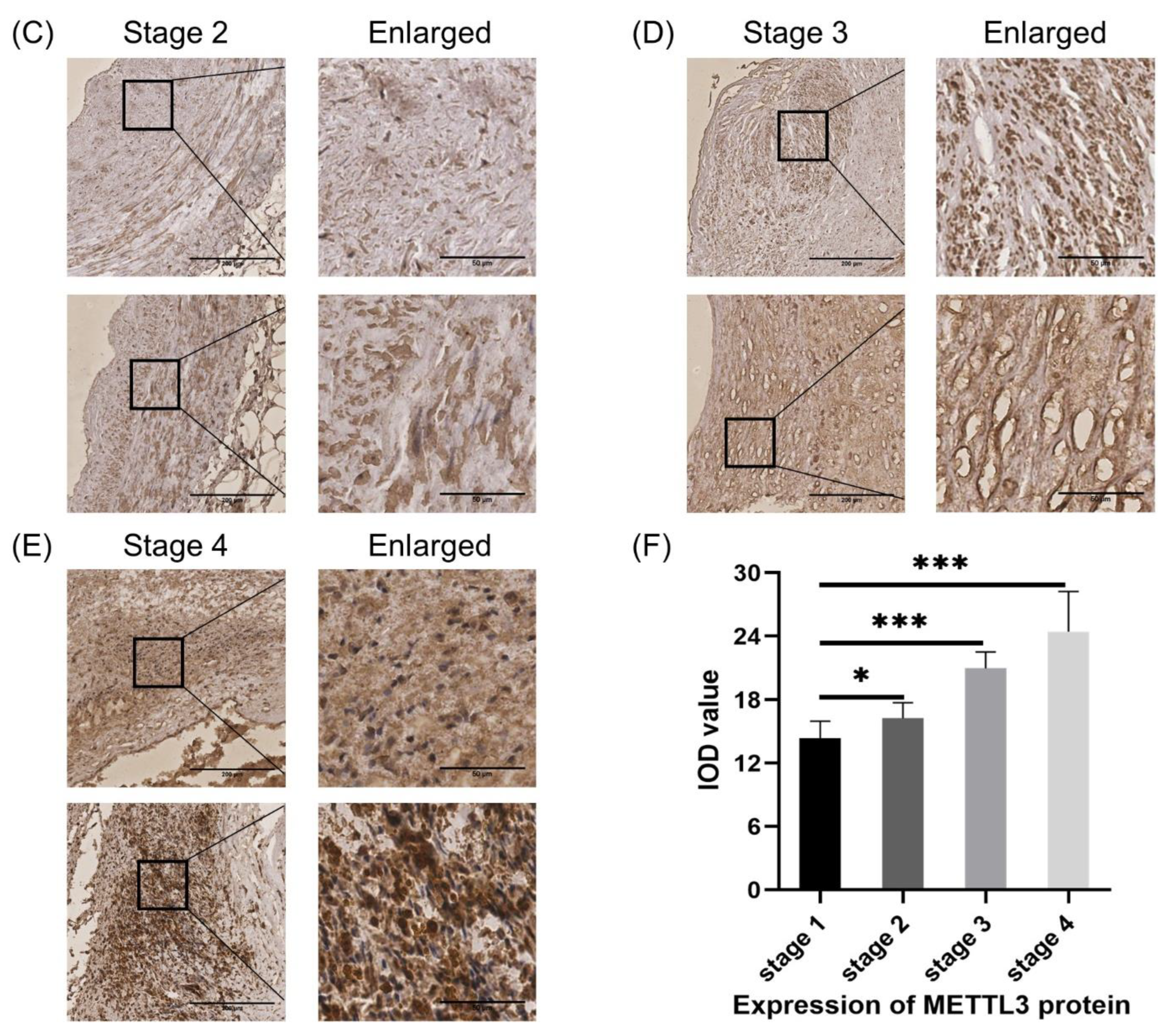
3.7. Validation of the Expression of Methylases in HCASMCs and the Expression of METTL3 in Coronary Specimens
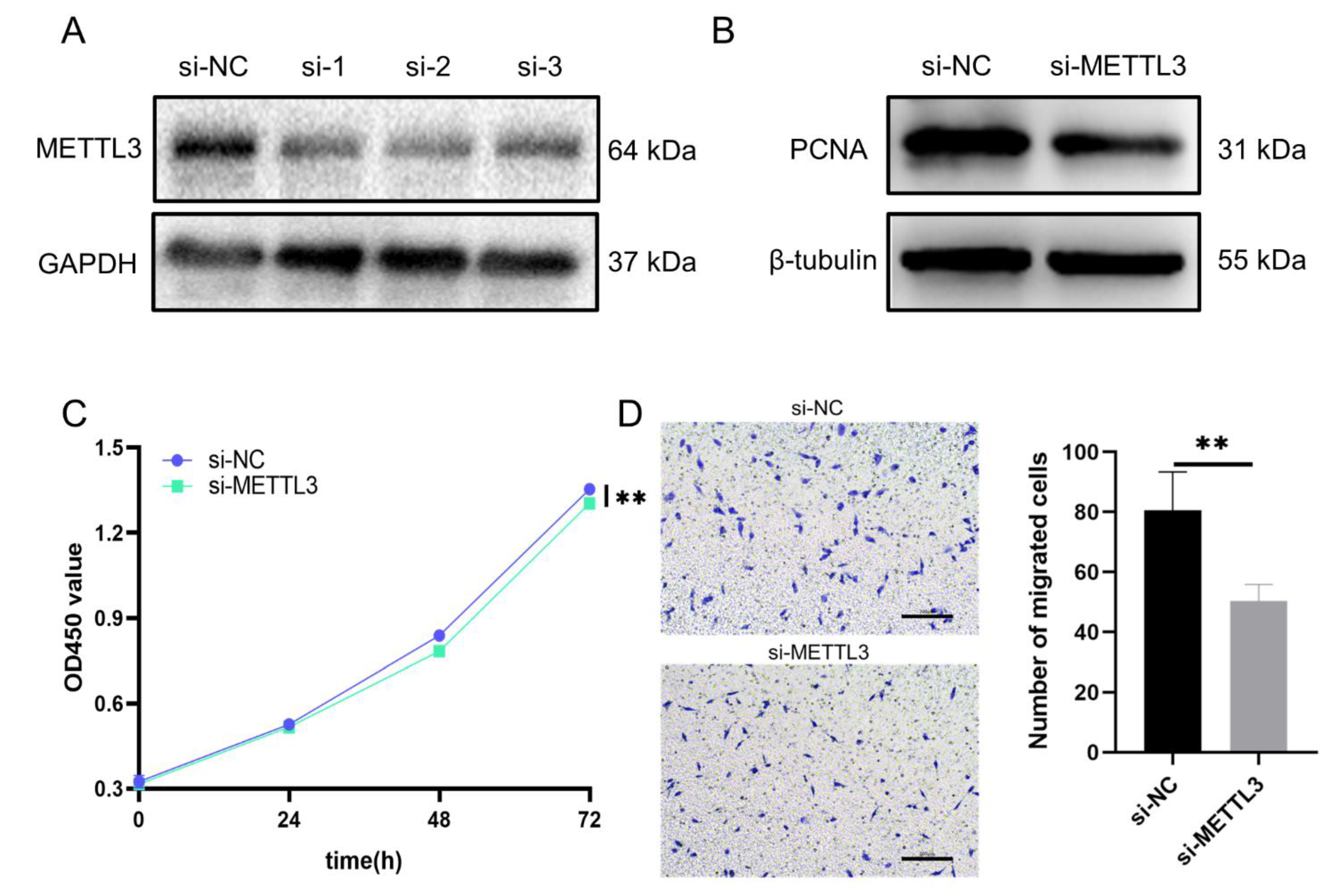
3.8. Regulatory Role of METTL3 in Proliferation and Migration Models of HCASMCs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kong, P.; Cui, Z.Y.; Huang, X.F.; Zhang, D.D.; Guo, R.J.; Han, M. Inflammation and atherosclerosis: Signaling pathways and therapeutic intervention. Signal Transduct. Target. Ther. 2022, 7, 131. [Google Scholar] [CrossRef]
- Gregersen, I.; Michelsen, A.E.; Lunde, N.N.; Åkerblom, A.; Lakic, T.G.; Skjelland, M.; Ryeng Skagen, K.; Becker, R.C.; Lindbäck, J.; Himmelmann, A.; et al. Legumain in Acute Coronary Syndromes: A Substudy of the PLATO (Platelet Inhibition and Patient Outcomes) Trial. J. Am. Heart Assoc. 2020, 9, e016360. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cui, X.; Wang, C.; Zhao, S. LncRNA HCG11 regulates proliferation and apoptosis of vascular smooth muscle cell through targeting miR-144-3p/FOXF1 axis in atherosclerosis. Biol. Res. 2020, 53, 44. [Google Scholar] [CrossRef]
- Orekhov, A.N.; Andreeva, E.R.; Mikhailova, I.A.; Gordon, D. Cell proliferation in normal and atherosclerotic human aorta: Proliferative splash in lipid-rich lesions. Atherosclerosis 1998, 139, 41–48. [Google Scholar] [CrossRef]
- Mulvihill, E.R.; Jaeger, J.; Sengupta, R.; Ruzzo, W.L.; Reimer, C.; Lukito, S.; Schwartz, S.M. Atherosclerotic plaque smooth muscle cells have a distinct phenotype. Arterioscler Thromb. Vasc. Biol. 2004, 24, 1283–1289. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, Y.; Chen, B.; Guo, X.; Gao, S.; Wang, M.; Duan, M.; Li, X. MOF Regulates TNK2 Transcription Expression to Promote Cell Proliferation in Thyroid Cancer. Front. Pharmacol. 2020, 11, 607605. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Zhao, M. An Updated Review of the Epigenetic Mechanism Underlying the Pathogenesis of Age-related Macular Degeneration. Aging Dis. 2020, 11, 1219–1234. [Google Scholar] [CrossRef]
- Shi, H.; Chai, P.; Jia, R.; Fan, X. Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol. Cancer 2020, 19, 78. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef]
- Mongelli, A.; Atlante, S.; Bachetti, T.; Martelli, F.; Farsetti, A.; Gaetano, C. Epigenetic Signaling and RNA Regulation in Cardiovascular Diseases. Int. J. Mol. Sci. 2020, 21, 509. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Gong, Y.; Wang, X.; He, W.; Wu, L.; Zhang, L.; Xiong, L.; Huang, Y.; Su, L.; Shi, P.; et al. METTL3-m(6)A-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease. Mol. Ther. 2022, 30, 932–946. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301–304. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Bell, J.L.; Wang, J.Y.; Hüttelmaier, S.; Zhang, X.D.; Zhang, L.; Liu, T. The Emerging Roles of RNA m(6)A Methylation and Demethylation as Critical Regulators of Tumorigenesis, Drug Sensitivity, and Resistance. Cancer Res. 2021, 81, 3431–3440. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, H.; Hua, L.; Hou, C.; Jia, Q.; Chen, J.; Zhang, S.; Wang, Y.; He, S.; Jia, E. Verification of ferroptosis and pyroptosis and identification of PTGS2 as the hub gene in human coronary artery atherosclerosis. Free Radic. Biol. Med. 2021, 171, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, S.; Ji, W.; Gan, X.; Hua, L.; Hou, C.; Chen, J.; Wang, Y.; He, S.; Zhou, H.; et al. LncRNA Landscape of Coronary Atherosclerosis Reveals Differentially Expressed LncRNAs in Proliferation and Migration of Coronary Artery Smooth Muscle Cells. Front. Cell Dev. Biol. 2021, 9, 656636. [Google Scholar] [CrossRef]
- Roy, P.; Orecchioni, M.; Ley, K. How the immune system shapes atherosclerosis: Roles of innate and adaptive immunity. Nat. Rev. Immunol. 2021, 22, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Chauhan, A.K. Role of Integrins in Modulating Smooth Muscle Cell Plasticity and Vascular Remodeling: From Expression to Therapeutic Implications. Cells 2022, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Garcia-Campos, M.A.; Edelheit, S.; Toth, U.; Safra, M.; Shachar, R.; Viukov, S.; Winkler, R.; Nir, R.; Lasman, L.; Brandis, A.; et al. Deciphering the “m(6)A Code” via Antibody-Independent Quantitative Profiling. Cell 2019, 178, 731–747.e16. [Google Scholar] [CrossRef]
- Wen, K.; Zhang, Y.; Li, Y.; Wang, Q.; Sun, J. Comprehensive analysis of transcriptome-wide m(6)A methylome in the anterior capsule of the lens of high myopia patients. Epigenetics 2021, 16, 955–968. [Google Scholar] [CrossRef]
- Li, P.; Yu, H.; Zhang, G.; Kang, L.; Qin, B.; Cao, Y.; Luo, J.; Chen, X.; Wang, Y.; Qin, M.; et al. Identification and Characterization of N6-Methyladenosine CircRNAs and Methyltransferases in the Lens Epithelium Cells From Age-Related Cataract. Investig. Ophthalmol. Vis. Sci. 2020, 61, 13. [Google Scholar] [CrossRef]
- Lv, Z.; Sun, L.; Xu, Q.; Xing, C.; Yuan, Y. Joint analysis of lncRNA m(6)A methylome and lncRNA/mRNA expression profiles in gastric cancer. Cancer Cell Int. 2020, 20, 464. [Google Scholar] [CrossRef]
- Deng, K.; Ning, X.; Ren, X.; Yang, B.; Li, J.; Cao, J.; Chen, J.; Lu, X.; Chen, S.; Wang, L. Transcriptome-wide N6-methyladenosine methylation landscape of coronary artery disease. Epigenomics 2021, 13, 793–808. [Google Scholar] [CrossRef]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014, 15, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Popova, N.V.; Jücker, M. The Functional Role of Extracellular Matrix Proteins in Cancer. Cancers 2022, 14, 238. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Du, R.; Liu, W.; Huang, G.; Dong, Z.; Li, X. PI3K/Akt/mTOR Signaling Pathway: Role in Esophageal Squamous Cell Carcinoma, Regulatory Mechanisms and Opportunities for Targeted Therapy. Front. Oncol. 2022, 12, 852383. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Zhou, Y.; Hou, C.; Chen, J.; Wang, Y.; Zhang, S.; Zhou, H.; He, S.; Jia, E. Shexiang Baoxin Pills Inhibited Proliferation and Migration of Human Coronary Artery Smooth Muscle Cells via PI3K/AKT/mTOR Pathway. Front. Cardiovasc. Med. 2021, 8, 700630. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell. 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wei, L.; Law, C.T.; Tsang, F.H.; Shen, J.; Cheng, C.L.; Tsang, L.H.; Ho, D.W.; Chiu, D.K.; Lee, J.M.; et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018, 67, 2254–2270. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Liu, L.L.; Wang, C.H.; Lu, S.X.; Yang, X.; He, Y.F.; Zhang, C.Z.; Yun, J.P. Loss of RDM1 enhances hepatocellular carcinoma progression via p53 and Ras/Raf/ERK pathways. Mol. Oncol. 2020, 14, 373–386. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, X.; Chen, Y.; Zhou, T.; Li, D.; Zheng, W.V. METTL3 facilitates the progression of hepatocellular carcinoma by modulating the m6A level of USP7. Am. J. Transl. Res. 2021, 13, 13423–13437. [Google Scholar]
- Qin, Y.; Qiao, Y.; Li, L.; Luo, E.; Wang, D.; Yao, Y.; Tang, C.; Yan, G. The m(6)A methyltransferase METTL3 promotes hypoxic pulmonary arterial hypertension. Life Sci. 2021, 274, 119366. [Google Scholar] [CrossRef]


| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
| METTL3 | CTATCTCCTGGCACTCGCAAGA | GCTTGAACCGTGCAACCACATC |
| METTL14 | CTGAAAGTGCCGACAGCATTGG | CTCTCCTTCATCCAGATACTTACG |
| ZC3H13 | CGGACAGTGATGCCTACAACAG | TCTGTGAGGTGCGAGGGACTAA |
| KIAA1429 | CTCTTCCTAACCACAGTGAACC | AGCCTTTCTATTTCCCCTTCAC |
| WTAP | GCAACAACAGCAGGAGTCTGCA | CTGCTGGACTTGCTTGAGGTAC |
| RBM15 | GTTGTGGCTTATGTGGAGTTTAC | CACTTAAAACACCGGCATTGG |
| RBM15B | TGGTAACCTGGACCACAGCGTA | GGTTCTGGAACTTGAGGAAGGC |
| YTHDC1 | TCAGGAGTTCGCCGAGATGTGT | AGGATGGTGTGGAGGTTGTTCC |
| YTHDC2 | GAAAGCTCCTGAACCTCCACCA | GGTTCTACTGGCAAGTCAGCCA |
| HNRNPC | CCAGCAACGTTACCAACAAGACA | CCTCCACATCAGATTTCTTGACC |
| YTHDF1 | CAAGCACACAACCTCCATCTTCG | GTAAGAAACTGGTTCGCCCTCAT |
| YTHDF2 | TAGCCAGCTACAAGCACACCAC | CAACCGTTGCTGCAGTCTGTGT |
| YTHDF3 | GTTCCTCAGCTCTTTTCTCCAG | TGGATCAAGGCCATATTTTCAAAG |
| FTO | GGTATCTCGCATCCTCATTGG | GAGGAAGGTCTCACAAGCAG |
| ALKBH5 | CCAGCTATGCTTCAGATCGCCT | GGTTCTCTTCCTTGTCCATCTCC |
| si-METTL3-1 | GCUGCACUUCAGACGAAUUTT | AAUUCGUCUGAAGUGCAGCTT |
| si-METTL3-2 | GCUCAACAUACCCGUACUATT | UAGUACGGGUAUGUUGAGCCT |
| si-METTL3-3 | GCAAGAAUUCUGUGACUAUTT | AUAGUCACAGAAUUCUUGCAC |
| Regulation | Gene | Fold Change | Chromosome | Start | End | Peak Length | p-Value |
|---|---|---|---|---|---|---|---|
| Up | CTB-133G6.1 | 87.9 | Chr19 | 7445841 | 7445880 | 39 | 8.11 × 10–9 |
| PSMF1 | 87.2 | Chr20 | 1093905 | 1093920 | 15 | 3.80 × 10−7 | |
| TCAF2 | 75.6 | Chr7 | 143400621 | 143400710 | 89 | 2.58 × 10−6 | |
| SLA2 | 69.6 | Chr20 | 35242781 | 35242840 | 59 | 5.05 × 10−6 | |
| SEPT9 | 69.2 | Chr17 | 75447561 | 75447610 | 49 | 1.90 × 10−6 | |
| OR2T8 | 68.5 | Chr1 | 248084361 | 248084560 | 199 | 7.86 × 10−6 | |
| THAP6 | 66.7 | Chr4 | 76474814 | 76474930 | 116 | 1.04 × 10−5 | |
| TCAF2 | 62.5 | Chr7 | 143318541 | 143318596 | 55 | 1.81 × 10−5 | |
| TMF1 | 59.7 | Chr3 | 69068977 | 69069160 | 183 | 2.38 × 10−6 | |
| ZNF774 | 56.6 | Chr15 | 90908721 | 90908940 | 219 | 4.74 × 10−6 | |
| Down | DHRS3 | 308.2 | Chr1 | 12632755 | 12632840 | 85 | 1.36 × 10−9 |
| ARSI | 139.8 | Chr5 | 149677321 | 149677860 | 539 | 3.40 × 10−9 | |
| MDGA1 | 99.7 | Chr6 | 37665261 | 37665766 | 505 | 9.99 × 10−10 | |
| TULP2 | 96.6 | Chr19 | 49398254 | 49398419 | 165 | 8.49 × 10−8 | |
| XYLT1 | 95.6 | Chr16 | 17202541 | 17202874 | 333 | 1.77 × 10−7 | |
| CLEC18A | 93 | Chr16 | 69993641 | 69993749 | 108 | 3.82 × 10−9 | |
| AL953854.2-002 | 92.9 | Chr9 | 65661850 | 65662160 | 310 | 1.11 × 10−7 | |
| HYDIN | 90.2 | Chr16 | 70969854 | 70970017 | 163 | 7.61 × 10−9 | |
| MRGPRG | 87.2 | Chr11 | 3239601 | 3239980 | 379 | 1.65 × 10−7 | |
| C10orf54 | 83.3 | Chr10 | 73512731 | 73512759 | 28 | 5.26 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Jiang, R.; Jiang, Y.; Fu, Y.; Manafhan, Y.; Zhu, J.; Jia, E. Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis. Cells 2022, 11, 2980. https://doi.org/10.3390/cells11192980
Zhou Y, Jiang R, Jiang Y, Fu Y, Manafhan Y, Zhu J, Jia E. Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis. Cells. 2022; 11(19):2980. https://doi.org/10.3390/cells11192980
Chicago/Turabian StyleZhou, Yaqing, Rongli Jiang, Yali Jiang, Yahong Fu, Yerbolat Manafhan, Jinfu Zhu, and Enzhi Jia. 2022. "Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis" Cells 11, no. 19: 2980. https://doi.org/10.3390/cells11192980
APA StyleZhou, Y., Jiang, R., Jiang, Y., Fu, Y., Manafhan, Y., Zhu, J., & Jia, E. (2022). Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis. Cells, 11(19), 2980. https://doi.org/10.3390/cells11192980








