Effect of Hypoxia in the Transcriptomic Profile of Lung Fibroblasts from Idiopathic Pulmonary Fibrosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Lung Fibroblasts
2.2. Hypoxia
2.3. Total RNA Extraction
2.4. Microarray Hybridization
2.5. Analysis of Differential Gene Expression
2.6. Data Visualization
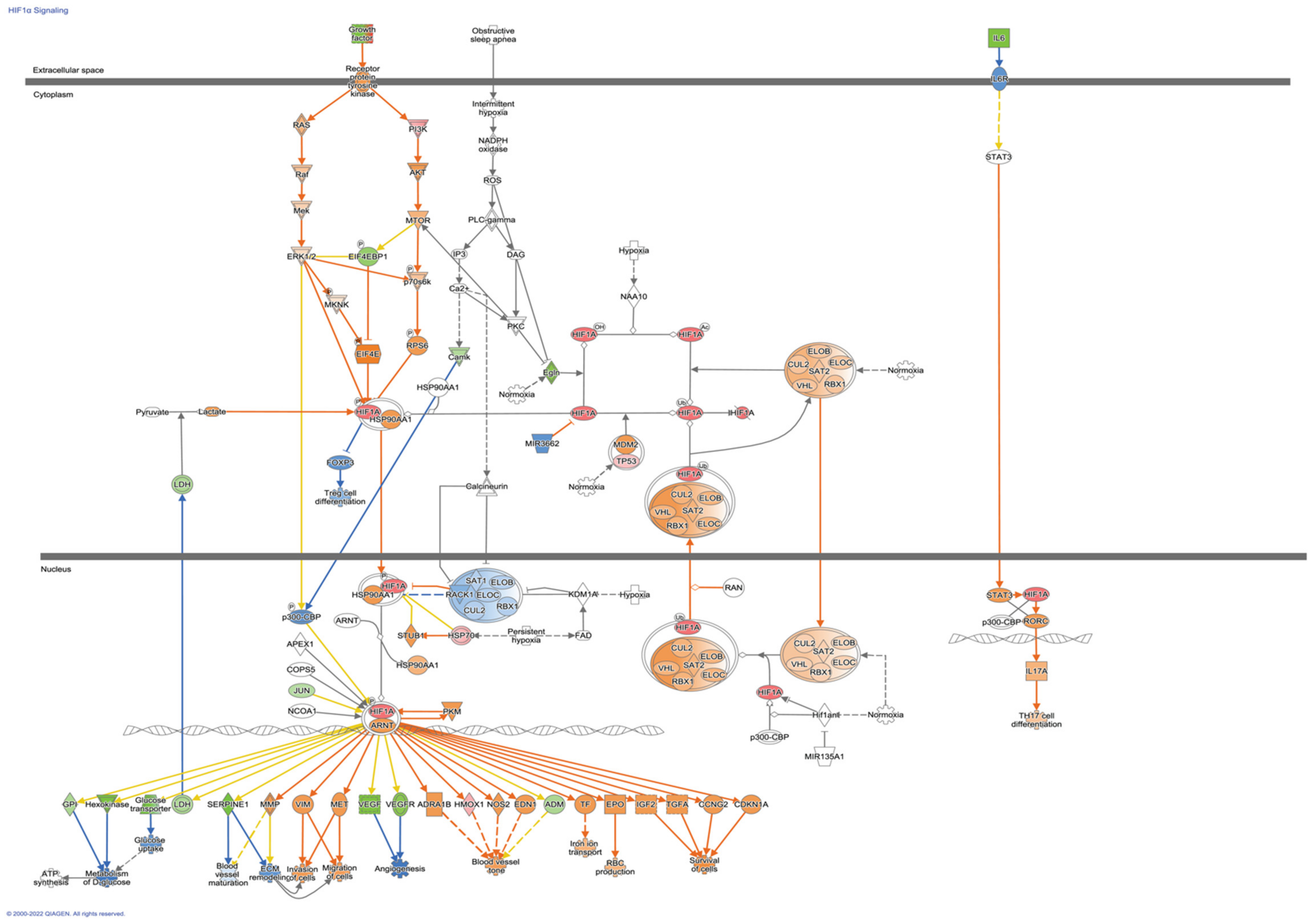
2.7. Enrichment Pathways Analysis and Networks Generation
3. Results
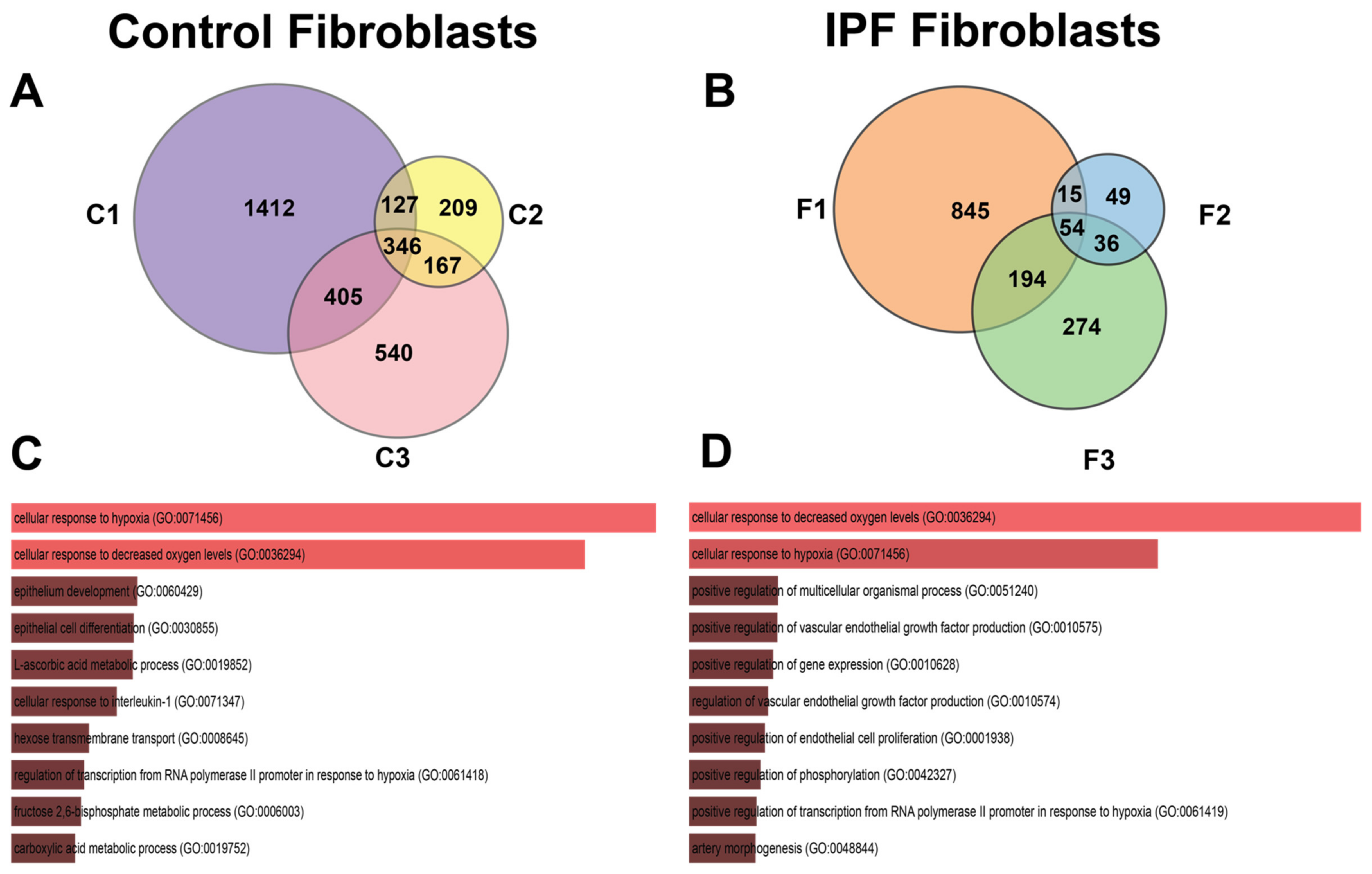
3.1. Transcriptional Response to Hypoxia in Control and IPF-Derived Lung Fibroblasts
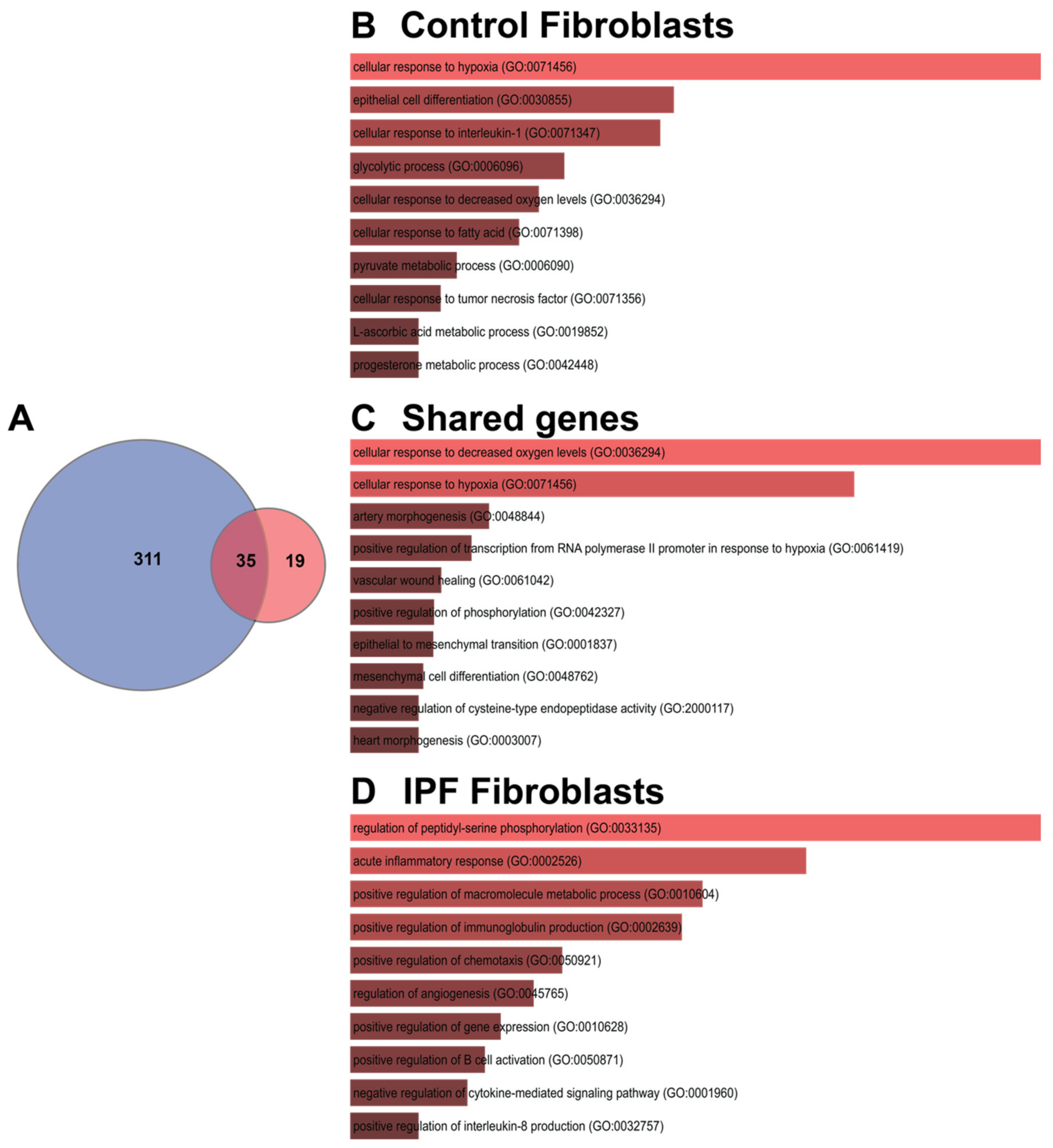
3.2. Shared Hypoxic Genes Float in a Sea of Doubts
3.3. The Transcription Signature of Hypoxia in IPF Fibroblasts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selman, M.; Pardo, A. Revealing the Pathogenic and Aging-Related Mechanisms of the Enigmatic Idiopathic Pulmonary Fibrosis. an Integral Model. Am. J. Respir. Crit. Care Med. 2014, 189, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- King, T.E., Jr.; Pardo, A.; Selman, M. Idiopathic Pulmonary Fibrosis. Lancet 2011, 378, 1949–1961. [Google Scholar] [CrossRef]
- Selman, M.; King, T.E.; Pardo, A.; American Thoracic Society; European Respiratory Society; American College of Chest Physicians. Idiopathic Pulmonary Fibrosis: Prevailing and Evolving Hypotheses about Its Pathogenesis and Implications for Therapy. Ann. Intern. Med. 2001, 134, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Bodempudi, V.; Hergert, P.; Smith, K.; Xia, H.; Herrera, J.; Peterson, M.; Khalil, W.; Kahm, J.; Bitterman, P.B.; Henke, C.A. miR-210 Promotes IPF Fibroblast Proliferation in Response to Hypoxia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L283–L294. [Google Scholar] [CrossRef]
- Weng, T.; Poth, J.M.; Karmouty-Quintana, H.; Garcia-Morales, L.J.; Melicoff, E.; Luo, F.; Chen, N.-Y.; Evans, C.M.; Bunge, R.R.; Bruckner, B.A.; et al. Hypoxia-Induced Deoxycytidine Kinase Contributes to Epithelial Proliferation in Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2014, 190, 1402–1412. [Google Scholar] [CrossRef]
- Tzouvelekis, A.; Harokopos, V.; Paparountas, T.; Oikonomou, N.; Chatziioannou, A.; Vilaras, G.; Tsiambas, E.; Karameris, A.; Bouros, D.; Aidinis, V. Comparative Expression Profiling in Pulmonary Fibrosis Suggests a Role of Hypoxia-Inducible Factor-1α in Disease Pathogenesis. Am. J. Respir. Crit. Care Med. 2007, 176, 1108–1119. [Google Scholar] [CrossRef]
- Romero, Y.; Aquino-Gálvez, A. Hypoxia in Cancer and Fibrosis: Part of the Problem and Part of the Solution. Int. J. Mol. Sci. 2021, 22, 8335. [Google Scholar] [CrossRef]
- Senavirathna, L.K.; Huang, C.; Yang, X.; Munteanu, M.C.; Sathiaseelan, R.; Xu, D.; Henke, C.A.; Liu, L. Hypoxia Induces Pulmonary Fibroblast Proliferation through NFAT Signaling. Sci. Rep. 2018, 8, 2709. [Google Scholar] [CrossRef]
- Aquino-Gálvez, A.; González-Ávila, G.; Jiménez-Sánchez, L.L.; Maldonado-Martínez, H.A.; Cisneros, J.; Toscano-Marquez, F.; Castillejos-López, M.; Torres-Espíndola, L.M.; Velázquez-Cruz, R.; Rodríguez, V.H.O.; et al. Dysregulated Expression of Hypoxia-Inducible Factors Augments Myofibroblasts Differentiation in Idiopathic Pulmonary Fibrosis. Respir. Res. 2019, 20, 130. [Google Scholar] [CrossRef]
- Villaseñor-Altamirano, A.B.; Moretto, M.; Maldonado, M.; Zayas-Del Moral, A.; Munguía-Reyes, A.; Romero, Y.; García-Sotelo, J.S.; Aguilar, L.A.; Aldana-Assad, O.; Engelen, K.; et al. PulmonDB: A Curated Lung Disease Gene Expression Database. Sci. Rep. 2020, 10, 514. [Google Scholar] [CrossRef] [Green Version]
- Raghu, G.; Rochwerg, B.; Zhang, Y.; Garcia, C.A.C.; Azuma, A.; Behr, J.; Brozek, J.L.; Collard, H.R.; Cunningham, W.; Homma, S.; et al. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline: Treatment of Idiopathic Pulmonary Fibrosis. An Update of the 2011 Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2015, 192, e3–e19. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; GBIF: Copenhagen, Denmark, 2013. [Google Scholar]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- MacDonald, J.W. Functions Useful for Those Doing Repetitive Analyses with Affymetrix GeneChips. R Package Version 1.68.1. Affycoretools 2022. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data Bioinformatics. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust Hyperparameter Estimation Protects against Hypervariable Genes and Improves Power to Detect Differential Expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef]
- MacDonald, J.W. Pd.clariom.d.human: Platform Design Info for Affymetrix Clariom_D_Human. R package version 3.14.1. Bioconductor 2022. [Google Scholar]
- Wickham, H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. R Package Version 1.14.0. EnhancedVolcano 2022. [Google Scholar] [CrossRef]
- Hinder, L.M.; Park, M.; Rumora, A.E.; Hur, J.; Eichinger, F.; Pennathur, S.; Kretzler, M.; Brosius, F.C., 3rd; Feldman, E.L. Comparative RNA-Seq Transcriptome Analyses Reveal Distinct Metabolic Pathways in Diabetic Nerve and Kidney Disease. J. Cell. Mol. Med. 2017, 21, 2140–2152. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Bouros, E.; Filidou, E.; Arvanitidis, K.; Mikroulis, D.; Steiropoulos, P.; Bamias, G.; Bouros, D.; Kolios, G. Lung Fibrosis-Associated Soluble Mediators and Bronchoalveolar Lavage from Idiopathic Pulmonary Fibrosis Patients Promote the Expression of Fibrogenic Factors in Subepithelial Lung Myofibroblasts. Pulm. Pharmacol. Ther. 2017, 46, 78–87. [Google Scholar] [CrossRef]
- Bolaños, A.L.; Milla, C.M.; Lira, J.C.; Ramírez, R.; Checa, M.; Barrera, L.; García-Alvarez, J.; Carbajal, V.; Becerril, C.; Gaxiola, M.; et al. Role of Sonic Hedgehog in Idiopathic Pulmonary Fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 303, L978–L990. [Google Scholar] [CrossRef]
- Selman, M.; López-Otín, C.; Pardo, A. Age-Driven Developmental Drift in the Pathogenesis of Idiopathic Pulmonary Fibrosis. Eur. Respir. J. 2016, 48, 538–552. [Google Scholar] [CrossRef]
- Epstein Shochet, G.; Brook, E.; Bardenstein-Wald, B.; Shitrit, D. TGF-β Pathway Activation by Idiopathic Pulmonary Fibrosis (IPF) Fibroblast Derived Soluble Factors Is Mediated by IL-6 Trans-Signaling. Respir. Res. 2020, 21, 56. [Google Scholar] [CrossRef]
- Le, T.-T.T.; Karmouty-Quintana, H.; Melicoff, E.; Le, T.-T.T.; Weng, T.; Chen, N.-Y.; Pedroza, M.; Zhou, Y.; Davies, J.; Philip, K.; et al. Blockade of IL-6 Trans Signaling Attenuates Pulmonary Fibrosis. J. Immunol. 2014, 193, 3755–3768. [Google Scholar] [CrossRef]
- Sullivan, D.I.; Jiang, M.; Hinchie, A.M.; Roth, M.G.; Bahudhanapati, H.; Nouraie, M.; Liu, J.; McDyer, J.F.; Mallampalli, R.K.; Zhang, Y.; et al. Transcriptional and Proteomic Characterization of Telomere-Induced Senescence in a Human Alveolar Epithelial Cell Line. Front. Med. 2021, 8, 600626. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guo, Y.; Wang, J.; Ai, L.; Ma, L.; He, W.; Li, Z.; Yu, X.; Li, J.; Fan, X.; et al. LncRNA CTD-2528L19.6 Prevents the Progression of IPF by Alleviating Fibroblast Activation. Cell Death Dis. 2021, 12, 600. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.K.; Kim, R.Y.; Brown, A.C.; Donovan, C.; Vanka, K.S.; Mayall, J.R.; Liu, G.; Pillar, A.L.; Jones-Freeman, B.; Xenaki, D.; et al. Critical Role for Iron Accumulation in the Pathogenesis of Fibrotic Lung Disease. J. Pathol. 2020, 251, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Allden, S.J.; Ogger, P.P.; Ghai, P.; McErlean, P.; Hewitt, R.; Toshner, R.; Walker, S.A.; Saunders, P.; Kingston, S.; Molyneaux, P.L.; et al. The Transferrin Receptor CD71 Delineates Functionally Distinct Airway Macrophage Subsets during Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 200, 209–219. [Google Scholar] [CrossRef]
- Ashley, S.L.; Wilke, C.A.; Kim, K.K.; Moore, B.B. Periostin Regulates Fibrocyte Function to Promote Myofibroblast Differentiation and Lung Fibrosis. Mucosal Immunol. 2017, 10, 341–351. [Google Scholar] [CrossRef]
- Leppäranta, O.; Sens, C.; Salmenkivi, K.; Kinnula, V.L.; Keski-Oja, J.; Myllärniemi, M.; Koli, K. Regulation of TGF-β Storage and Activation in the Human Idiopathic Pulmonary Fibrosis Lung. Cell Tissue Res. 2012, 348, 491–503. [Google Scholar] [CrossRef]
- Jain, R.; Shaul, P.W.; Borok, Z.; Willis, B.C. Endothelin-1 Induces Alveolar Epithelial-Mesenchymal Transition through Endothelin Type A Receptor-Mediated Production of TGF-beta1. Am. J. Respir. Cell Mol. Biol. 2007, 37, 38–47. [Google Scholar] [CrossRef]
- Park, S.H.; Saleh, D.; Giaid, A.; Michel, R.P. Increased Endothelin-1 in Bleomycin-Induced Pulmonary Fibrosis and the Effect of an Endothelin Receptor Antagonist. Am. J. Respir. Crit. Care Med. 1997, 156, 600–608. [Google Scholar] [CrossRef]
- Gordan, J.D.; Bertout, J.A.; Hu, C.-J.; Diehl, J.A.; Simon, M.C. HIF-2α Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell 2007, 11, 335–347. [Google Scholar] [CrossRef]
- Mizuno, S.; Bogaard, H.J.; Voelkel, N.F.; Umeda, Y.; Kadowaki, M.; Ameshima, S.; Miyamori, I.; Ishizaki, T. Hypoxia Regulates Human Lung Fibroblast Proliferation via p53-Dependent and -Independent Pathways. Respir. Res. 2009, 10, 17. [Google Scholar] [CrossRef]
- Cisneros, J.; Hagood, J.; Checa, M.; Ortiz-Quintero, B.; Negreros, M.; Herrera, I.; Ramos, C.; Pardo, A.; Selman, M. Hypermethylation-Mediated Silencing of p14ARF in Fibroblasts from Idiopathic Pulmonary Fibrosis. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2012, 303, L295–L303. [Google Scholar] [CrossRef]
- Romero, Y.; Bueno, M.; Ramirez, R.; Álvarez, D.; Sembrat, J.C.; Goncharova, E.A.; Rojas, M.; Selman, M.; Mora, A.L.; Pardo, A. mTORC1 Activation Decreases Autophagy in Aging and Idiopathic Pulmonary Fibrosis and Contributes to Apoptosis Resistance in IPF Fibroblasts. Aging Cell 2016, 15, 1103–1112. [Google Scholar] [CrossRef]
- Luis-García, E.R.; Becerril, C.; Salgado-Aguayo, A.; Aparicio-Trejo, O.E.; Romero, Y.; Flores-Soto, E.; Mendoza-Milla, C.; Montaño, M.; Chagoya, V.; Pedraza-Chaverri, J.; et al. Mitochondrial Dysfunction and Alterations in Mitochondrial Permeability Transition Pore (mPTP) Contribute to Apoptosis Resistance in Idiopathic Pulmonary Fibrosis Fibroblasts. Int. J. Mol. Sci. 2021, 22, 7870. [Google Scholar] [CrossRef]
- Hu, C.-J.; Wang, L.-Y.; Chodosh, L.A.; Keith, B.; Simon, M.C. Differential Roles of Hypoxia-Inducible Factor 1α (HIF-1α) and HIF-2α in Hypoxic Gene Regulation. Mol. Cell. Biol. 2003, 23, 9361–9374. [Google Scholar] [CrossRef]
- Loboda, A.; Jozkowicz, A.; Dulak, J. HIF-1 versus HIF-2—Is One More Important than the Other? Vasc. Pharmacol. 2012, 56, 245–251. [Google Scholar] [CrossRef]
- Le Bras, A.; Lionneton, F.; Mattot, V.; Lelièvre, E.; Caetano, B.; Spruyt, N.; Soncin, F. HIF-2α Specifically Activates the VE-Cadherin Promoter Independently of Hypoxia and in Synergy with Ets-1 through Two Essential ETS-Binding Sites. Oncogene 2007, 26, 7480–7489. [Google Scholar] [CrossRef]
- Coulet, F.; Nadaud, S.; Agrapart, M.; Soubrier, F. Identification of Hypoxia-Response Element in the Human Endothelial Nitric-Oxide Synthase Gene Promoter. J. Biol. Chem. 2003, 278, 46230–46240. [Google Scholar] [CrossRef]
- Lin, Q.; Cong, X.; Yun, Z. Differential Hypoxic Regulation of Hypoxia-Inducible Factors 1α and 2α. Mol. Cancer Res. 2011, 9, 757–765. [Google Scholar] [CrossRef]
- Holmquist-Mengelbier, L.; Fredlund, E.; Löfstedt, T.; Noguera, R.; Navarro, S.; Nilsson, H.; Pietras, A.; Vallon-Christersson, J.; Borg, A.; Gradin, K.; et al. Recruitment of HIF-1α and HIF-2α to Common Target Genes Is Differentially Regulated in Neuroblastoma: HIF-2α Promotes an Aggressive Phenotype. Cancer Cell 2006, 10, 413–423. [Google Scholar] [CrossRef]
- Adamovich, Y.; Ladeuix, B.; Golik, M.; Koeners, M.P.; Asher, G. Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1α. Cell Metab. 2017, 25, 93–101. [Google Scholar] [CrossRef]
- Selman, M.; Pardo, A.; Kaminski, N. Idiopathic Pulmonary Fibrosis: Aberrant Recapitulation of Developmental Programs? PLoS Med. 2008, 5, e62. [Google Scholar] [CrossRef] [Green Version]
- Novianti, T.; Juniantito, V.; Jusuf, A.A.; Arida, E.A.; Jusman, S.W.A.; Sadikin, M. Expression and Role of HIF-1α and HIF-2α in Tissue Regeneration: A Study of Hypoxia in House Gecko Tail Regeneration. Organogenesis 2019, 15, 69–84. [Google Scholar] [CrossRef]
- Scheerer, N.; Dehne, N.; Stockmann, C.; Swoboda, S.; Baba, H.A.; Neugebauer, A.; Johnson, R.S.; Fandrey, J. Myeloid Hypoxia-Inducible Factor-1α Is Essential for Skeletal Muscle Regeneration in Mice. J. Immunol. 2013, 191, 407–414. [Google Scholar] [CrossRef] [Green Version]




| ID | Age | Condition | Passage Number | Primary or Commercial |
|---|---|---|---|---|
| C1 | 7 weeks | Healthy | 12 | ATCC CCL-215 |
| C2 | >50 years | Healthy | 12 | Primary |
| C3 | 79 years | Healthy | 14 | ATCC PCS-201-013 |
| F1 | >50 years | IPF | 6 | Primary |
| F2 | >50 years | IPF | 7 | Primary |
| F3 | 58 years | IPF | 7 | Primary |
| Gene | Also Known as | N | H | OS | Description | Models | References |
|---|---|---|---|---|---|---|---|
| F3 | TF; AGT; CD142 | NS | Down | Up | Coagulation factor III is a surface receptor, and it participates in the coagulation cascade | IPF-HLF | [27] |
| HHIP | HIP | NS | Down | Up | Hedgehog interacting protein is involved with Hedgehog signaling pathway in embryonic development | IPF-HLF | [28,29] |
| IL6 | CDF; HGF; HSF; BSF2; IL-6; BSF-2; IFNB2; IFN-β-2 | NS | Down | Up | Interleukin 6 encodes for a cytokine with inflammatory functions | IPF-HLF and BMM | [30,31] |
| STC1 | STC | NS | Down | Up | Stanniocalcin 1 encodes for homodimeric glycoprotein with paracrine and autocrine functions | Increased in plasma of patients with IPF | [32] |
| DDIT4 | Dig2; REDD1; REDD-1 | NS | Down | - | DNA Damage Inducible Transcript 4. Gene related to response to virus, hypoxia, DNA damage, and tumor regulation | Expression associated with lncRNAs in IPF | [33] |
| CCNG2 | - | NS | Down | - | Encodes for cyclin-G2 involved in cell cycle | NS-IPF | - |
| KCTD16 | - | NS | Down | - | Potassium channel tetramerization domain containing 16 regulates GABA receptor signaling | NS-IPF | - |
| BHLHE41 | DEC2; FNSS1; hDEC2; BHLHB3; SHARP1 | NS | Down | - | Basic helix-loop-helix family member e41, involved in circadian rhythm and cell differentiation | NS-IPF | - |
| STXBP6 | amisyn; HSPC156 | Up | Down | - | Syntaxin binding protein 6 is involved in regulating SNARE complex formation | NS-IPF | - |
| SERPINB7 | PPKN; TP55; MEGSIN | NS | Down | - | Serpin family B member 7 encodes for a protein that functions as a protease inhibitor | NS-IPF | - |
| Gene | Also Known as | N | H | OS | Description | Models | References |
|---|---|---|---|---|---|---|---|
| EPAS1 | HLF; MOP2; ECYT4; HIF2A; PASD2; bHLHe73 | Down | Up | Up | Endothelial PAS domain protein 1 is a gene that encodes a transcription factor involved in signaling pathway in hypoxia | IPF-HLF | [9] |
| TFRC | CD71, IMD46, T9, TFR, TFR1, TR, TRFR, p90 | Down | Up | Up | Transferrin receptor encodes a cell surface receptor associated with cellular iron uptake and is required for erythropoiesis | IPF-HLF, BMM and BAL of IPF patients | [34,35] |
| POSTN | PN; OSF2; OSF-2; PDLPOSTN | Down | Up | Up | Gen encodes for periostin protein with functions in tissue development and regeneration. Expression related to IPF progression | IPF-HLF and BMM | [36,37] |
| EDNRA | ETA; ET-A; ETAR; ETRA; MFDA; ETA-R; hET-AR | NS | Up | Up | Endothelin receptor type A encodes an endothelin-1 receptor with vasoconstriction properties | Primary rat alveolar type II cells | [38,39] |
| HOXA5 | HOX1; HOX1C; HOX1.3 | NS | Up | Up | Homeobox A5 encodes for transcription factors called homeobox genes spatially and temporally regulated during embryonic development | NS-IPF | - |
| PCDH18 | PCDH68L | NS | Up | Down | Protocadherin 18 encodes for a protein member of the subfamily of cadherin superfamily related to cell–cell connections | NS-IPF | - |
| ADH1B | ADH2; HEL-S-117 | NS | Up | - | Alcohol dehydrogenase 1B encodes for a protein member of the alcohol dehydrogenase family | NS-IPF | - |
| SLC14A1 | JK; UT1; UTE; HUT11; Jk(a); Jk(b); RACH1; RACH2; UT-B1; HUT11A; HsT1341 | NS | Up | - | Solute carrier family 14-member 1 encodes for a protein membrane transporter that mediates urea transport in erythrocytes | NS-IPF | - |
| USP18 | ISG43; UBP43; PTORCH2 | NS | Up | - | Ubiquitin specific peptidase 18 encodes for an enzyme that belongs to ubiquitin-specific proteases family | NS-IPF | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero, Y.; Balderas-Martínez, Y.I.; Vargas-Morales, M.A.; Castillejos-López, M.; Vázquez-Pérez, J.A.; Calyeca, J.; Torres-Espíndola, L.M.; Patiño, N.; Camarena, A.; Carlos-Reyes, Á.; et al. Effect of Hypoxia in the Transcriptomic Profile of Lung Fibroblasts from Idiopathic Pulmonary Fibrosis. Cells 2022, 11, 3014. https://doi.org/10.3390/cells11193014
Romero Y, Balderas-Martínez YI, Vargas-Morales MA, Castillejos-López M, Vázquez-Pérez JA, Calyeca J, Torres-Espíndola LM, Patiño N, Camarena A, Carlos-Reyes Á, et al. Effect of Hypoxia in the Transcriptomic Profile of Lung Fibroblasts from Idiopathic Pulmonary Fibrosis. Cells. 2022; 11(19):3014. https://doi.org/10.3390/cells11193014
Chicago/Turabian StyleRomero, Yair, Yalbi Itzel Balderas-Martínez, Miguel Angel Vargas-Morales, Manuel Castillejos-López, Joel Armando Vázquez-Pérez, Jazmín Calyeca, Luz María Torres-Espíndola, Nelly Patiño, Angel Camarena, Ángeles Carlos-Reyes, and et al. 2022. "Effect of Hypoxia in the Transcriptomic Profile of Lung Fibroblasts from Idiopathic Pulmonary Fibrosis" Cells 11, no. 19: 3014. https://doi.org/10.3390/cells11193014






