Modulatory Role of TPPP3 in Microtubule Organization and Its Impact on Alpha-Synuclein Pathology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Comparison
2.2. Prediction of Unstructured Regions
2.3. Prediction of Protein Structure
2.4. Antibodies
2.5. Production of Mouse TPPP3 (Clone No.8/B3-B7) Monoclonal Antibody
2.6. Antibody Capture Indirect ELISA
2.7. Plasmids
2.8. Expression and Purification of Proteins
2.9. Brain Sample
2.10. Preparation of Cytosolic Brain Extract
2.11. ELISA Experiments
2.12. Turbidimetry Measurements and Pelleting Experiments
2.13. Affinity Chromatography
2.14. Cell Culture, Transfection and Manipulation
2.15. Preparation of HeLa Cell Extract
2.16. ELISA on Cells (cELISA) for Detection of the Transfected EGFP-TPPP1 and EGFP-TPPP3
2.17. Western Blot
2.18. Immunofluorescence Microscopy
2.19. Statistical Analysis
3. Results and Discussion
3.1. Characteristics of the Human TPPP3 Protein
3.2. Quantification of the TPPP Levels in Human Brain
3.3. TPPP3, like TPPP1, Is a Microtubule-Associated Protein
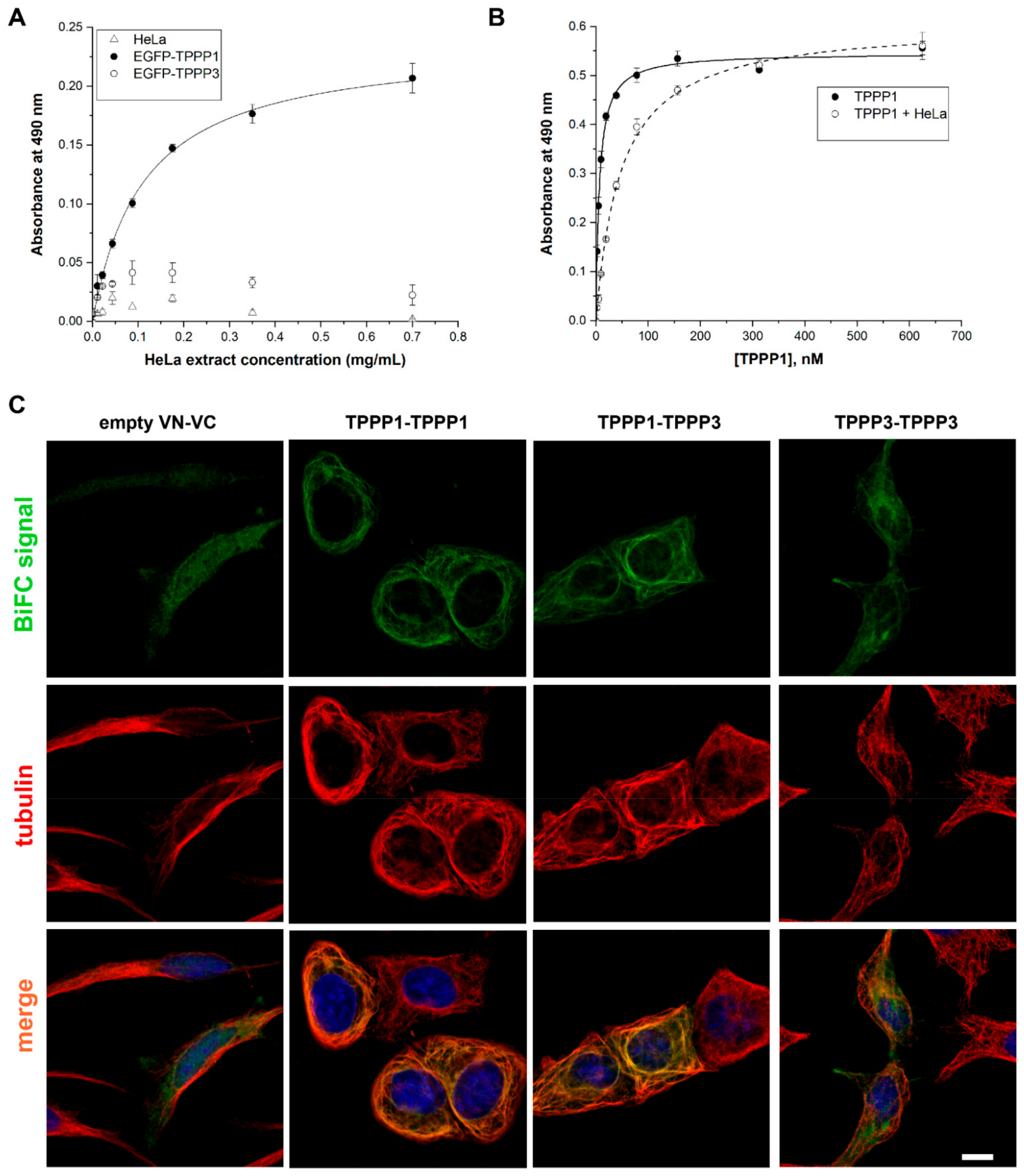
3.4. Homo- and Hetero-Association of TPPP3
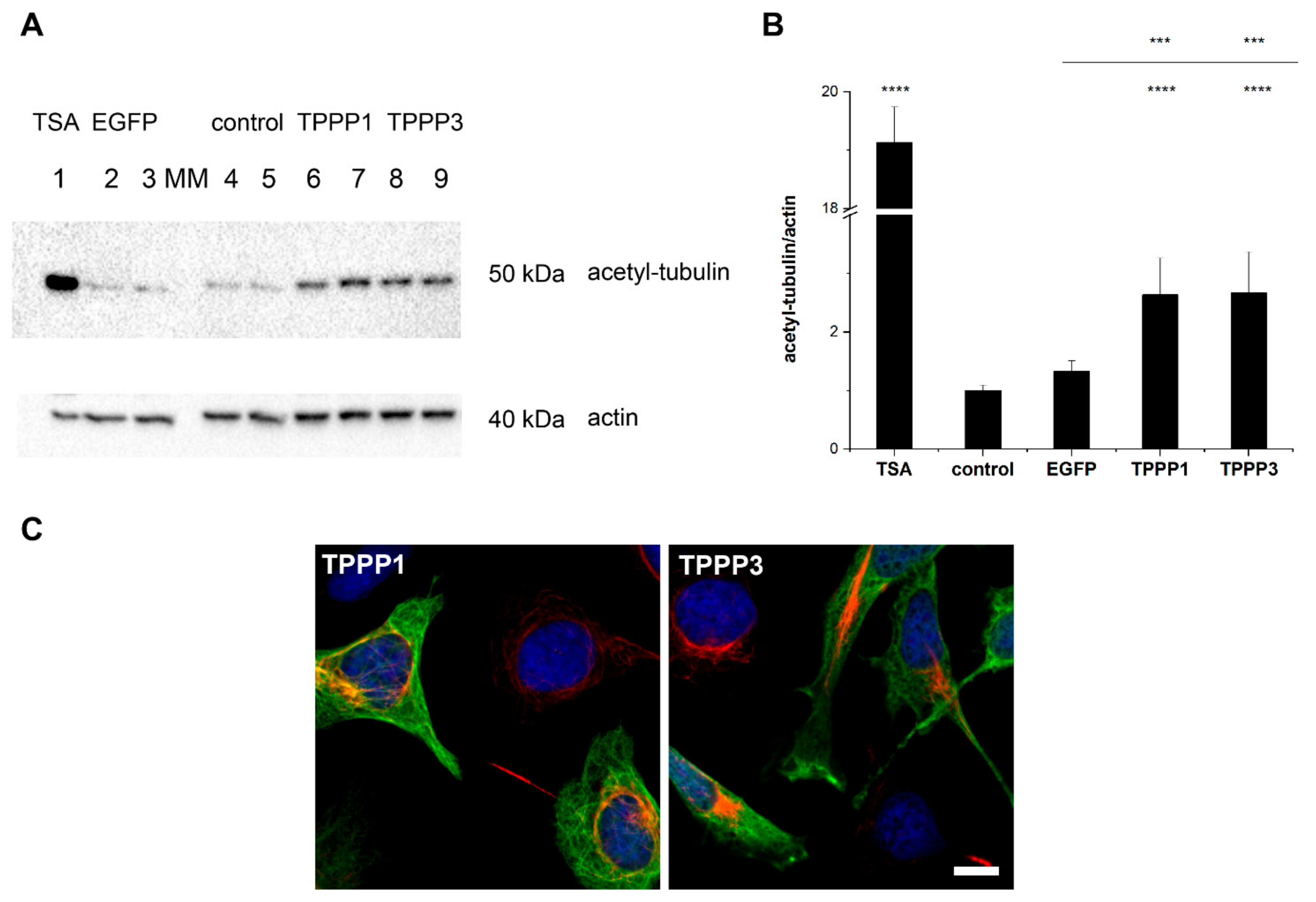
3.5. Effect of TPPP3 on the Acetylation Level of the Microtubule Network
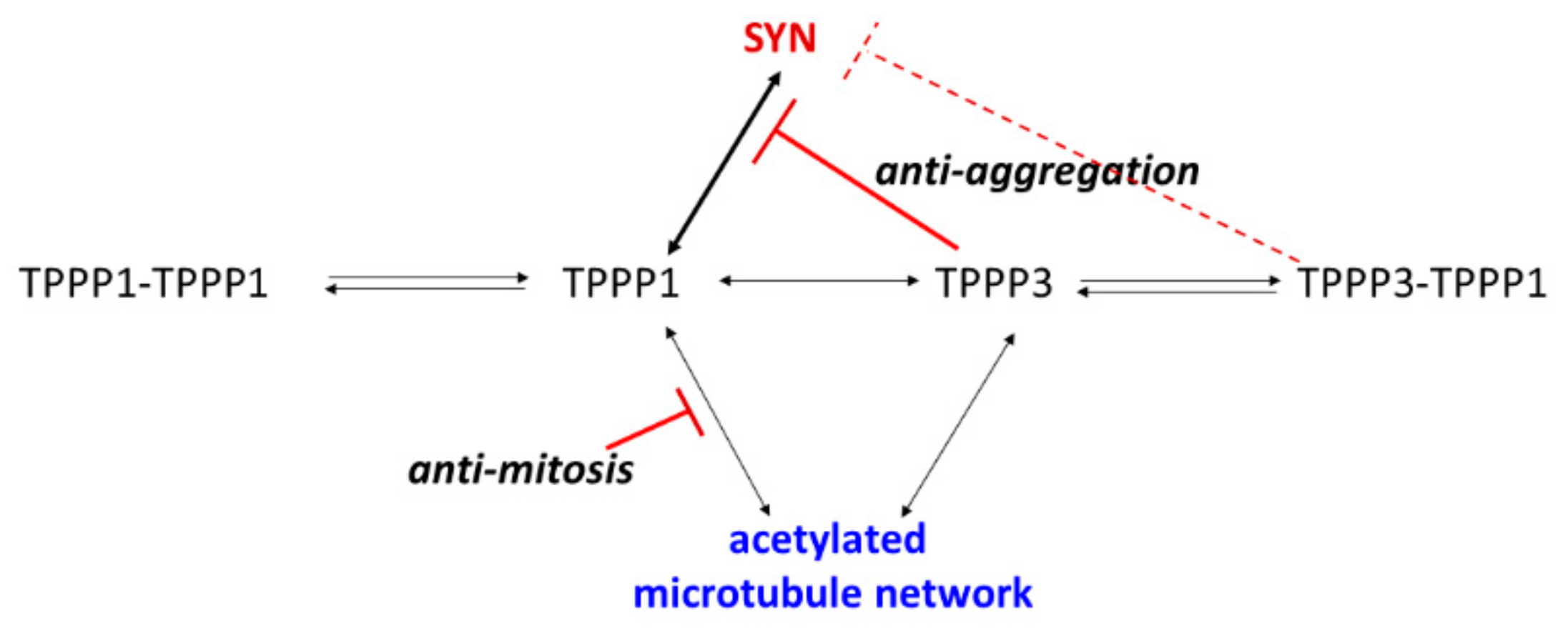
3.6. Distinct Pathological Functions of the TPPPs Proteins
3.7. Anti-Aggregative Potency of TPPP3
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jellinger, K.A. Dementia with Lewy bodies and Parkinson’s disease-dementia: Current concepts and controversies. J. Neural Transm. 2018, 125, 615–650. [Google Scholar] [CrossRef] [PubMed]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Poewe, W.; Stankovic, I.; Halliday, G.; Meissner, W.G.; Wenning, G.K.; Pellecchia, M.T.; Seppi, K.; Palma, J.A.; Kaufmann, H. Multiple system atrophy. Nat. Rev. Dis. Primers 2022, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G.; Laszlo, L.; Kovacs, J.; Jensen, P.H.; Lindersson, E.; Botond, G.; Molnar, T.; Perczel, A.; Hudecz, F.; Mezo, G.; et al. Natively unfolded tubulin polymerization promoting protein TPPP/p25 is a common marker of alpha-synucleinopathies. Neurobiol. Dis. 2004, 17, 155–162. [Google Scholar] [CrossRef]
- Oláh, J.; Lehotzky, A.; Szunyogh, S.; Szénási, T.; Orosz, F.; Ovádi, J. Microtubule-Associated Proteins with Regulatory Functions by Day and Pathological Potency at Night. Cells 2020, 9, 357. [Google Scholar] [CrossRef]
- Bernal-Conde, L.D.; Ramos-Acevedo, R.; Reyes-Hernandez, M.A.; Balbuena-Olvera, A.J.; Morales-Moreno, I.D.; Arguero-Sanchez, R.; Schule, B.; Guerra-Crespo, M. Alpha-Synuclein Physiology and Pathology: A Perspective on Cellular Structures and Organelles. Front. Neurosci. 2019, 13, 1399. [Google Scholar] [CrossRef]
- Cheng, F.; Vivacqua, G.; Yu, S. The role of alpha-synuclein in neurotransmission and synaptic plasticity. J. Chem. Neuroanat. 2011, 42, 242–248. [Google Scholar] [CrossRef]
- Lehotzky, A.; Lau, P.; Tőkési, N.; Muja, N.; Hudson, L.D.; Ovádi, J. Tubulin polymerization-promoting protein (TPPP/p25) is critical for oligodendrocyte differentiation. Glia 2010, 58, 157–168. [Google Scholar] [CrossRef]
- Fu, M.M.; McAlear, T.S.; Nguyen, H.; Oses-Prieto, J.A.; Valenzuela, A.; Shi, R.D.; Perrino, J.J.; Huang, T.T.; Burlingame, A.L.; Bechstedt, S.; et al. The Golgi Outpost Protein TPPP Nucleates Microtubules and Is Critical for Myelination. Cell 2019, 179, 132–146.e14. [Google Scholar] [CrossRef]
- Vincze, O.; Tőkési, N.; Oláh, J.; Hlavanda, E.; Zotter, A.; Horváth, I.; Lehotzky, A.; Tirián, L.; Medzihradszky, K.F.; Kovács, J.; et al. Tubulin polymerization promoting proteins (TPPPs): Members of a new family with distinct structures and functions. Biochemistry 2006, 45, 13818–13826. [Google Scholar] [CrossRef]
- Xie, J.; Chen, S.; Bopassa, J.C.; Banerjee, S. Drosophila tubulin polymerization promoting protein mutants reveal pathological correlates relevant to human Parkinson’s disease. Sci. Rep. 2021, 11, 13614. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, X.; Li, L.; Feng, X.; Yang, Z.; Zhang, W.; Hu, R. Depletion of tubulin polymerization promoting protein family member 3 suppresses HeLa cell proliferation. Mol. Cell. Biochem. 2010, 333, 91–98. [Google Scholar] [CrossRef]
- Zhou, W.; Li, J.; Wang, X.; Hu, R. Stable knockdown of TPPP3 by RNA interference in Lewis lung carcinoma cell inhibits tumor growth and metastasis. Mol. Cell. Biochem. 2010, 343, 231–238. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Y.; Ye, K.; Wu, N.; Li, J.; Liu, N.; He, M.; Lu, B.; Zhou, W.; Hu, R. Knockdown of Tubulin Polymerization Promoting Protein Family Member 3 Suppresses Proliferation and Induces Apoptosis in Non-Small-Cell Lung Cancer. J. Cancer 2016, 7, 1189–1196. [Google Scholar] [CrossRef]
- Li, Y.; Bai, M.; Xu, Y.; Zhao, W.; Liu, N.; Yu, J. TPPP3 Promotes Cell Proliferation, Invasion and Tumor Metastasis via STAT3/ Twist1 Pathway in Non-Small-Cell Lung Carcinoma. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 50, 2004–2016. [Google Scholar] [CrossRef]
- Pastor, M.D.; Nogal, A.; Molina-Pinelo, S.; Melendez, R.; Salinas, A.; Gonzalez De la Pena, M.; Martin-Juan, J.; Corral, J.; Garcia-Carbonero, R.; Carnero, A.; et al. Identification of proteomic signatures associated with lung cancer and COPD. J. Proteom. 2013, 89, 227–237. [Google Scholar] [CrossRef]
- Ye, K.; Li, Y.; Zhao, W.; Wu, N.; Liu, N.; Li, R.; Chen, L.; He, M.; Lu, B.; Wang, X.; et al. Knockdown of Tubulin Polymerization Promoting Protein Family Member 3 inhibits cell proliferation and invasion in human colorectal cancer. J. Cancer 2017, 8, 1750–1758. [Google Scholar] [CrossRef]
- King, E.R.; Tung, C.S.; Tsang, Y.T.; Zu, Z.; Lok, G.T.; Deavers, M.T.; Malpica, A.; Wolf, J.K.; Lu, K.H.; Birrer, M.J.; et al. The anterior gradient homolog 3 (AGR3) gene is associated with differentiation and survival in ovarian cancer. Am. J. Surg. Pathol. 2011, 35, 904–912. [Google Scholar] [CrossRef]
- Komura, S.; Ito, K.; Ohta, S.; Ukai, T.; Kabata, M.; Itakura, F.; Semi, K.; Matsuda, Y.; Hashimoto, K.; Shibata, H.; et al. Cell-type dependent enhancer binding of the EWS/ATF1 fusion gene in clear cell sarcomas. Nat. Commun. 2019, 10, 3999. [Google Scholar] [CrossRef]
- Pang, L.; Hu, J.; Li, F.; Yuan, H.; Yan, M.; Liao, G.; Xu, L.; Pang, B.; Ping, Y.; Xiao, Y.; et al. Discovering Rare Genes Contributing to Cancer Stemness and Invasive Potential by GBM Single-Cell Transcriptional Analysis. Cancers 2019, 11, 2025. [Google Scholar] [CrossRef] [Green Version]
- Ren, Q.; Hou, Y.; Li, X.; Fan, X. Silence of TPPP3 suppresses cell proliferation, invasion and migration via inactivating NF-kappaB/COX2 signal pathway in breast cancer cell. Cell Biochem. Funct. 2020, 38, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Wan, H.; Zhang, X. Requirement of splicing factor hnRNP A2B1 for tumorigenesis of melanoma stem cells. Stem Cell Res. Ther. 2021, 12, 90. [Google Scholar] [CrossRef] [PubMed]
- Shen, A.; Tong, X.; Li, H.; Chu, L.; Jin, X.; Ma, H.; Ouyang, Y. TPPP3 inhibits the proliferation, invasion and migration of endometrial carcinoma targeted with miR-1827. Clin. Exp. Pharmacol. Physiol. 2021, 48, 890–901. [Google Scholar] [CrossRef]
- Hu, D.; Ansari, D.; Pawlowski, K.; Zhou, Q.; Sasor, A.; Welinder, C.; Kristl, T.; Bauden, M.; Rezeli, M.; Jiang, Y.; et al. Proteomic analyses identify prognostic biomarkers for pancreatic ductal adenocarcinoma. Oncotarget 2018, 9, 9789–9807. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Yang, Z.; Guo, X.; Mo, W.; Li, X. Tubulin polymerization promoting protein family member 3 (TPPP3) overexpression inhibits cell proliferation and invasion in nasopharyngeal carcinoma. Bioengineered 2021, 12, 8485–8495. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, X.; Li, J.; Su, Q.; Qiu, Y.; Zhang, Z.; Zhang, L.; Mo, W. TPPP3 Associated with Prognosis and Immune Infiltrates in Head and Neck Squamous Carcinoma. BioMed Res. Int. 2020, 2020, 3962146. [Google Scholar] [CrossRef]
- Staverosky, J.A.; Pryce, B.A.; Watson, S.S.; Schweitzer, R. Tubulin polymerization-promoting protein family member 3, Tppp3, is a specific marker of the differentiating tendon sheath and synovial joints. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2009, 238, 685–692. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, M.; Zhao, X.; Bin, E.; Hu, Y.; Tang, N.; Dai, H.; Wang, C. Telomere shortening impairs alveolar regeneration. Cell Prolif. 2022, 55, e13211. [Google Scholar] [CrossRef]
- Huang, R.; Chen, M.; Yang, L.; Wagle, M.; Guo, S.; Hu, B. MicroRNA-133b Negatively Regulates Zebrafish Single Mauthner-Cell Axon Regeneration through Targeting tppp3 in Vivo. Front. Mol. Neurosci. 2017, 10, 375. [Google Scholar] [CrossRef]
- Liu, N.; Li, Y.; Nan, W.; Zhou, W.; Huang, J.; Li, R.; Zhou, L.; Hu, R. Interaction of TPPP3 with VDAC1 Promotes Endothelial Injury through Activation of Reactive Oxygen Species. Oxid. Med. Cell. Longev. 2020, 2020, 5950195. [Google Scholar] [CrossRef]
- Shukla, V.; Kaushal, J.B.; Sankhwar, P.; Manohar, M.; Dwivedi, A. Inhibition of TPPP3 attenuates beta-catenin/NF-kappaB/COX-2 signaling in endometrial stromal cells and impairs decidualization. J. Endocrinol. 2019, 240, 417–429. [Google Scholar] [CrossRef]
- Shukla, V.; Popli, P.; Kaushal, J.B.; Gupta, K.; Dwivedi, A. Uterine TPPP3 plays important role in embryo implantation via modulation of beta-catenin. Biol. Reprod. 2018, 99, 982–999. [Google Scholar] [CrossRef]
- Szénási, T.; Oláh, J.; Szabó, A.; Szunyogh, S.; Lang, A.; Perczel, A.; Lehotzky, A.; Uversky, V.N.; Ovádi, J. Challenging drug target for Parkinson’s disease: Pathological complex of the chameleon TPPP/p25 and alpha-synuclein proteins. Biochim. Biophys. Acta. Mol. Basis Dis. 2017, 1863, 310–323. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Li, X.; Romero, P.; Rani, M.; Dunker, A.K.; Obradovic, Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. 1999, 10, 30–40. [Google Scholar]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Hoftberger, R.; Fink, S.; Aboul-Enein, F.; Botond, G.; Oláh, J.; Berki, T.; Ovádi, J.; Lassmann, H.; Budka, H.; Kovacs, G.G. Tubulin polymerization promoting protein (TPPP/p25) as a marker for oligodendroglial changes in multiple sclerosis. Glia 2010, 58, 1847–1857. [Google Scholar] [CrossRef]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Oláh, J.; Szénási, T.; Szunyogh, S.; Szabó, A.; Lehotzky, A.; Ovádi, J. Further evidence for microtubule-independent dimerization of TPPP/p25. Sci. Rep. 2017, 7, 40594. [Google Scholar] [CrossRef] [PubMed]
- Tőkesi, N.; Lehotzky, A.; Horváth, I.; Szabó, B.; Oláh, J.; Lau, P.; Ovádi, J. TPPP/p25 promotes tubulin acetylation by inhibiting histone deacetylase 6. J. Biol. Chem. 2010, 285, 17896–17906. [Google Scholar] [CrossRef] [PubMed]
- Tőkési, N.; Oláh, J.; Hlavanda, E.; Szunyogh, S.; Szabó, A.; Babos, F.; Magyar, A.; Lehotzky, A.; Vass, E.; Ovádi, J. Identification of motives mediating alternative functions of the neomorphic moonlighting TPPP/p25. Biochim. Biophys. Acta 2014, 1842, 547–557. [Google Scholar] [CrossRef]
- Paik, S.R.; Lee, J.H.; Kim, D.H.; Chang, C.S.; Kim, J. Aluminum-induced structural alterations of the precursor of the non-A beta component of Alzheimer’s disease amyloid. Arch Biochem. Biophys. 1997, 344, 325–334. [Google Scholar] [CrossRef]
- Na, C.N.; Timasheff, S.N. Interaction of vinblastine with calf brain tubulin: Multiple equilibria. Biochemistry 1986, 25, 6214–6222. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hlavanda, E.; Kovács, J.; Oláh, J.; Orosz, F.; Medzihradszky, K.F.; Ovádi, J. Brain-specific p25 protein binds to tubulin and microtubules and induces aberrant microtubule assemblies at substoichiometric concentrations. Biochemistry 2002, 41, 8657–8664. [Google Scholar] [CrossRef]
- Aramini, J.M.; Rossi, P.; Shastry, R.; Nwosu, C.; Cunningham, K.; Xiao, R.; Liu, J.; Baran, M.C.; Rajan, P.K.; Acton, T.B. Solution NMR Structure of TUBULIN Polymerization-Promoting Protein Family Member 3 from Homo Sapiens. 2007. Available online: https://www.rcsb.org/structure/2JRF (accessed on 10 January 2022).
- Takahashi, M.; Tomizawa, K.; Fujita, S.C.; Sato, K.; Uchida, T.; Imahori, K. A brain-specific protein p25 is localized and associated with oligodendrocytes, neuropil, and fiber-like structures of the CA hippocampal region in the rat brain. J. Neurochem. 1993, 60, 228–235. [Google Scholar] [CrossRef]
- Oláh, J.; Vincze, O.; Virok, D.; Simon, D.; Bozso, Z.; Tőkési, N.; Horváth, I.; Hlavanda, E.; Kovács, J.; Magyar, A.; et al. Interactions of pathological hallmark proteins: Tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J. Biol. Chem. 2011, 286, 34088–34100. [Google Scholar] [CrossRef]
- Zotter, A.; Oláh, J.; Hlavanda, E.; Bodor, A.; Perczel, A.; Szigeti, K.; Fidy, J.; Ovádi, J. Zn(2)+-induced rearrangement of the disordered TPPP/p25 affects its microtubule assembly and GTPase activity. Biochemistry 2011, 50, 9568–9578. [Google Scholar] [CrossRef] [PubMed]
- Lehotzky, A.; Oláh, J.; Szunyogh, S.; Szabó, A.; Berki, T.; Ovádi, J. Zinc-induced structural changes of the disordered tppp/p25 inhibits its degradation by the proteasome. Biochim. Biophys. Acta 2015, 1852, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Oláh, J.; Zotter, A.; Hlavanda, E.; Szunyogh, S.; Orosz, F.; Szigeti, K.; Fidy, J.; Ovádi, J. Microtubule assembly-derived by dimerization of TPPP/p25. Evaluation of thermodynamic parameters for multiple equilibrium system from ITC data. Biochim. Biophys. Acta 2012, 1820, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Szabó, A.; Oláh, J.; Szunyogh, S.; Lehotzky, A.; Szénási, T.; Csaplár, M.; Schiedel, M.; Low, P.; Jung, M.; Ovádi, J. Modulation Of Microtubule Acetylation By The Interplay Of TPPP/p25, SIRT2 And New Anticancer Agents With Anti-SIRT2 Potency. Sci. Rep. 2017, 7, 17070. [Google Scholar] [CrossRef]
- Furumai, R.; Komatsu, Y.; Nishino, N.; Khochbin, S.; Yoshida, M.; Horinouchi, S. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc. Natl. Acad. Sci. USA 2001, 98, 87–92. [Google Scholar] [CrossRef]
- Dean, D.N.; Lee, J.C. Linking Parkinson’s Disease and Melanoma: Interplay Between alpha-Synuclein and Pmel17 Amyloid Formation. Mov. Disord. Off. J. Mov. Disord. Soc. 2021, 36, 1489–1498. [Google Scholar] [CrossRef]
- Lee, J.Y.S.; Ng, J.H.; Saffari, S.E.; Tan, E.K. Parkinson’s disease and cancer: A systematic review and meta-analysis on the influence of lifestyle habits, genetic variants, and gender. Aging 2022, 14, 2148–2173. [Google Scholar] [CrossRef]
- Szunyogh, S.; Oláh, J.; Szénási, T.; Szabó, A.; Ovádi, J. Targeting the interface of the pathological complex of alpha-synuclein and TPPP/p25. Biochim. Biophys. Acta 2015, 1852, 2653–2661. [Google Scholar] [CrossRef]
- Lehotzky, A.; Oláh, J.; Fekete, J.T.; Szénási, T.; Szabó, E.; Győrffy, B.; Várady, G.; Ovádi, J. Co-Transmission of Alpha-Synuclein and TPPP/p25 Inhibits Their Proteolytic Degradation in Human Cell Models. Front. Mol. Biosci. 2021, 8, 666026. [Google Scholar] [CrossRef]







| Antibody | Company | Catalog Number | Dilution |
|---|---|---|---|
| Rat polyclonal anti-TPPP1 | [4] | 1:5000 | |
| Mouse monoclonal anti-TPPP1 | [40] | 1:1000 | |
| Mouse monoclonal anti-TPPP3 | cf. 2.5 and 2.6 | 1:2000 | |
| Mouse monoclonal anti-alpha-tubulin, clone DM1A | Merck (Darmstadt, Germany) | T9026 | 1:5000 |
| Mouse monoclonal anti-acetyl-tubulin, clone 6-11B-1 | Merck (Darmstadt, Germany) | T6793 | 1:1000 cELISA; 1:5000 Western blot |
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific (Waltham, MA, USA) | A11122 | 1:2000 |
| Mouse monoclonal anti-SYN | Merck (Darmstadt, Germany) | S5566 | 1:5000 |
| Mouse monoclonal anti-beta-actin | Thermo Fisher Scientific (Waltham, MA, USA) | MA1-140 | 1:5000 |
| Anti-rat IgG, HRP-linked | Merck (Darmstadt, Germany) | A9037 | 1:5000 |
| Anti-mouse IgG, HRP-linked | Merck (Darmstadt, Germany) | A2554 | 1:2000 cELISA; 1:5000 other |
| Anti-rabbit IgG, HRP-linked | Thermo Fisher Scientific (Waltham, MA, USA) | 32260 | 1:2000 cELISA |
| Anti-mouse IgG, Alexa-546 linked | Thermo Fisher Scientific (Waltham, MA, USA) | A11003 | 1:1000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oláh, J.; Lehotzky, A.; Szénási, T.; Berki, T.; Ovádi, J. Modulatory Role of TPPP3 in Microtubule Organization and Its Impact on Alpha-Synuclein Pathology. Cells 2022, 11, 3025. https://doi.org/10.3390/cells11193025
Oláh J, Lehotzky A, Szénási T, Berki T, Ovádi J. Modulatory Role of TPPP3 in Microtubule Organization and Its Impact on Alpha-Synuclein Pathology. Cells. 2022; 11(19):3025. https://doi.org/10.3390/cells11193025
Chicago/Turabian StyleOláh, Judit, Attila Lehotzky, Tibor Szénási, Tímea Berki, and Judit Ovádi. 2022. "Modulatory Role of TPPP3 in Microtubule Organization and Its Impact on Alpha-Synuclein Pathology" Cells 11, no. 19: 3025. https://doi.org/10.3390/cells11193025






