Recent Progress and Future Prospect of CRISPR/Cas-Derived Transcription Activation (CRISPRa) System in Plants
Abstract
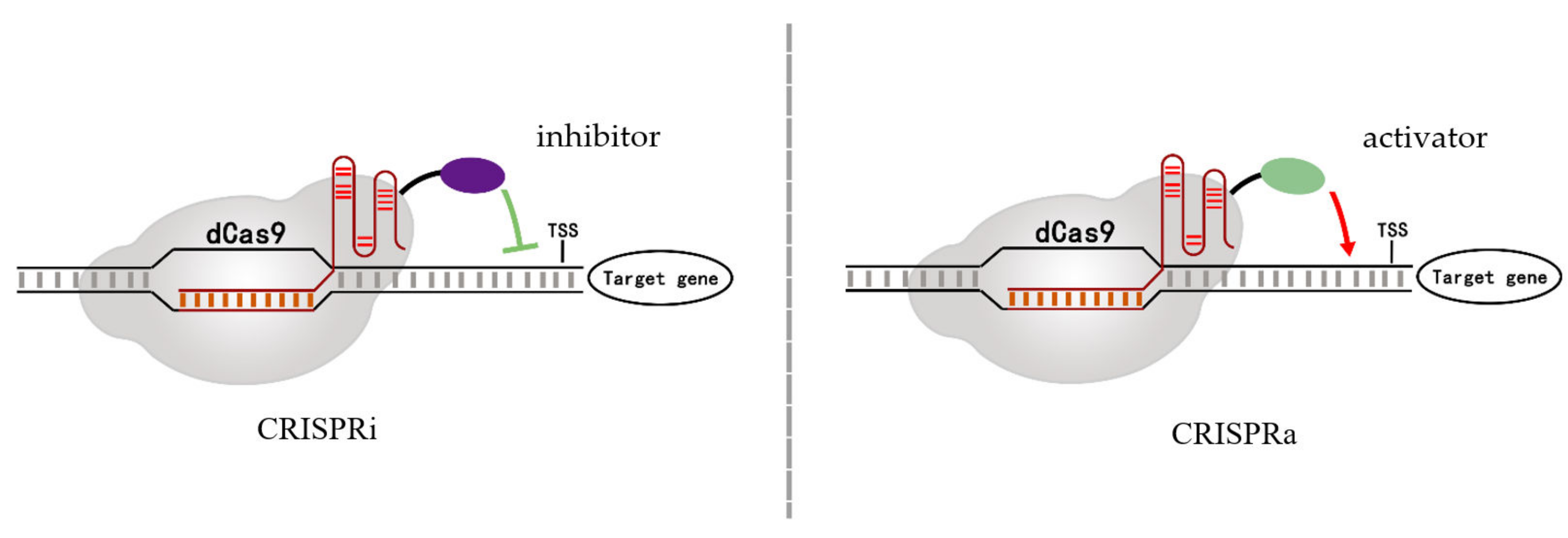
:1. Introduction
2. Composition of the CRISPR/Cas-Derived Activation System
2.1. CRISPR/Cas System
2.2. Cas9 and dCas9
2.3. Guide RNA (gRNA)
2.4. Transcriptional Regulators
3. Strategies for Achieving Transcriptional Activation using the CRISPR/dCas9 System
3.1. Fusion of Transcriptional Activation Effectors with dCas9
3.1.1. dCas9- SunTag System
3.1.2. dCas9-VPR System
3.1.3. dCas9-TV System
3.2. Modification of gRNA into a Scaffold and Recruitment of Transcriptional Activators
3.2.1. scRNA System
3.2.2. SAM System
3.2.3. CRISPR-Act 2.0 System
3.2.4. CRISPR-Act3.0 System
4. Application and Limitation of CRISPRa System in Plants
5. Concluding Remarks and Perspective of Transcription Activation System in Plants
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Spitz, F.; Furlong, E.E.M. Transcription Factors: From Enhancer Binding to Developmental Control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Alabed, D.; Whitteck, J.T.; Chen, W.; Bennett, S.; Asberry, A.; Wang, X.; Desloover, D.; Rangasamy, M.; Wright, T.R.; et al. A combinatorial bidirectional and bicistronic approach for coordinated multi-gene expression in corn. Plant Mol. Biol. 2015, 87, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Weeks, D.P.; Spalding, M.H.; Yang, B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 2016, 14, 483–495. [Google Scholar] [CrossRef]
- Wang, Q.; Alariqi, M.; Wang, F.; Li, B.; Ding, X.; Rui, H.; Li, Y.; Xu, Z.; Qin, L.; Sun, L.; et al. The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol. J. 2020, 18, 2436–2443. [Google Scholar] [CrossRef]
- Zhang, D.; Hussain, A.; Manghwar, H.; Xie, K.; Xie, S.; Zhao, S.; Larkin, R.M.; Qing, P.; Jin, S.; Ding, F. Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnol. J. 2020, 18, 1651–1669. [Google Scholar] [CrossRef]
- Wang, G.; Xu, Z.; Wang, F.; Huang, Y.; Xin, Y.; Liang, S.; Li, B.; Si, H.; Sun, L.; Wang, Q.; et al. Development of an efficient and precise adenine base editor (ABE) with expanded target range in allotetraploid cotton (Gossypium hirsutum). BMC Biol. 2022, 20, 45. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Lowder, L.; Malzahn, A.; Qi, Y. Rapid Evolution of Manifold CRISPR Systems for Plant Genome Editing. Front. Plant Sci. 2016, 7, 1683. [Google Scholar] [CrossRef] [PubMed]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/dCas9 platforms in plants: Strategies and applications beyond genome editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Zhang, F.; Koonin, E.V. SnapShot: Class 1 CRISPR-Cas Systems. Cell 2017, 168, 946.e1. [Google Scholar] [CrossRef]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef]
- Li, B.; Rui, H.; Li, Y.; Wang, Q.; Alariqi, M.; Qin, L.; Sun, L.; Ding, X.; Wang, F.; Zou, J.; et al. Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnol. J. 2019, 17, 1862–1864. [Google Scholar] [CrossRef]
- Cobb, R.E.; Wang, Y.; Zhao, H. High-Efficiency Multiplex Genome Editing of Streptomyces Species Using an Engineered CRISPR/Cas System. ACS Synth. Biol. 2015, 4, 723–728. [Google Scholar] [CrossRef] [Green Version]
- Apel, A.R.; D’Espaux, L.; Wehrs, M.; Sachs, D.; Li, R.A.; Tong, G.J.; Garber, M.; Nnadi, O.; Zhuang, W.; Hillson, N.J.; et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2017, 45, 496–508. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Yang, Y. RNA-Guided Genome Editing in Plants Using a CRISPR–Cas System. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Chylinski, K.; Le Rhun, A.; Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013, 10, 726–737. [Google Scholar] [CrossRef]
- Nishimasu, H.; Cong, L.; Yan, W.X.; Ran, F.A.; Zetsche, B.; Li, Y.; Kurabayashi, A.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal Structure of Staphylococcus aureus Cas9. Cell 2015, 162, 1113–1126. [Google Scholar] [CrossRef]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Zalatan, J.; Lee, M.E.; Almeida, R.; Gilbert, L.; Whitehead, E.H.; La Russa, M.; Tsai, J.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.-T.; Seo, S.-O.; Lynn, P.; Lu, T.; Jin, Y.-S.; Blaschek, H.P. Gene transcription repression in Clostridium beijerinckii using CRISPR-dCas9. Biotechnol. Bioeng. 2016, 113, 2739–2743. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. Correction: A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17103. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Vilar, M.; Bernabé-Orts, J.M.; Fernandez-Del-Carmen, A.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. A modular toolbox for gRNA–Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 2016, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Haslam, R.P.; Silvestre, S.; Lu, C.; Napier, J.A. Enhancing the accumulation of eicosapentaenoic acid and docosahexaenoic acid in transgenic Camelina through the CRISPR-Cas9 inactivation of the competing FAE1 pathway. Plant Biotechnol. J. 2022, 20, 1444–1446. [Google Scholar] [CrossRef] [PubMed]
- Gentzel, I.N.; Park, C.H.; Bellizzi, M.; Xiao, G.; Gadhave, K.R.; Murphree, C.; Yang, Q.; LaMantia, J.; Redinbaugh, M.G.; Balint-Kurti, P.; et al. A CRISPR/dCas9 toolkit for functional analysis of maize genes. Plant Methods 2020, 16, 133. [Google Scholar] [CrossRef]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2015, 13, 578–589. [Google Scholar] [CrossRef]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef]
- Didovyk, A.; Borek, B.; Tsimring, L.; Hasty, J. Transcriptional regulation with CRISPR-Cas9: Principles, advances, and applications. Curr. Opin. Biotechnol. 2016, 40, 177–184. [Google Scholar] [CrossRef]
- Li, Z.; Xiong, X.; Li, J.-F. The working dead: Repurposing inactive CRISPR-associated nucleases as programmable transcriptional regulators in plants. aBIOTECH 2020, 1, 32–40. [Google Scholar] [CrossRef]
- Collins, T.; Stone, J.R.; Williams, A.J. All in the Family: The BTB/POZ, KRAB, and SCAN Domains. Mol. Cell. Biol. 2001, 21, 3609–3615. [Google Scholar] [CrossRef] [Green Version]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pinera, P.; Ousterout, D.G.; Brunger, J.M.; Farin, A.M.; Glass, K.A.; Guilak, F.; Crawford, G.E.; Hartemink, A.J.; Gersbach, C.A. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods 2013, 10, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Belachew, A.; Ma, S.F.; Young, M.; Ade, J.; Shen, Y.; Marion, C.M.; Holtan, H.E.; Bailey, A.; Stone, J.K.; et al. The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 2012, 70, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Azzeme, A.M.; Abdullah, S.N.A.; Aziz, M.A.; Wahab, P.E.M. Oil palm drought inducible DREB1 induced expression of DRE/CRT- and non-DRE/CRT-containing genes in lowland transgenic tomato under cold and PEG treatments. Plant Physiol. Biochem. 2017, 112, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.N.A.; Ho, C.-L.; Carol, W. Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2017; pp. 71–99. [Google Scholar]
- Ebrahimi, M.; Abdullah, S.N.A.; Aziz, M.A.; Namasivayam, P. Oil palm EgCBF3 conferred stress tolerance in transgenic tomato plants through modulation of the ethylene signaling pathway. J. Plant Physiol. 2016, 202, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Çevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.; Holub, E.B.; Cahill, D.; Manners, J.M.; Schenk, P.; et al. MEDIATOR25 Acts as an Integrative Hub for the Regulation of Jasmonate-Responsive Gene Expression in Arabidopsis. Plant Physiol. 2012, 160, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Paul, J.W.; Qi, Y. Plant Gene Regulatory Networks; Springer: Berlin/Heidelberg, Germany, 2017; pp. 167–184. [Google Scholar]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.W.; Qi, Y. CRISPR/Cas9 for plant genome editing: Accomplishments, problems and prospects. Plant Cell Rep. 2016, 35, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Fusco, D.; Accornero, N.; Lavoie, B.; Shenoy, S.M.; Blanchard, J.-M.; Singer, R.H.; Bertrand, E. Single mRNA Molecules Demonstrate Probabilistic Movement in Living Mammalian Cells. Curr. Biol. 2003, 13, 161–167. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 2013, 159, 635–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, S.; Horii, T.; Kimura, M.; Hatada, I. Synergistic Upregulation of Target Genes by TET1 and VP64 in the dCas9–SunTag Platform. Int. J. Mol. Sci. 2020, 21, 1574. [Google Scholar] [CrossRef] [PubMed]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Van Essen, D.; Engist, B.; Natoli, G.; Saccani, S. Two Modes of Transcriptional Activation at Native Promoters by NF-κB p65. PLoS Biol. 2009, 7, e1000073. [Google Scholar] [CrossRef] [PubMed]
- Gwack, Y.; Baek, H.J.; Nakamura, H.; Lee, S.H.; Meisterernst, M.; Roeder, R.G.; Jung, J.U. Principal Role of TRAP/Mediator and SWI/SNF Complexes in Kaposi's Sarcoma-Associated Herpesvirus RTA-Mediated Lytic Reactivation. Mol. Cell. Biol. 2003, 23, 2055–2067. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.-F. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 2017, 3, 930–936. [Google Scholar] [CrossRef]
- Delebecque, C.J.; Lindner, A.B.; Silver, P.A.; Aldaye, F.A. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science 2011, 333, 470–474. [Google Scholar] [CrossRef]
- Spitale, R.C.; Tsai, M.-C.; Chang, H.Y. RNA templating the epigenome. Epigenetics 2011, 6, 539–543. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.; Habib, N.; Gootenberg, J.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef]
- Lemon, B.; Tjian, R. Orchestrated response: A symphony of transcription factors for gene control. Genes Dev. 2000, 14, 2551–2569. [Google Scholar] [CrossRef] [Green Version]
- Hunt, C.; Hartford, S.A.; White, D.; Pefanis, E.; Hanna, T.; Herman, C.; Wiley, J.; Brown, H.; Su, Q.; Xin, Y.; et al. Tissue-specific activation of gene expression by the Synergistic Activation Mediator (SAM) CRISPRa system in mice. Nat. Commun. 2021, 12, 2770. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.-F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust Transcriptional Activation in Plants Using Multiplexed CRISPR-Act2.0 and mTALE-Act Systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 2021, 7, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Li, H.; Liu, Y.; Li, S.; Liang, Z. Highly efficient activation of endogenous gene in grape using CRISPR/dCas9-based transcriptional activators. Hortic. Res. 2022, 9, uhab037. [Google Scholar] [CrossRef]
- Xiong, X.; Liang, J.; Li, Z.; Gong, B.; Li, J. Multiplex and optimization of dCas9-TV-mediated gene activation in plants. J. Integr. Plant Biol. 2021, 63, 634–645. [Google Scholar] [CrossRef]
- Lee, D.; Hua, L.; Khoshravesh, R.; Giuliani, R.; Kumar, I.; Cousins, A.; Sage, T.L.; Hibberd, J.M.; Brutnell, T.P. Engineering chloroplast development in rice through cell-specific control of endogenous genetic circuits. Plant Biotechnol. J. 2021, 19, 2291–2303. [Google Scholar] [CrossRef]
- Selma, S.; Sanmartín, N.; Espinosa-Ruiz, A.; Gianoglio, S.; Lopez-Gresa, M.P.; Vázquez-Vilar, M.; Flors, V.; Granell, A.; Orzaez, D. Custom-made design of metabolite composition in N. benthamiana leaves using CRISPR activators. Plant Biotechnol. J. 2022, 20, 1578–1590. [Google Scholar] [CrossRef]
- Park, J.; Dempewolf, E.; Zhang, W.; Wang, Z.-Y. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS ONE 2017, 12, e0179410. [Google Scholar] [CrossRef]
- Papikian, A.; Liu, W.; Gallego-Bartolomé, J.; Jacobsen, S.E. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019, 10, 729. [Google Scholar] [CrossRef]
- Zorrilla-López, U.; Masip, G.; Arjó, G.; Bai, C.; Banakar, R.; Bassie, L.; Berman, J.; Farré, G.; Miralpeix, B.; Pérez-Massot, E.; et al. Engineering metabolic pathways in plants by multigene transformation. Int. J. Dev. Biol. 2013, 57, 565–576. [Google Scholar] [CrossRef] [Green Version]
- Dzivenu, O.K.; Park, H.H.; Wu, H. General co-expression vectors for the overexpression of heterodimeric protein complexes in Escherichia coli. Protein Expr. Purif. 2004, 38, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wally, O.; Punja, Z.K. Genetic engineering for increasing fungal and bacterial disease resistance in crop plants. GM Crops 2010, 1, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Zare, K.; Negahdaripour, M.; Barekati-Mowahed, M.; Ghasemi, Y. CRISPR Cpf1 proteins: Structure, function and implications for genome editing. Cell Biosci. 2019, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Chiarella, A.M.; Butler, K.V.; Gryder, B.E.; Lu, D.; Wang, T.A.; Yu, X.; Pomella, S.; Khan, J.; Jin, J.; Hathaway, N.A. Dose-dependent activation of gene expression is achieved using CRISPR and small molecules that recruit endogenous chromatin machinery. Nat. Biotechnol. 2020, 38, 50–55. [Google Scholar] [CrossRef]
- Gamboa, L.; Phung, E.V.; Li, H.; Meyers, J.P.; Hart, A.C.; Miller, I.C.; Kwong, G.A. Heat-Triggered Remote Control of CRISPR-dCas9 for Tunable Transcriptional Modulation. ACS Chem. Biol. 2020, 15, 533–542. [Google Scholar] [CrossRef]
- Ochoa-Fernandez, R.; Abel, N.B.; Wieland, F.-G.; Schlegel, J.; Koch, L.-A.; Miller, J.B.; Engesser, R.; Giuriani, G.; Brandl, S.M.; Timmer, J.; et al. Optogenetic control of gene expression in plants in the presence of ambient white light. Nat. Methods 2020, 17, 717–725. [Google Scholar] [CrossRef]
- Li, J.; Manghwar, H.; Sun, L.; Wang, P.; Wang, G.; Sheng, H.; Zhang, J.; Liu, H.; Qin, L.; Rui, H.; et al. Whole genome sequencing reveals rare off-target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR /Cas9-edited cotton plants. Plant Biotechnol. J. 2017, 17, 858–868. [Google Scholar] [CrossRef]
- Mccarty, N.S.; Graham, A.E.; Studená, L.; Ledesma-Amaro, R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020, 11, 1281. [Google Scholar] [CrossRef]
- Zhang, S.; Voigt, C.A. Engineered dCas9 with reduced toxicity in bacteria: Implications for genetic circuit design. Nucleic Acids Res. 2018, 46, 11115–11125. [Google Scholar] [CrossRef]
- Mao, Y.; Yang, X.; Zhou, Y.; Zhang, Z.; Botella, J.R.; Zhu, J.-K. Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol. 2018, 19, 149. [Google Scholar] [CrossRef]
- Hu, D.; Yu, Y.; Wang, C.; Long, Y.; Liu, Y.; Feng, L.; Lu, D.; Liu, B.; Jia, J.; Xia, R.; et al. Multiplex CRISPR-Cas9 editing of DNA methyltransferases in rice uncovers a class of non-CG methylation specific for GC-rich regions. Plant Cell. 2021, 33, 2950. [Google Scholar] [CrossRef] [PubMed]
- DeNizio, J.E.; Schutsky, E.K.; Berrios, K.N.; Liu, M.Y.; Kohli, R.M. Harnessing natural DNA modifying activities for editing of the genome and epigenome. Curr. Opin. Chem. Biol. 2018, 45, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Sun, M.; Li, W.; Xu, M.; Shao, L.; Liu, Y.; Zhao, G.; Liu, Z.; Xu, Z.; You, J.; et al. Single-cell RNA -seq reveals fate determination control of an individual fiber cell initiation in cotton (Gossypium hirsutum). Plant Biotechnol. J. 2022. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, Q.; Zhu, X.; Wang, G.; Qin, Y.; Ding, F.; Tu, L.; Daniell, H.; Zhang, X.; Jin, S. Plant Single Cell Transcriptome Hub (PsctH): An integrated online tool to explore the plant single-cell transcriptome landscape. Plant Biotechnol. J. 2021, 20, 10–12. [Google Scholar] [CrossRef] [PubMed]


| Repressor | Plant Species | Target Gene | Highest Repression Level (%) | References |
|---|---|---|---|---|
| dCas9 | N. benthamiana | PDS | 20 | (Piatek et al., 2015) |
| pNOS::LUC reporter | 80 | (Vazquez-Vilar et al., 2016) | ||
| dCas9-SRDX | N. benthamiana | PDS | 33 | (Piatek et al., 2015) |
| pNOS::LUC reporter | 50 | (Vazquez-Vilar et al., 2016) | ||
| Z. mays | ChlH | 75 | (Irene et al., 2020) | |
| Z. mays | PDS | 60 | ||
| dCas9-BRD | N. benthamiana | pNOS::LUC reporter | 60 | (Vazquez-Vilar et al., 2016) |
| dCas9-3 × SRDX | A. thaliana | CSTF64 | 60 | (Lowder et al., 2015) |
| miR159a | 80 | |||
| miR159b | 70 | |||
| dLbCpf1-SRDX | A. thaliana | miR159b | 90 | (Tang et al., 2017) |
| dAsCpf1-SRDX | A. thaliana | miR159b | 90 |
| Activator | Plant Species | Target Gene | Fold Change (Highest Level) | References |
|---|---|---|---|---|
| dCas9-EDLL | N. benthamiana | NbPDS | 3.5 | (Piatek et al., 2015) |
| pNOS::LUC reporter | 2.2 | (Vazquez-vilar et al., 2016) | ||
| dCas9-TAL | N. benthamiana | AtPDS | 4 | (Piatek et al., 2015) |
| dCas9-VP64 | A.thaliana | AtPAP1 | 7 | (Lowder et al., 2015) |
| miR319 | 7.5 | |||
| AtFIS2 | 400 | |||
| N. benthamiana | pNOS::LUC reporter | 2.3 | (Vazquez-vilar et al., 2016) | |
| A.thaliana | AtWRKY30 | 2.1 | (Li et al., 2017) | |
| AtRLP23 | 0.9 | |||
| AtCDG1 | 4.3 | |||
| O. sativa | OsGW7 | 2.7 | ||
| OsER1 | 0.3 | |||
| Os03g01240 | 2.1 | (Lowder et al., 2018) | ||
| Os04g39780 | 1.1 | |||
| Os11g35410 | 2.2 | |||
| Z. mays | PDS | 2.5 | (Irene et al., 2020) | |
| TrxH | 2.0 | |||
| dCas9-VP64 + MS2-p65-HSF1 (SAM) | A.thaliana | AtAVP1 | 5 | (Park et al., 2017) |
| AtPAP1 | 7 | |||
| dCas9-4 × EE-2 × VP64 | A.thaliana | pWRKY30::LUC reporter | 12.6 | (Li et al., 2017) |
| dCas9-6 × TAL-2 × VP64 (dCas9-TV) | A.thaliana | AtWRKY30 | 138.8 | (Li et al., 2017) |
| AtRLP23 | 32.3 | |||
| AtCDG1 | 92.2 | |||
| Oryza sativa | OsGW7 | 78.8 | ||
| OsER1 | 62 | |||
| dCpf1-TV | A.thaliana | pWRKY30::LUC reporter | 4.7 | (Li et al., 2017) |
| dCas9-VP64-EDLL | A.thaliana | AtPAP1 | 4 | (Lowder et al., 2018) |
| AtFIS2 | 3 | |||
| O. sativa | OsCGA1 | 5 | (Lee et al., 2021) | |
| dCas9-VP64 + MS2-EDLL | A.thaliana | AtPAP1 | 30 | (Lowder et al., 2018) |
| AtFIS2 | 30 | |||
| dCas9-VP64 + MS2-VP64 (CRISPR-Act2.0) | A.thaliana | AtPAP1 | 45 | (Lowder et al., 2018) |
| AtFIS2 | 1500 | |||
| AtULC1 | 40 | |||
| miR319 | 6 | |||
| O. sativa | Os03g01240 | 3 | ||
| Os04g39780 | 4 | |||
| Os11g35410 | 2.8 | |||
| dCas9-2 × GCN4 + scFv-sfGFP-VP64 (SunTag) | A.thaliana | AtFWA | 140 | (Papikian et al., 2019) |
| AtEVD | 4000 | |||
| AtAP3 | 350 | |||
| AtCLV3 | 130 | |||
| dCasEV2.1 (EDLL-MS2:VPR/gRNA2.1) | N. benthamiana | NbAN2 | 4000 | (Selma et al., 2019) |
| NbDFR | 10000 | |||
| NbPAL | 400 | (Selma et al., 2022) | ||
| NbC4H | 4 | |||
| Nb4CL | 15 | |||
| NbCHS | 18000 | |||
| NbCHI | 45 | |||
| NbF3H | 140 | |||
| NbFLS | 40 | |||
| dCas9-TV | O. sativa | OsER1 OsGW7 | 4000 200 | (Xiong et al., 2021) |
| Gossypium hirsutum | Ghpapid | 41.7 | (unpublished data) | |
| Ghaepsp | 16 | |||
| CRISPR-Act3.0 gR2.0 4xGCN4 | O. sativa | OsGW7 | 45 | (Pan et al., 2021) |
| OsER1 | 90 | |||
| CRISPR-Act3.0 gR2.0 10xGCN4 | OsGW7 | 70 | ||
| OsER1 | 95 | |||
| CRISPR-Act3.0 gR8xMS2 4xGCN4 | OsGW7 | 45 | ||
| OsER1 | 40 | |||
| CRISPR-Act3.0 gR8xMS210xGCN4 | OsGW7 | 15 | ||
| OsER1 | 10 | |||
| CRISPR-Act3.0 VP64 4xGCN4 | OsER1 | 30 | ||
| CRISPR-Act3.0 VP64 10xGCN4 | 90 | |||
| CRISPR-Act3.0 2xTAD 4xGCN4 | 140 | |||
| CRISPR-Act3.0 2xTAD 10xGCN4 | 250 | |||
| CRISPR-Act3.0 2xTAD–VP64 4xGCN4 | 120 | |||
| CRISPR-Act3.0 2xTAD–VP64 10xGCN4 | 50 | |||
| CRISPR-Act3.0 TV 4xGCN4 | 35 | |||
| CRISPR-Act3.0 TV 10xGCN4 | 25 | |||
| CRISPR-Act3.0 VPR 4xGCN4 | 30 | |||
| CRISPR-Act3.0 VPR 10xGCN4 | 45 | |||
| M-Act3.0 (Multiple sgRNAs) | OsDXS | 9 | ||
| OsPDS | 6 | |||
| OsPSY | 17 | |||
| OsCRTISO | 3 | |||
| OsZISO | 23 | |||
| OsZDS | 3.5 | |||
| OsCYB | 11 | |||
| OsCHS | 30 | |||
| OsCHI | 2 | |||
| OsF3H | 130 | |||
| OsDFR | 20 | |||
| OsLAR | 70 | |||
| A.thaliana | AtFT | 240 | ||
| AtTCL1 | 8 | |||
| AtEVD | 4000 | |||
| AtAP3 | 350 | |||
| AtCLV3 | 130 | |||
| Lycopersicon esculentum | LeSFT | 240 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, X.; Yu, L.; Chen, L.; Li, Y.; Zhang, J.; Sheng, H.; Ren, Z.; Li, Y.; Yu, X.; Jin, S.; et al. Recent Progress and Future Prospect of CRISPR/Cas-Derived Transcription Activation (CRISPRa) System in Plants. Cells 2022, 11, 3045. https://doi.org/10.3390/cells11193045
Ding X, Yu L, Chen L, Li Y, Zhang J, Sheng H, Ren Z, Li Y, Yu X, Jin S, et al. Recent Progress and Future Prospect of CRISPR/Cas-Derived Transcription Activation (CRISPRa) System in Plants. Cells. 2022; 11(19):3045. https://doi.org/10.3390/cells11193045
Chicago/Turabian StyleDing, Xiao, Lu Yu, Luo Chen, Yujie Li, Jinlun Zhang, Hanyan Sheng, Zhengwei Ren, Yunlong Li, Xiaohan Yu, Shuangxia Jin, and et al. 2022. "Recent Progress and Future Prospect of CRISPR/Cas-Derived Transcription Activation (CRISPRa) System in Plants" Cells 11, no. 19: 3045. https://doi.org/10.3390/cells11193045
APA StyleDing, X., Yu, L., Chen, L., Li, Y., Zhang, J., Sheng, H., Ren, Z., Li, Y., Yu, X., Jin, S., & Cao, J. (2022). Recent Progress and Future Prospect of CRISPR/Cas-Derived Transcription Activation (CRISPRa) System in Plants. Cells, 11(19), 3045. https://doi.org/10.3390/cells11193045









