ALG3 Promotes Peritoneal Metastasis of Ovarian Cancer through Increasing Interaction of α1,3-mannosylated uPAR and ADAM8
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Data Collection
2.3. Kaplan-Meier Survival Analysis
2.4. Cell Culture
2.5. Cell Transfection
2.6. Western Blot and Lectin Blot
2.7. Lectin Chip
2.8. Immunoprecipitation
2.9. Immunohistochemistry/Lectin Histochemistry
2.10. Lectin ELISA
2.11. Hematoxylin Eosin (HE) Staining
2.12. Immunofluorescence and Confocal Microscopy
2.13. Transwell Assay
2.14. Scratch Assay
2.15. 5-Ethynyl-2′-deoxyuridine (EdU) Incorporation Assay
2.16. Sphere Formation Assay
2.17. Adhesion to Mesothelial Cells
2.18. Molecular Docking
2.19. Peritoneal Metastasis Mouse Model
2.20. Statistical Analyses
3. Results
3.1. Increased α1,3-mannosylation of Glycoproteins in Ovarian Cancer Tissues
3.2. Expression Level of ALG3 Is Upregulated in Ovarian Cancer Tissues
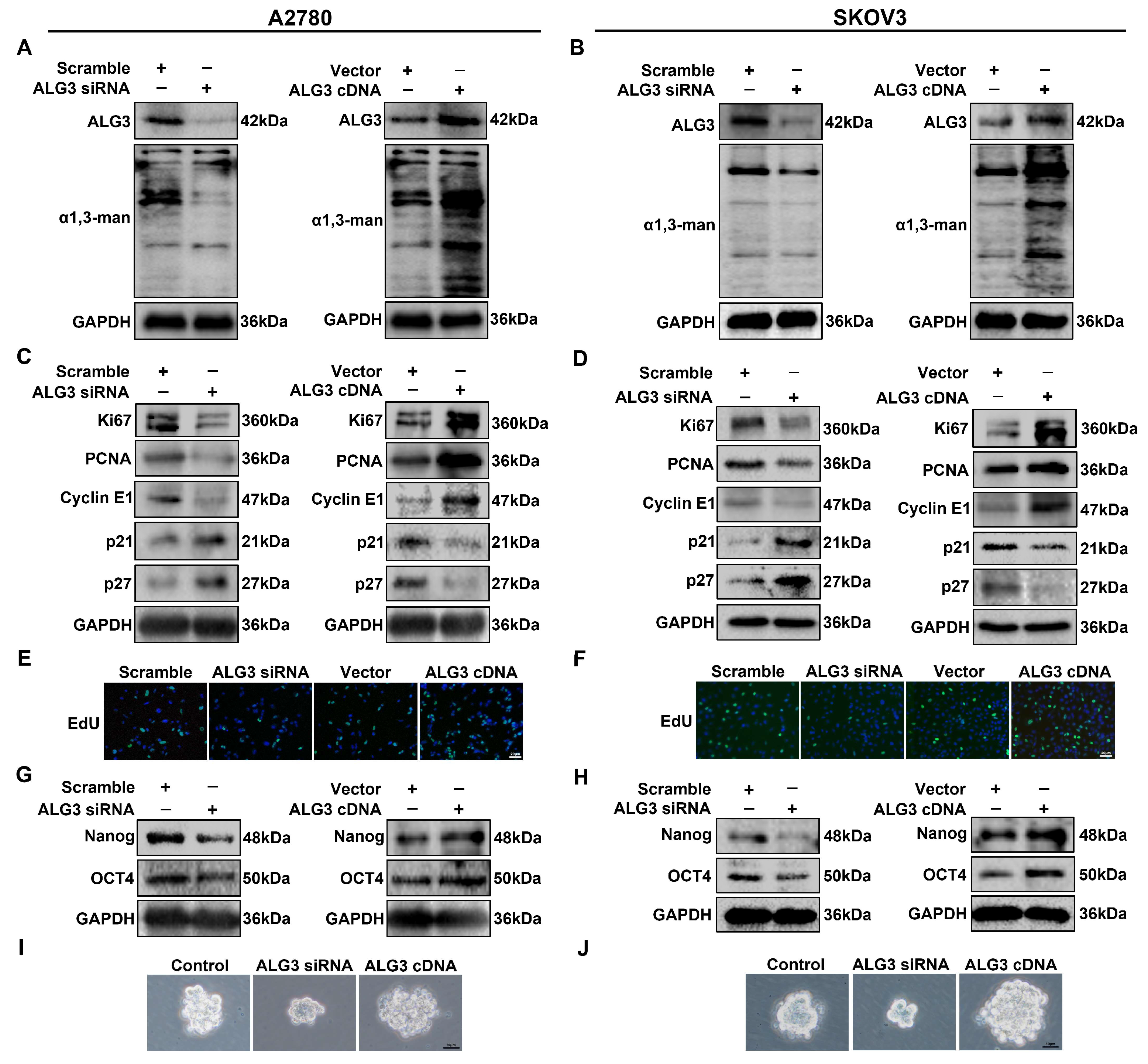
3.3. ALG3 Promotes Cancer Stemness and Proliferation of Ovarian Cancer Cells
3.4. ALG3 Enhances the Metastasis Capacity of Ovarian Cancer Cells
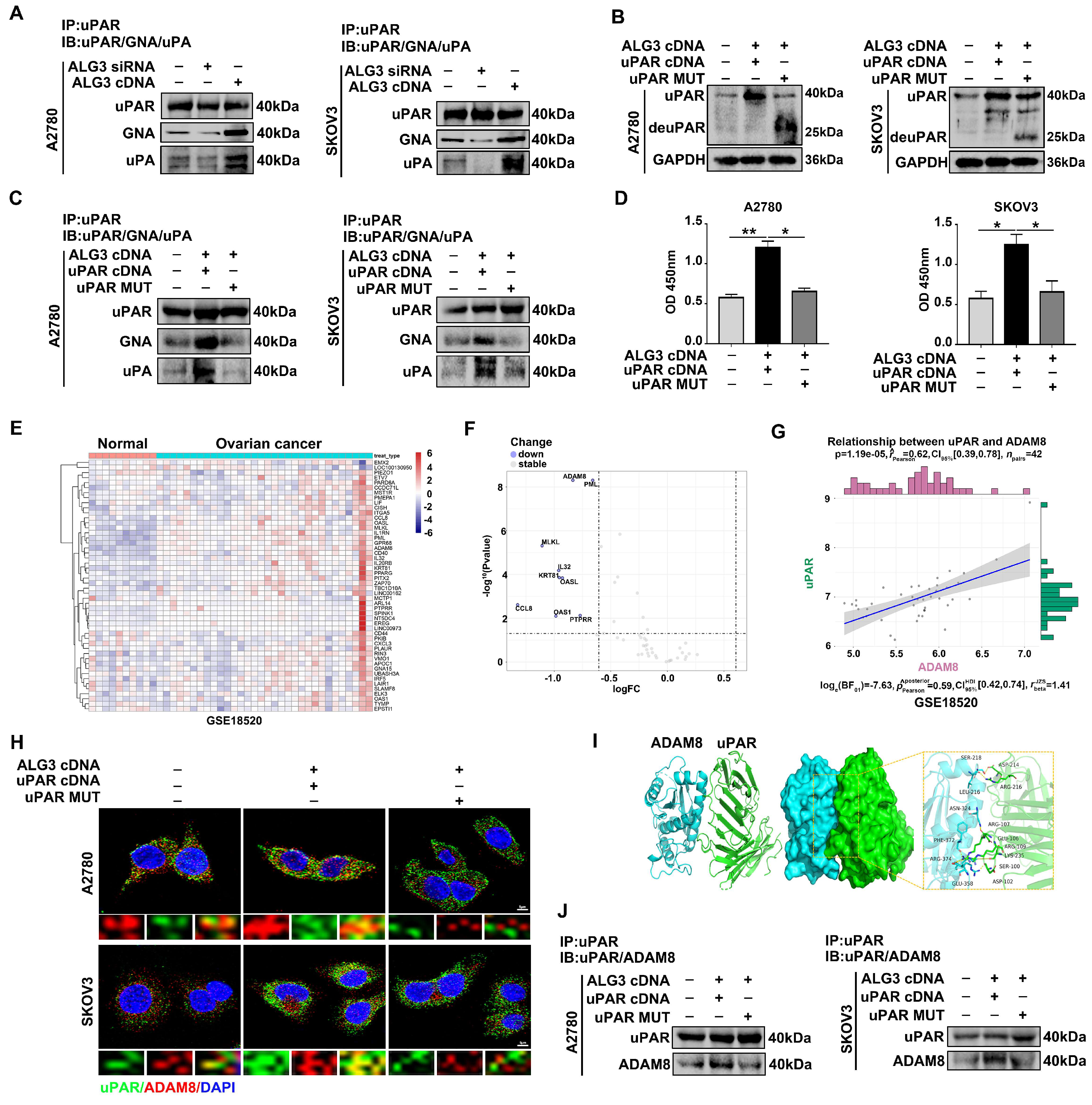
3.5. uPAR α1,3-mannosylation by ALG3 Increases Its Interaction with ADAM8
3.6. α1,3-mannosylated uPAR Binding to ADAM8 Stimulates Ovarian Metastasis by Activating Ras/ERK Signaling Pathway
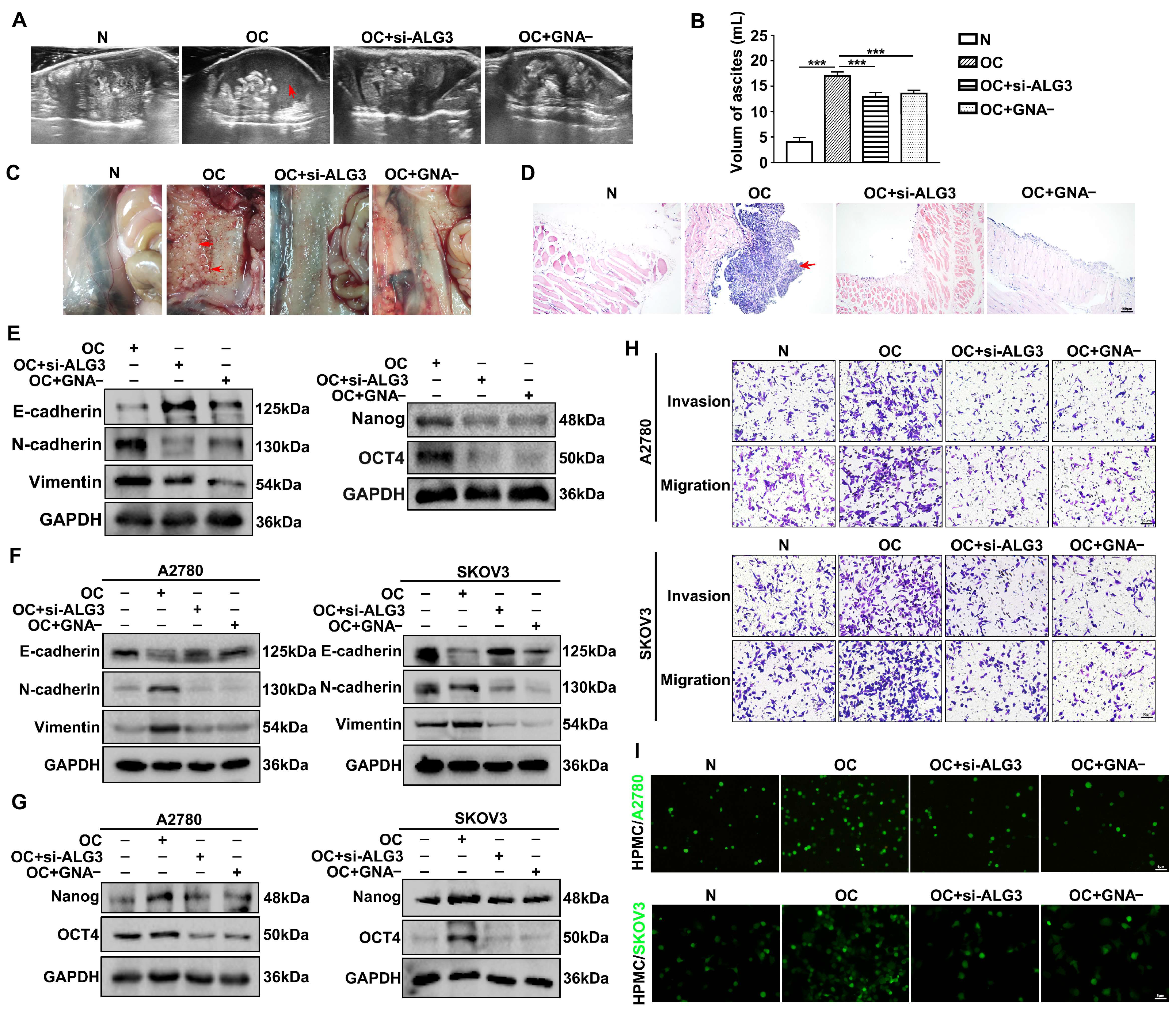
3.7. Decreased α1,3-mannosylation Inhibits Peritoneal Metastasis of Ovarian Cancer In Vivo
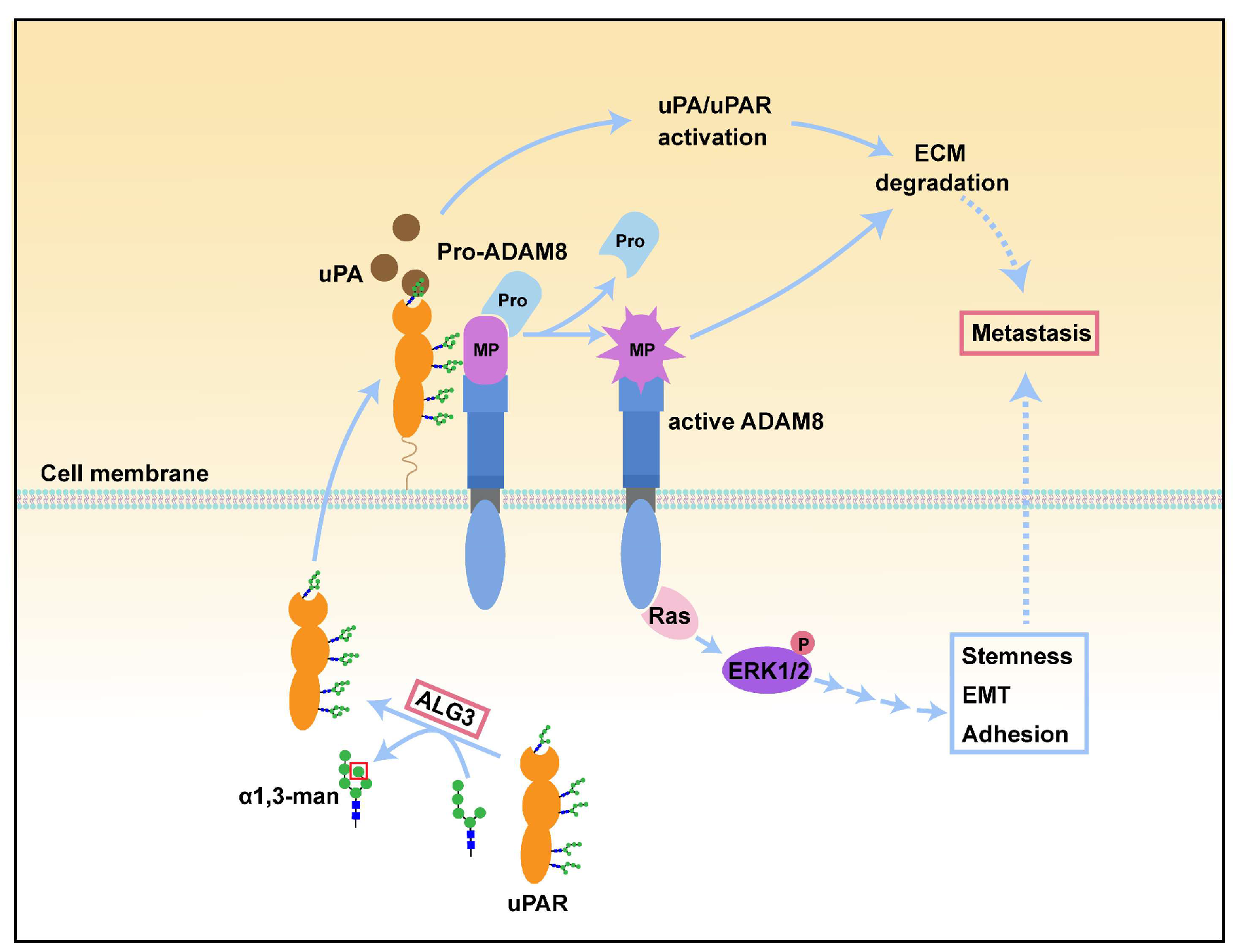
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Kuroki, L.; Guntupalli, S.R. Treatment of epithelial ovarian cancer. BMJ 2020, 371, m3773. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, B.; Song, Y.S. Ascites modulates cancer cell behavior, contributing to tumor heterogeneity in ovarian cancer. Cancer Sci. 2016, 107, 1173–1178. [Google Scholar] [CrossRef]
- Carey, P.; Low, E.; Harper, E.; Stack, M.S. Metalloproteinases in Ovarian Cancer. Int. J. Mol. Sci. 2021, 22, 3403. [Google Scholar] [CrossRef]
- Worzfeld, T.; Pogge von Strandmann, E.; Huber, M.; Adhikary, T.; Wagner, U.; Reinartz, S.; Müller, R. The Unique Molecular and Cellular Microenvironment of Ovarian Cancer. Front. Oncol. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Sawada, K.; Kinose, Y.; Yoshimura, A.; Toda, A.; Nakatsuka, E.; Hashimoto, K.; Mabuchi, S.; Morishige, K.I.; Kurachi, H.; et al. Exosomes Promote Ovarian Cancer Cell Invasion through Transfer of CD44 to Peritoneal Mesothelial Cells. Mol. Cancer Res. MCR 2017, 15, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Silsirivanit, A. Glycosylation markers in cancer. Adv. Clin. Chem. 2019, 89, 189–213. [Google Scholar] [CrossRef]
- Suzuki, N.; Abe, T.; Hanzawa, K.; Natsuka, S. Toward robust N-glycomics of various tissue samples that may contain glycans with unknown or unexpected structures. Sci. Rep. 2021, 11, 6334. [Google Scholar] [CrossRef]
- Planinc, A.; Dejaegher, B.; Vander Heyden, Y.; Viaene, J.; Van Praet, S.; Rappez, F.; Van Antwerpen, P.; Delporte, C. Batch-to-batch N-glycosylation study of infliximab, trastuzumab and bevacizumab, and stability study of bevacizumab. Eur. J. Hosp. Pharm. Sci. Pract. 2017, 24, 286–292. [Google Scholar] [CrossRef]
- Haeuptle, M.A.; Hennet, T. Congenital disorders of glycosylation: An update on defects affecting the biosynthesis of dolichol-linked oligosaccharides. Hum. Mutat. 2009, 30, 1628–1641. [Google Scholar] [CrossRef]
- Li, S.T.; Lu, T.T.; Xu, X.X.; Ding, Y.; Li, Z.; Kitajima, T.; Dean, N.; Wang, N.; Gao, X.D. Reconstitution of the lipid-linked oligosaccharide pathway for assembly of high-mannose N-glycans. Nat. Commun. 2019, 10, 1813. [Google Scholar] [CrossRef] [PubMed]
- Stanley, P.; Taniguchi, N.; Aebi, M. N-Glycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2015; pp. 99–111. [Google Scholar]
- Park, D.D.; Phoomak, C.; Xu, G.; Olney, L.P.; Tran, K.A.; Park, S.S.; Haigh, N.E.; Luxardi, G.; Lert-Itthiporn, W.; Shimoda, M.; et al. Metastasis of cholangiocarcinoma is promoted by extended high-mannose glycans. Proc. Natl. Acad. Sci. USA 2020, 117, 7633–7644. [Google Scholar] [CrossRef]
- Oliveira-Ferrer, L.; Legler, K.; Milde-Langosch, K. Role of protein glycosylation in cancer metastasis. Semin. Cancer Biol. 2017, 44, 141–152. [Google Scholar] [CrossRef]
- Biskup, K.; Braicu, E.I.; Sehouli, J.; Fotopoulou, C.; Tauber, R.; Berger, M.; Blanchard, V. Serum glycome profiling: A biomarker for diagnosis of ovarian cancer. J. Proteome Res. 2013, 12, 4056–4063. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Z.; Felder, M.A.; Yu, Q.; Shi, X.; Peng, Y.; Cao, Q.; Wang, B.; Puglielli, L.; Patankar, M.S.; et al. Mass Spectrometry Imaging of N-Glycans from Formalin-Fixed Paraffin-Embedded Tissue Sections Using a Novel Subatmospheric Pressure Ionization Source. Anal. Chem. 2019, 91, 12942–12947. [Google Scholar] [CrossRef]
- Choi, Y.W.; Bae, S.M.; Kim, Y.W.; Lee, H.N.; Kim, Y.W.; Park, T.C.; Ro, D.Y.; Shin, J.C.; Shin, S.J.; Seo, J.S.; et al. Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int. J. Gynecol. Cancer 2007, 17, 687–696. [Google Scholar] [CrossRef]
- Sun, X.; He, Z.; Guo, L.; Wang, C.; Lin, C.; Ye, L.; Wang, X.; Li, Y.; Yang, M.; Liu, S.; et al. ALG3 contributes to stemness and radioresistance through regulating glycosylation of TGF-β receptor II in breast cancer. J. Exp. Clin. Cancer Res. 2021, 40, 149. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef]
- Alfano, D.; Franco, P.; Stoppelli, M.P. Modulation of Cellular Function by the Urokinase Receptor Signalling: A Mechanistic View. Front. Cell Dev. Biol. 2022, 10, 818616. [Google Scholar] [CrossRef]
- Metrangolo, V.; Ploug, M.; Engelholm, L.H. The Urokinase Receptor (uPAR) as a "Trojan Horse" in Targeted Cancer Therapy: Challenges and Opportunities. Cancers 2021, 13, 5376. [Google Scholar] [CrossRef]
- Wang, L.; Madigan, M.C.; Chen, H.; Liu, F.; Patterson, K.I.; Beretov, J.; O’Brien, P.M.; Li, Y. Expression of urokinase plasminogen activator and its receptor in advanced epithelial ovarian cancer patients. Gynecol. Oncol. 2009, 114, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Borgfeldt, C.; Hansson, S.R.; Gustavsson, B.; Måsbäck, A.; Casslén, B. Dedifferentiation of serous ovarian cancer from cystic to solid tumors is associated with increased expression of mRNA for urokinase plasminogen activator (uPA), its receptor (uPAR) and its inhibitor (PAI-1). Int. J. Cancer 2001, 92, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Dass, K.; Ahmad, A.; Azmi, A.S.; Sarkar, S.H.; Sarkar, F.H. Evolving role of uPA/uPAR system in human cancers. Cancer Treat. Rev. 2008, 34, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Al-Hassan, N.N.; Behzadian, A.; Caldwell, R.; Ivanova, V.S.; Syed, V.; Motamed, K.; Said, N.A. Differential roles of uPAR in peritoneal ovarian carcinomatosis. Neoplasia 2012, 14, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Llinas, P.; Le Du, M.H.; Gårdsvoll, H.; Danø, K.; Ploug, M.; Gilquin, B.; Stura, E.A.; Ménez, A. Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide. EMBO J. 2005, 24, 1655–1663. [Google Scholar] [CrossRef]
- Srinivasan, S.; Romagnoli, M.; Bohm, A.; Sonenshein, G.E. N-glycosylation regulates ADAM8 processing and activation. J. Biol. Chem. 2014, 289, 33676–33688. [Google Scholar] [CrossRef]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef]
- Barkeer, S.; Chugh, S.; Batra, S.K.; Ponnusamy, M.P. Glycosylation of Cancer Stem Cells: Function in Stemness, Tumorigenesis, and Metastasis. Neoplasia 2018, 20, 813–825. [Google Scholar] [CrossRef]
- Magalhães, A.; Duarte, H.O.; Reis, C.A. Aberrant Glycosylation in Cancer: A Novel Molecular Mechanism Controlling Metastasis. Cancer Cell 2017, 31, 733–735. [Google Scholar] [CrossRef]
- Li, Q.; Li, G.; Zhou, Y.; Zhang, X.; Sun, M.; Jiang, H.; Yu, G. Comprehensive N-Glycome Profiling of Cells and Tissues for Breast Cancer Diagnosis. J. Proteome Res. 2019, 18, 2559–2570. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Qian, Y.; Wu, X.; Zhang, Z.; Liu, X.; Zhao, R.; Zhou, L.; Ruan, Y.; Xu, J.; et al. Discovery of specific metastasis-related N-glycan alterations in epithelial ovarian cancer based on quantitative glycomics. PLoS ONE 2014, 9, e87978. [Google Scholar] [CrossRef]
- Anugraham, M.; Jacob, F.; Nixdorf, S.; Everest-Dass, A.V.; Heinzelmann-Schwarz, V.; Packer, N.H. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: Glycan structures reflect gene expression and DNA methylation status. Mol. Cell. Proteom. MCP 2014, 13, 2213–2232. [Google Scholar] [CrossRef]
- Carvalho, S.; Oliveira, T.; Bartels, M.F.; Miyoshi, E.; Pierce, M.; Taniguchi, N.; Carneiro, F.; Seruca, R.; Reis, C.A.; Strahl, S.; et al. O-mannosylation and N-glycosylation: Two coordinated mechanisms regulating the tumour suppressor functions of E-cadherin in cancer. Oncotarget 2016, 7, 65231–65246. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, J.; Ruan, Y.; Sun, L.; Xu, C.; Jiang, H. Sialyltransferase ST3GAL1 promotes cell migration, invasion, and TGF-β1-induced EMT and confers paclitaxel resistance in ovarian cancer. Cell Death Dis. 2018, 9, 1102. [Google Scholar] [CrossRef]
- Escrevente, C.; Machado, E.; Brito, C.; Reis, C.A.; Stoeck, A.; Runz, S.; Marmé, A.; Altevogt, P.; Costa, J. Different expression levels of alpha3/4 fucosyltransferases and Lewis determinants in ovarian carcinoma tissues and cell lines. Int. J. Oncol. 2006, 29, 557–566. [Google Scholar]
- Shao, P.; Wei, C.; Wang, Y. ALG3 contributes to the malignant properties of OSCC cells by regulating CDK-Cyclin pathway. Oral Dis. 2021, 27, 1426–1434. [Google Scholar] [CrossRef]
- Ke, S.B.; Qiu, H.; Chen, J.M.; Shi, W.; Han, C.; Gong, Y.; Chen, Y.S. ALG3 contributes to the malignancy of non-small cell lung cancer and is negatively regulated by MiR-98-5p. Pathol. Res. Pract. 2020, 216, 152761. [Google Scholar] [CrossRef]
- Magnussen, S.N.; Hadler-Olsen, E.; Costea, D.E.; Berg, E.; Jacobsen, C.C.; Mortensen, B.; Salo, T.; Martinez-Zubiaurre, I.; Winberg, J.O.; Uhlin-Hansen, L.; et al. Cleavage of the urokinase receptor (uPAR) on oral cancer cells: Regulation by transforming growth factor—β1 (TGF-β1) and potential effects on migration and invasion. BMC Cancer 2017, 17, 350. [Google Scholar] [CrossRef] [PubMed]
- Alfano, M.; Mariani, S.A.; Elia, C.; Pardi, R.; Blasi, F.; Poli, G. Ligand-engaged urokinase-type plasminogen activator receptor and activation of the CD11b/CD18 integrin inhibit late events of HIV expression in monocytic cells. Blood 2009, 113, 1699–1709. [Google Scholar] [CrossRef] [PubMed]
- Yepes, M. Urokinase-type plasminogen activator is a modulator of synaptic plasticity in the central nervous system: Implications for neurorepair in the ischemic brain. Neural Regen. Res. 2020, 15, 620–624. [Google Scholar] [CrossRef]
- Jo, M.; Thomas, K.S.; O’Donnell, D.M.; Gonias, S.L. Epidermal growth factor receptor-dependent and -independent cell-signaling pathways originating from the urokinase receptor. J. Biol. Chem. 2003, 278, 1642–1646. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.; Benzel, J.; Dorzweiler, K.; Cook, L.; Schlomann, U.; Zarbock, A.; Slater, E.P.; Nimsky, C.; Bartsch, J.W. ADAM8 in invasive cancers: Links to tumor progression, metastasis, and chemoresistance. Clin. Sci. 2019, 133, 83–99. [Google Scholar] [CrossRef]
- Das, S.G.; Romagnoli, M.; Mineva, N.D.; Barillé-Nion, S.; Jézéquel, P.; Campone, M.; Sonenshein, G.E. miR-720 is a downstream target of an ADAM8-induced ERK signaling cascade that promotes the migratory and invasive phenotype of triple-negative breast cancer cells. Breast Cancer Res. BCR 2016, 18, 40. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, X.; Pei, X.; Wang, H.; Feng, P.; Qin, H.; Liu, S.; Yan, Q.; Liu, J. ALG3 Promotes Peritoneal Metastasis of Ovarian Cancer through Increasing Interaction of α1,3-mannosylated uPAR and ADAM8. Cells 2022, 11, 3141. https://doi.org/10.3390/cells11193141
Cui X, Pei X, Wang H, Feng P, Qin H, Liu S, Yan Q, Liu J. ALG3 Promotes Peritoneal Metastasis of Ovarian Cancer through Increasing Interaction of α1,3-mannosylated uPAR and ADAM8. Cells. 2022; 11(19):3141. https://doi.org/10.3390/cells11193141
Chicago/Turabian StyleCui, Xinyuan, Xiaosong Pei, Hao Wang, Ping Feng, Huamin Qin, Shuai Liu, Qiu Yan, and Jiwei Liu. 2022. "ALG3 Promotes Peritoneal Metastasis of Ovarian Cancer through Increasing Interaction of α1,3-mannosylated uPAR and ADAM8" Cells 11, no. 19: 3141. https://doi.org/10.3390/cells11193141
APA StyleCui, X., Pei, X., Wang, H., Feng, P., Qin, H., Liu, S., Yan, Q., & Liu, J. (2022). ALG3 Promotes Peritoneal Metastasis of Ovarian Cancer through Increasing Interaction of α1,3-mannosylated uPAR and ADAM8. Cells, 11(19), 3141. https://doi.org/10.3390/cells11193141







