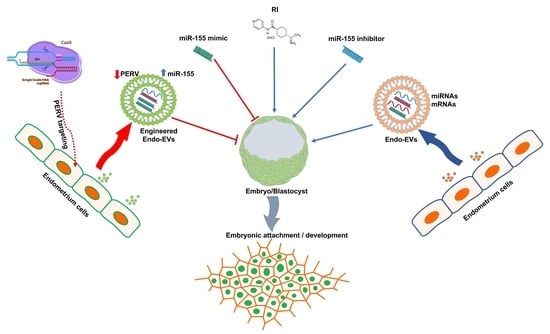
ROCK Inhibitor (Y-27632) Abolishes the Negative Impacts of miR-155 in the Endometrium-Derived Extracellular Vesicles and Supports Embryo Attachment
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Generation of Porcine Parthenogenetic Embryos
2.3. Endometrium Culture
2.4. Extracellular Vesicles Isolation and Characterization
2.5. Embryo Attachment Model
2.6. Experimental Design
2.6.1. Effects of Endometrial-EVs and ROCK Inhibitor on Embryo Development and Attachment
2.6.2. Effect of miR-155 on Embryo Development and Attachment
2.6.3. Effect of Targeting PERV on Embryo Development and Attachment
2.7. EVs Labeling and Uptake
2.8. Immunofluorescence
2.9. TdT-Mediated dUTP-X Nick End Labeling (TUNEL) Assay
2.10. MiR-155 Mimic, miR-155 Inhibitor, and CRISPR/Cas9 Transfection
2.11. Conventional and Real-Time Polymerase Chain Reaction
2.12. Preparation of Endo-EVs Protein Fraction, In-Gel Digestion and Proteomic Analysis by LC–MS/MS
2.13. Statistical Analysis
3. Results
3.1. Endo-EVs Isolation, Characteristics, and Cargo Contents
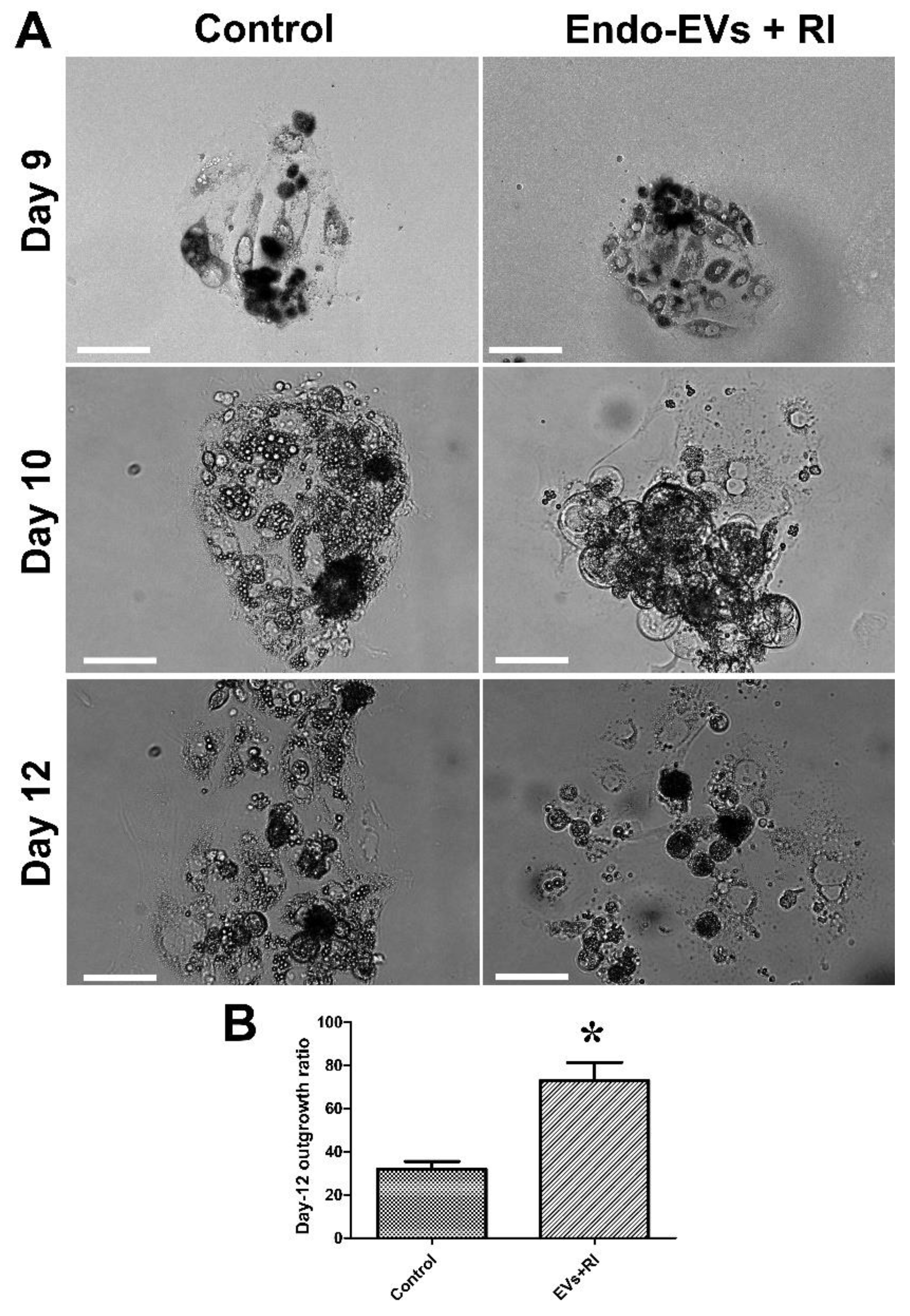
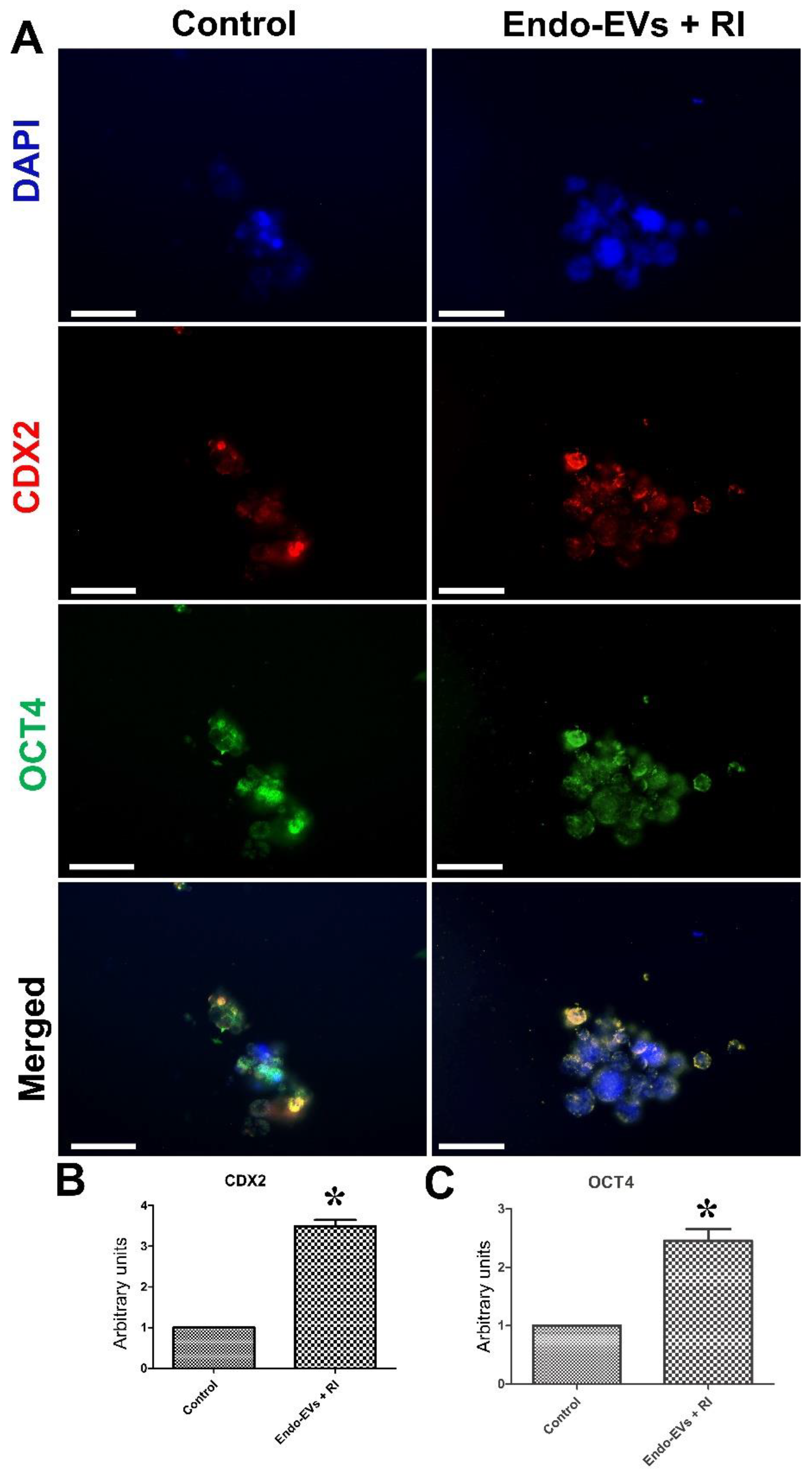
3.2. Effect of Endo-EVs and ROCK-Inhibitor (RI) on Embryo Attachment and Outgrowths
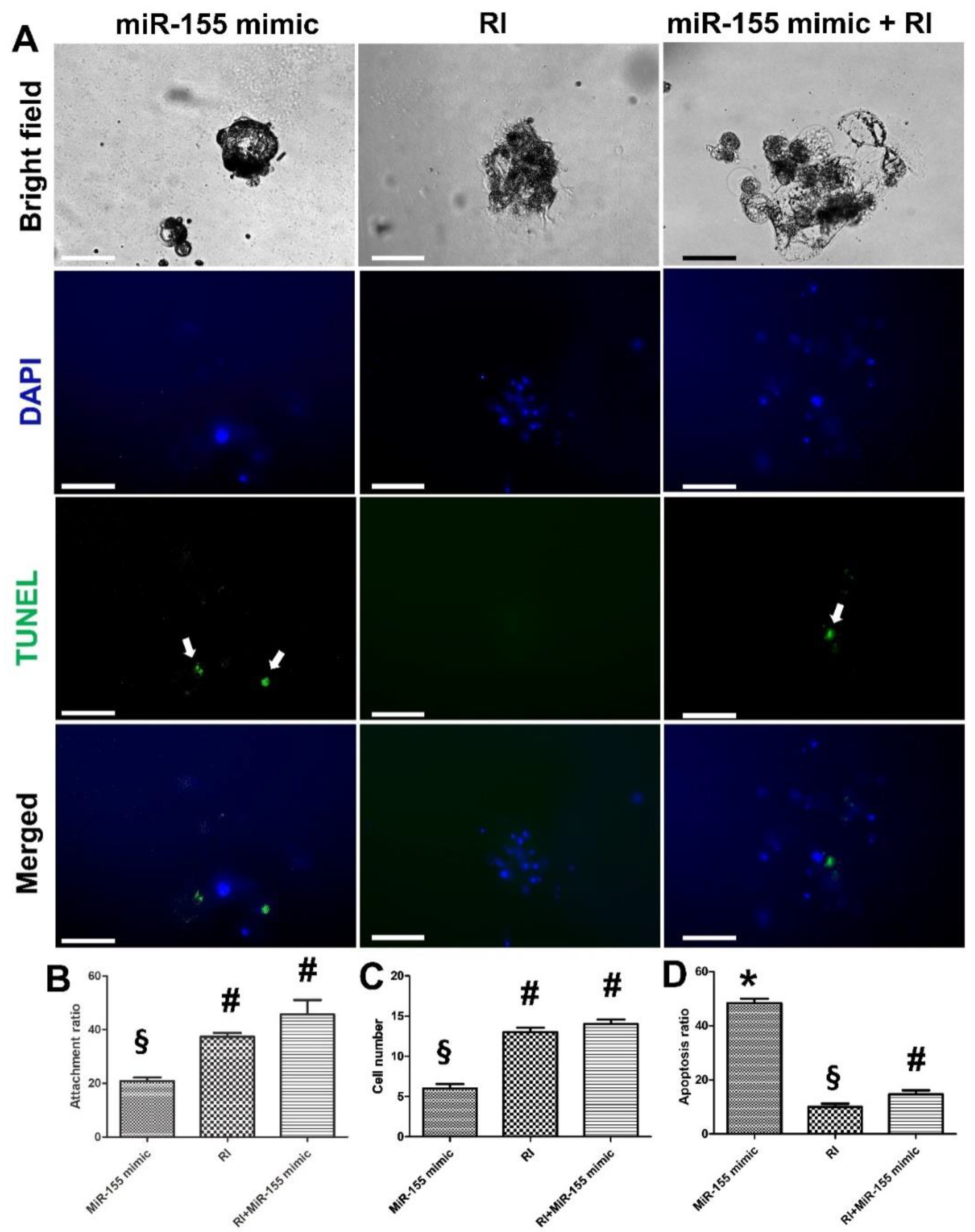
3.3. Impact of miR-155 on Embryo Attachment and Development
3.4. Effects of Endo-EVs PERV Depletion by CRISPR/Cas9 on Embryo Attachment and Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, N.; Du, X.; Wu, S. Advances in pig models of human diseases. Anim. Model. Exp. Med. 2022, 5, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Eisenson, D.L.; Hisadome, Y.; Yamada, K. Progress in Xenotransplantation: Immunologic Barriers, Advances in Gene Editing, and Successful Tolerance Induction Strategies in Pig-To-Primate Transplantation. Front. Immunol. 2022, 13, 2308. [Google Scholar] [CrossRef] [PubMed]
- Stokes, P.J.; Abeydeera, L.R.; Leese, H.J. Development of porcine embryos in vivo and in vitro; evidence for embryo ‘cross talk’ in vitro. Dev. Biol. 2005, 284, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.M.; Kim, S.J.; Bin Choi, Y.; Lee, B.C. Improvement of Cloned Embryos Development by Co-Culturing with Parthenotes: A Possible Role of Exosomes/Microvesicles for Embryos Paracrine Communication. Cell Reprogram. 2014, 16, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Tanga, B.M.; Bang, S.; Seong, G.; Saadeldin, I.M.; Lee, S.; Cho, J. Oviduct epithelial cells-derived extracellular vesicles improve preimplantation developmental competence of in vitro produced porcine parthenogenetic and cloned embryos. Mol. Reprod. Dev. 2021, 89, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Kashiwazaki, N.; Kikuchi, K.; Suzuki, K.; Noguchi, J.; Nagai, T.; Kaneko, H.; Shino, M. Development In Vivo and In Vitro to Blastocysts of Porcine Oocytes Matured and Fertilized In Vitro. J. Reprod. Dev. 2001, 47, 303–310. [Google Scholar] [CrossRef]
- van der Weijden, V.A.; Schmidhauser, M.; Kurome, M.; Knubben, J.; Flöter, V.L.; Wolf, E.; Ulbrich, S.E. Transcriptome dynamics in early in vivo developing and in vitro produced porcine embryos. BMC. Genom. 2021, 22, 1–13. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Tanga, B.M.; Bang, S.; Fang, X.; Yoon, K.-Y.; Lee, S.; Cho, S.L.A.J. The theranostic roles of extracellular vesicles in pregnancy disorders. J. Anim. Reprod. Biotechnol. 2022, 37, 2–12. [Google Scholar] [CrossRef]
- Saadeldin, I.; Oh, H.J.; Lee, B. Embryonic–maternal cross-talk via exosomes: Potential implications. Stem. Cells Clon. Adv. Appl. 2015, 8, 103–107. [Google Scholar] [CrossRef]
- Godakumara, K.; Ord, J.; Lättekivi, F.; Dissanayake, K.; Viil, J.; Boggavarapu, N.R.; Faridani, O.R.; Jääger, K.; Velthut-Meikas, A.; Jaakma, Ü.; et al. Trophoblast derived extracellular vesicles specifically alter the transcriptome of endometrial cells and may constitute a critical component of embryo-maternal communication. Reprod. Biol. Endocrinol. 2021, 19, 1–14. [Google Scholar] [CrossRef]
- Greening, D.W.; Nguyen, H.P.; Elgass, K.; Simpson, R.J.; Salamonsen, L.A. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions. Biol. Reprod. 2016, 94, 38. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Tan, Q.; Liang, J.; Cao, D.; Wang, S.; Liang, J.; Chen, K.; Wang, Z. Placental trophoblast cell-derived exosomal microRNA-1290 promotes the interaction between endometrium and embryo by targeting LHX6. Mol. Ther.-Nucleic. Acids 2021, 26, 760–772. [Google Scholar] [CrossRef]
- Ruiz-González, I.; Xu, J.; Wang, X.; Burghardt, R.; Dunlap, K.A.; Bazer, F.W. Exosomes, endogenous retroviruses and toll-like receptors: Pregnancy recognition in ewes. Reproduction 2015, 149, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Hirata, M.; Wittayarat, M.; Hirano, T.; Nguyen, N.T.; Le, Q.A.; Namula, Z.; Fahrudin, M.; Tanihara, F.; Otoi, T. The Relationship between Embryonic Development and the Efficiency of Target Mutations in Porcine Endogenous Retroviruses (PERVs) Pol Genes in Porcine Embryos. Animals 2019, 9, 593. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.E.; Johnson, G.A.; Bazer, F.W.; Burghardt, R.C.; Palmarini, M. Pregnancy recognition and conceptus implantation in domestic ruminants: Roles of progesterone, interferons and endogenous retroviruses. Reprod. Fertil. Dev. 2007, 19, 65–78. [Google Scholar] [CrossRef]
- Baek, S.-K.; Cho, Y.-S.; Kim, I.-S.; Jeon, S.-B.; Moon, D.-K.; Hwangbo, C.; Choi, J.-W.; Kim, T.-S.; Lee, J.-H. A Rho-Associated Coiled-Coil Containing Kinase Inhibitor, Y-27632, Improves Viability of Dissociated Single Cells, Efficiency of Colony Formation, and Cryopreservation in Porcine Pluripotent Stem Cells. Cell Reprogram. 2019, 21, 37–50. [Google Scholar] [CrossRef]
- Kurosawa, H. Application of Rho-associated protein kinase (ROCK) inhibitor to human pluripotent stem cells. J. Biosci. Bioeng. 2012, 114, 577–581. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Tukur, H.A.; Aljumaah, R.S.; Sindi, R.A. Rocking the Boat: The Decisive Roles of Rho Kinases During Oocyte, Blastocyst, and Stem Cell Development. Front. Cell Dev. Biol. 2021, 8, 616762. [Google Scholar] [CrossRef]
- Motomura, K.; Okada, N.; Morita, H.; Hara, M.; Tamari, M.; Orimo, K.; Matsuda, G.; Imadome, K.-I.; Matsuda, A.; Nagamatsu, T.; et al. A Rho-associated coiled-coil containing kinases (ROCK) inhibitor, Y-27632, enhances adhesion, viability and differentiation of human term placenta-derived trophoblasts in vitro. PLoS ONE 2017, 12, e0177994. [Google Scholar] [CrossRef]
- Hou, D.; Su, M.; Li, X.; Li, Z.; Yun, T.; Zhao, Y.; Zhang, M.; Zhao, L.; Li, R.; Yu, H.; et al. The Efficient Derivation of Trophoblast Cells from Porcine In Vitro Fertilized and Parthenogenetic Blastocysts and Culture with ROCK Inhibitor Y-27632. PLoS ONE 2015, 10, e0142442. [Google Scholar] [CrossRef]
- Tanga, B.M.; Fang, X.; Bang, S.; Seong, G.; De Zoysa, M.; Saadeldin, I.M.; Lee, S.; Cho, J. MiRNA-155 inhibition enhances porcine embryo preimplantation developmental competence by upregulating ZEB2 and downregulating ATF4. Theriogenology 2022, 183, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.; Kim, S.; Choi, Y.; Lee, B. Post-maturation zona perforation improves porcine parthenogenetic trophoblast culture. Placenta 2014, 35, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Paria, B.C.; Davis, D.L. Pig endometrial cells in primary culture: Morphology, secretion of prostaglandins and proteins, and effects of pregnancy. J. Anim. Sci. 1991, 69, 3005–3015. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial Exosomes/Microvesicles in the Uterine Microenvironment: A New Paradigm for Embryo-Endometrial Cross Talk at Implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef] [PubMed]
- Abumaghaid, M.M.; Abdelazim, A.M.; Belali, T.M.; Alhujaily, M.; Saadeldin, I.M. Shuttle transfer of mRNA transcripts via extracellular vesicles from male reproductive tract cells to the cumulus–oocyte complex in rabbits (Oryctolagus cuniculus). Front. Veter. Sci. 2022. [Google Scholar] [CrossRef]
- Mehdiani, A.; Maier, A.; Pinto, A.; Barth, M.; Akhyari, P.; Lichtenberg, A. An Innovative Method for Exosome Quantification and Size Measurement. J. Vis. Exp. 2015, 95, e50974. [Google Scholar] [CrossRef]
- Lee, H.; Yun, S.H.; Hyon, J.-Y.; Lee, S.-Y.; Yi, Y.-S.; Choi, C.-W.; Jun, S.; Park, E.C.; Kim, S.I. Streptococcus equi-derived Extracellular Vesicles as a Vaccine Candidate against Streptococcus equi Infection. Veter. Microbiol. 2021, 259, 109165. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Kim, S.J.; Lee, B.C. Blastomeres aggregation as an efficient alternative for trophoblast culture from porcine parthenogenetic embryos. Dev. Growth Differ. 2015, 57, 362–368. [Google Scholar] [CrossRef]
- Tukur, H.A.; Aljumaah, R.S.; Swelum, A.A.-A.; Alowaimer, A.N.; Abdelrahman, M.; Saadeldin, I.M. Effects of Short-Term Inhibition of Rho Kinase on Dromedary Camel Oocyte In Vitro Maturation. Animals 2020, 10, 750. [Google Scholar] [CrossRef]
- Ferraz, M.; Fujihara, M.; Nagashima, J.B.; Noonan, M.J.; Inoue-Murayama, M.; Songsasen, N. Follicular extracellular vesicles enhance meiotic resumption of domestic cat vitrified oocytes. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Simonsen, J.B. Pitfalls associated with lipophilic fluorophore staining of extracellular vesicles for uptake studies. J. Extracell. Vesicles 2019, 8, 1582237. [Google Scholar] [CrossRef] [PubMed]
- Takov, K.; Yellon, D.; Davidson, S.M. Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. J. Extracell. Vesicles 2017, 6, 1388731. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.; Koo, O.J.; Kang, J.-T.; Kwon, D.K.; Park, S.J.; Kim, S.J.; Moon, J.H.; Oh, H.J.; Jang, G.; Lee, B.C. Paradoxical effects of kisspeptin: It enhances oocyte in vitro maturation but has an adverse impact on hatched blastocysts during in vitro culture. Reprod. Fertil. Dev. 2012, 24, 656–668. [Google Scholar] [CrossRef]
- Zheng, Z.; Sun, R.; Zhao, H.-J.; Fu, D.; Zhong, H.-J.; Weng, X.-Q.; Qu, B.; Zhao, Y.; Wang, L.; Zhao, W.-L. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol. Cancer 2019, 18, 1–13. [Google Scholar] [CrossRef]
- Qin, A.; Zhou, Y.; Sheng, M.; Fei, G.; Ren, T.; Xu, L. Effects of microRNA-155 on the growth of human lung cancer cell line 95D in vitro. Zhongguo Fei Ai Za Zhi 2011, 14, 575–580. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Swelum, A.A.-A.; Elsafadi, M.; Mahmood, A.; Osama, A.; Shikshaky, H.; Alfayez, M.; Alowaimer, A.N.; Magdeldin, S. Thermotolerance and plasticity of camel somatic cells exposed to acute and chronic heat stress. J. Adv. Res. 2020, 22, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef]
- McKenzie, P.P.; Foster, J.S.; House, S.; Bukovsky, A.; Caudle, M.R.; Wimalasena, J. Expression of G1 Cyclins and Cyclin-Dependent Kinase-2 Activity during Terminal Differentiation of Cultured Human Trophoblast1. Biol. Reprod. 1998, 58, 1283–1289. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, L.; Wang, J.; Fan, X.; Zhang, Y.; Zhoua, M.; Li, X.; Wang, L. LASP2 inhibits trophoblast cell migration and invasion in preeclampsia through inactivation of the Wnt/β-catenin signaling pathway. J. Recept. Signal Transduct. 2020, 41, 67–73. [Google Scholar] [CrossRef]
- Han, Q.; Zheng, L.; Liu, Z.; Luo, J.; Chen, R.; Yan, J. Expression of β-catenin in human trophoblast and its role in placenta accreta and placenta previa. J. Int. Med. Res. 2018, 47, 206–214. [Google Scholar] [CrossRef]
- Johns, D.N.; Lucas, C.G.; Pfeiffer, A.C.; Chen, P.R.; Meyer, E.A.; Perry, S.D.; Spate, L.D.; Cecil, R.F.; Fudge, A.M.; Samuel, M.S.; et al. Conceptus interferon gamma is essential for establishment of pregnancy in the pig. Biol. Reprod. 2021, 105, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Arregui, C.O.; González, Á.; Burdisso, J.E.; González Wusener, A.E. Protein tyrosine phosphatase PTP1B in cell adhesion and migration. Cell Adhes. Migr. 2013, 7, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Wang, N.; Lv, H.; Yang, J.; Liu, J.; Li, W.-P.; Zhang, C.; Chen, Z.-J. The Attenuation of Trophoblast Invasion Caused by the Downregulation of EZH2 Is Involved in the Pathogenesis of Human Recurrent Miscarriage. Mol. Ther.-Nucleic. Acids 2018, 14, 377–387. [Google Scholar] [CrossRef]
- Hong, L.; Han, K.; Wu, K.; Liu, R.; Huang, J.; Lunney, J.K.; Zhao, S.; Yu, M. E-cadherin and ZEB2 modulate trophoblast cell differentiation during placental development in pigs. Reproduction 2017, 154, 765–775. [Google Scholar] [CrossRef] [PubMed][Green Version]
- DaSilva-Arnold, S.C.; Kuo, C.-Y.; Davra, V.; Remache, Y.; Kim, P.C.W.; Fisher, J.P.; Zamudio, S.; Al-Khan, A.; Birge, R.B.; Illsley, N.P. ZEB2, a master regulator of the epithelial–mesenchymal transition, mediates trophoblast differentiation. Mol. Hum. Reprod. 2018, 25, 61–75. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Yun, S.H.; Lee, H.; Yi, Y.-S.; Park, E.C.; Kim, W.; Kim, H.-Y.; Lee, J.C.; Kim, G.-H.; Kim, S.I. Analysis of the Extracellular Proteome of Colistin-Resistant Korean Acinetobacter baumannii Strains. ACS Omega 2020, 5, 5713–5720. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Mishra, A.; Ashary, N.; Sharma, R.; Modi, D. Extracellular vesicles in embryo implantation and disorders of the endometrium. Am. J. Reprod. Immunol. 2020, 85, e13360. [Google Scholar] [CrossRef]
- Evans, J.; Rai, A.; Nguyen, H.P.T.; Poh, Q.H.; Elglass, K.; Simpson, R.J.; Salamonsen, L.; Greening, D.W. Human Endometrial Extracellular Vesicles Functionally Prepare Human Trophectoderm Model for Implantation: Understanding Bidirectional Maternal-Embryo Communication. Proteomics 2019, 19, e1800423. [Google Scholar] [CrossRef]
- Gurung, S.; Greening, D.; Catt, S.; Salamonsen, L.; Evans, J. Exosomes and soluble secretome from hormone-treated endometrial epithelial cells direct embryo implantation. Mol. Hum. Reprod. 2020, 26, 510–520. [Google Scholar] [CrossRef]
- O’Neil, E.V.; Burns, G.W.; Ferreira, C.R.; Spencer, T.E. Characterization and regulation of extracellular vesicles in the lumen of the ovine uterusdagger. Biol. Reprod. 2020, 102, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Senol, S.; Sayar, I.; Ceyran, A.B.; Ibiloglu, I.; Akalin, I.; Firat, U.; Kosemetin, D.; Zerk, P.E.; Aydin, A. Stromal Clues in Endometrial Carcinoma: Loss of Expression of β-Catenin, Epithelial-Mesenchymal Transition Regulators, and Estrogen-Progesterone Receptor. Int. J. Gynecol. Pathol. 2016, 35, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, M.; Cao, F.; Han, J.; Han, P.; Wu, Y.; Deng, R.; Zhang, G.; An, X.; Zhang, L.; et al. Effect of MiR-100-5p on proliferation and apoptosis of goat endometrial stromal cell in vitro and embryo implantation in vivo. J. Cell Mol. Med. 2022, 26, 2543–2556. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Shi, S.; Liang, J.; Cao, D.; Wang, S.; Wang, Z. Endometrial cell-derived small extracellular vesicle miR-100-5p promotes functions of trophoblast during embryo implantation. Mol. Ther.-Nucleic. Acids 2020, 23, 217–231. [Google Scholar] [CrossRef]
- Su, L.; Liu, R.; Cheng, W.; Zhu, M.; Li, X.; Zhao, S.; Yu, M. Expression Patterns of MicroRNAs in Porcine Endometrium and Their Potential Roles in Embryo Implantation and Placentation. PLoS ONE 2014, 9, e87867. [Google Scholar] [CrossRef]
- Wang, Y.-P.; Zhao, P.; Liu, J.-Y.; Liu, S.-M.; Wang, Y.-X. MicroRNA-132 stimulates the growth and invasiveness of trophoblasts by targeting DAPK-1. Eur. Rev. Med. Pharm. Sci. 2020, 24, 9837–9843. [Google Scholar]
- Saadeldin, I.M.; Kim, B.; Lee, B.; Jang, G. Effect of different culture media on the temporal gene expression in the bovine developing embryos. Theriogenology 2011, 75, 995–1004. [Google Scholar] [CrossRef]
- du Puy, L.; Lopes, S.M.C.D.S.; Haagsman, H.P.; Roelen, B.A. Analysis of co-expression of OCT4, NANOG and SOX2 in pluripotent cells of the porcine embryo, in vivo and in vitro. Theriogenology 2011, 75, 513–526. [Google Scholar] [CrossRef]
- Ezashi, T.; Matsuyama, H.; Telugu, B.P.V.; Roberts, R.M. Generation of Colonies of Induced Trophoblast Cells During Standard Reprogramming of Porcine Fibroblasts to Induced Pluripotent Stem Cells1. Biol. Reprod. 2011, 85, 779–787. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Dong, X.; Gou, W. MicroRNA-155 inhibits migration of trophoblast cells and contributes to the pathogenesis of severe preeclampsia by regulating endothelial nitric oxide synthase. Mol. Med. Rep. 2014, 10, 550–554. [Google Scholar] [CrossRef]
- Dai, Y.; Qiu, Z.; Diao, Z.; Shen, L.; Xue, P.; Sun, H.; Hu, Y. MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta 2012, 33, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Shan, Y.; Yang, Y.; Wang, T.; Guo, Z. MicroRNA-155 is upregulated in the placentas of patients with preeclampsia and affects trophoblast apoptosis by targeting SHH/GLi1/BCL2. Hum. Exp. Toxicol. 2020, 40, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, A.; Tranguch, S.; Daikoku, T.; Jensen, K.; Furneaux, H.; Dey, S.K. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl. Acad. Sci. USA 2007, 104, 15144–15149. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-X.; Zeng, D.-Y.; Li, R.-T.; Pang, R.-P.; Yang, H.; Hu, Y.-L.; Zhang, Q.; Jiang, Y.; Huang, L.-Y.; Tang, Y.-B.; et al. Essential Role of MicroRNA-155 in Regulating Endothelium-Dependent Vasorelaxation by Targeting Endothelial Nitric Oxide Synthase. Hypertension 2012, 60, 1407–1414. [Google Scholar] [CrossRef]
- Alexy, T.; Rooney, K.; Weber, M.; Gray, W.D.; Searles, C.D. TNF-α alters the release and transfer of microparticle-encapsulated miRNAs from endothelial cells. Physiol. Genom. 2014, 46, 833–840. [Google Scholar] [CrossRef]
- Dakic, A.; DiVito, K.; Fang, S.; Suprynowicz, F.; Gaur, A.; Li, X.; Palechor-Ceron, N.; Simic, V.; Choudhury, S.; Yu, S.; et al. ROCK inhibitor reduces Myc-induced apoptosis and mediates immortalization of human keratinocytes. Oncotarget 2016, 7, 66740–66753. [Google Scholar] [CrossRef]
- Kim, K.; Min, S.; Kim, D.; Kim, H.; Roh, S. A Rho Kinase (ROCK) Inhibitor, Y-27632, Inhibits the Dissociation-Induced Cell Death of Salivary Gland Stem Cells. Molecules 2021, 26, 2658. [Google Scholar] [CrossRef]
- Street, C.A.; Bryan, B.A. Rho kinase proteins--pleiotropic modulators of cell survival and apoptosis. Anticancer Res. 2011, 31, 3645–3657. [Google Scholar]
- Shi, J.; Wei, L. Rho kinase in the regulation of cell death and survival. Arch. Immunol. Ther. Exp. 2007, 55, 61–75. [Google Scholar] [CrossRef]
- Shiokawa, S.; Iwashita, M.; Akimoto, Y.; Nagamatsu, S.; Sakai, K.; Hanashi, H.; Kabir-Salmani, M.; Nakamura, Y.; Uehata, M.; Yoshimura, Y. Small Guanosine TriphospataseRhoA and Rho-Associated Kinase as Regulators of Trophoblast Migration. J. Clin. Endocrinol. Metab. 2002, 87, 5808–5816. [Google Scholar] [CrossRef][Green Version]
- Lokossou, A.G.; Toudic, C.; Nguyen, P.T.; Elisseeff, X.; Vargas, A.; Rassart, É.; Lafond, J.; LeDuc, L.; Bourgault, S.; Gilbert, C.; et al. Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells†. Biol. Reprod. 2019, 102, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, K.A.; Palmarini, M.; Varela, M.; Burghardt, R.C.; Hayashi, K.; Farmer, J.L.; Spencer, T.E. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 14390–14395. [Google Scholar] [CrossRef]
- Fu, B.; Ma, H.; Liu, D. Endogenous Retroviruses Function as Gene Expression Regulatory Elements During Mammalian Pre-implantation Embryo Development. Int. J. Mol. Sci. 2019, 20, 790. [Google Scholar] [CrossRef]
- Prudhomme, S.; Bonnaud, B.; Mallet, F. Endogenous retroviruses and animal reproduction. Cytogenet. Genome. Res. 2005, 110, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.J.; Rosenkrantz, J.; Carbone, L.; Chavez, S. Endogenous Retroviruses: With Us and against Us. Front. Chem. 2017, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhu, W.; Chen, S.; Liu, Y.; Sun, Z.; Geng, T.; Wang, X.; Gao, B.; Song, C.; Qin, A.; et al. Expression of the env gene from the avian endogenous retrovirus ALVE and regulation by miR-155. Arch. Virol. 2016, 161, 1623–1632. [Google Scholar] [CrossRef]







| Name | Sequence (5′ → 3′) |
|---|---|
| miRNA-155 inhibitor | UUAAUGCUAAUCGUGAUAGGGG |
| miRNA-155 mimic sense | UGGUGCAGGUUUAAUGCUAAUCGUGAUAGGGGUUUA |
| miRNA-155 mimic anti-sense | GUGCUGAUGAACACCUAUGCUGUUAGCAUUAAUCUUGCGCUA |
| Name | Sequence 5′ → 3′ | Product Size | Accession No. | |
|---|---|---|---|---|
| Forward | Reverse | |||
| miR-100-p | AAACCCGTAGATCCGAACT | CAAGCTTGTGCGGACTAATA | 43 | NR_029515.1 |
| miR-132-p | GTCTCCAGGGCAACCGTG | CGACCATGGCTGTAGACTGT | 70 | LM608489.1 |
| miR-155 | GCGGTTAATGCTAATCGTGATA | CGAGGAAGAAGACGGAAGAAT | 65 | LM608611.1 |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT | 89 | NR_004394.1 |
| Bax | GAGAGACACCTGAGCTGG | AGTTCATCTCCAATGCGC | 165 | XM_013998624.2 |
| Bcl2 | GTTGACTTTCTCTCCTACAAG | GGTACCTCAGTTCAAACTCAT | 277 | NM_214285.1 |
| CDK2 | GCTTCAGGGGCTAGCTTTTT | AGCCCAGAAGGATTTCAGGT | 197 | NM_001285465.1 |
| CTNNB1 | CCATTCCATTGTTTGTGCAG | GTTGCCACACCTTCATTCCT | 175 | NM_214367.1 |
| GAPDH | ACACTCACTCTTCTACCTTTG | CAAATTCATTGTCGTACCAG | 90 | DQ845173.1 |
| IFNG | CCATTCAAAGGAGCATGGAT | TTCAGTTTCCCAGAGCTACCA | 76 | NM_213948.1 |
| PERV | TCCGTGCTTACGGGTTTTAC | TTTCTCCCAGAGCCTCCATA | 388 | XM_021074788.1 |
| PTPN1 | TCTCAAGAAACTCGAGAGAT | TCAGCCAGACAGAAGGTC | 194 | XM_021077277.1 |
| Zeb1 | ACGGATGCAGCAGATTGTGA | CCGGGTAACACTGTCTGGTC | 71 | XM_021064196.1 |
| Zeb2 | GACAATGTAGTGGACACGGGT | GGGGAGCACTCCTGGTT | 131 | XM_021076508.1 |
| Universal stem-loop primer | GAAAGAAGGCGAGGAGCAGATCGAGGAAGAAGACGGAAGAATGTGCGTCTCGCCTTCTTTCNNNNNNNN | |||
| UniProt Accession | Description | Mol % |
|---|---|---|
| A0A287B5W2 | Trypsinogen isoform X1 | 6.0325 |
| A0A287AEL2 | Keratin 14 | 5.0814 |
| F1SGG6 | Keratin 5 | 4.831 |
| A0A287A0Q6 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation Protein zeta | 4.6558 |
| A0A287ANZ8 | Thy-1 membrane glycoprotein | 4.1051 |
| A0A286ZT13 | Albumin | 4.03 |
| P20112 | SPARC | 3.5544 |
| Q28944 | Procathepsin | 3.0288 |
| A0A5G2QTF5 | Thioredoxin | 2.8536 |
| A0A287AHS0 | Calmodulin 3 | 2.8285 |
| A0A287BA49 | Keratin 5 | 2.7785 |
| A0A287A8S8 | Phosphopyruvate hydratase | 2.7284 |
| F1SGG3 | Keratin, type II cytoskeletal 1 | 2.7034 |
| I3LDS3 | Keratin 10 | 2.6783 |
| A0A287AHK1 | Vitamin D-binding protein | 2.6283 |
| K7GQ95 | S100 calcium binding protein A2 | 2.3529 |
| A0A5G2QSE8 | Keratin 3 | 2.3029 |
| A0A287BHY5 | Keratin 2 | 1.7772 |
| A0A5G2QUE0 | Antithrombin-III | 1.7522 |
| A0A287ATD0 | Keratin 75 | 1.577 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saadeldin, I.M.; Tanga, B.M.; Bang, S.; Seo, C.; Koo, O.; Yun, S.H.; Kim, S.I.; Lee, S.; Cho, J. ROCK Inhibitor (Y-27632) Abolishes the Negative Impacts of miR-155 in the Endometrium-Derived Extracellular Vesicles and Supports Embryo Attachment. Cells 2022, 11, 3178. https://doi.org/10.3390/cells11193178
Saadeldin IM, Tanga BM, Bang S, Seo C, Koo O, Yun SH, Kim SI, Lee S, Cho J. ROCK Inhibitor (Y-27632) Abolishes the Negative Impacts of miR-155 in the Endometrium-Derived Extracellular Vesicles and Supports Embryo Attachment. Cells. 2022; 11(19):3178. https://doi.org/10.3390/cells11193178
Chicago/Turabian StyleSaadeldin, Islam M., Bereket Molla Tanga, Seonggyu Bang, Chaerim Seo, Okjae Koo, Sung Ho Yun, Seung Il Kim, Sanghoon Lee, and Jongki Cho. 2022. "ROCK Inhibitor (Y-27632) Abolishes the Negative Impacts of miR-155 in the Endometrium-Derived Extracellular Vesicles and Supports Embryo Attachment" Cells 11, no. 19: 3178. https://doi.org/10.3390/cells11193178
APA StyleSaadeldin, I. M., Tanga, B. M., Bang, S., Seo, C., Koo, O., Yun, S. H., Kim, S. I., Lee, S., & Cho, J. (2022). ROCK Inhibitor (Y-27632) Abolishes the Negative Impacts of miR-155 in the Endometrium-Derived Extracellular Vesicles and Supports Embryo Attachment. Cells, 11(19), 3178. https://doi.org/10.3390/cells11193178