Phylogeny, Structure, Functions, and Role of AIRE in the Formation of T-Cell Subsets
Abstract
:1. Introduction
2. Evolutionary Prerequisites
3. Structure and Localization of AIRE
4. Molecular Mechanisms of AIRE Action
5. The Role of AIRE in the Formation of the TEC Transcriptome and Immunopeptidome
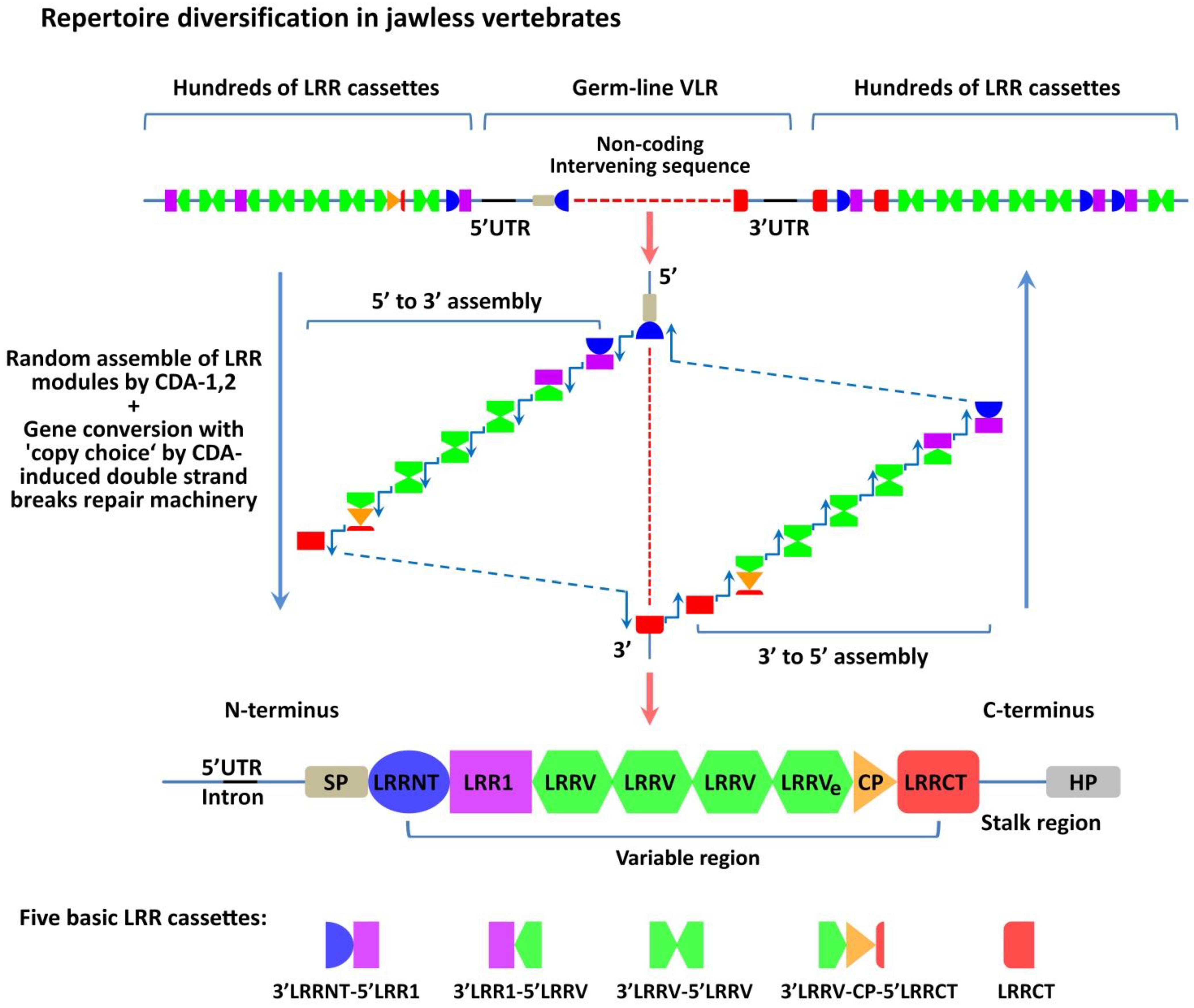
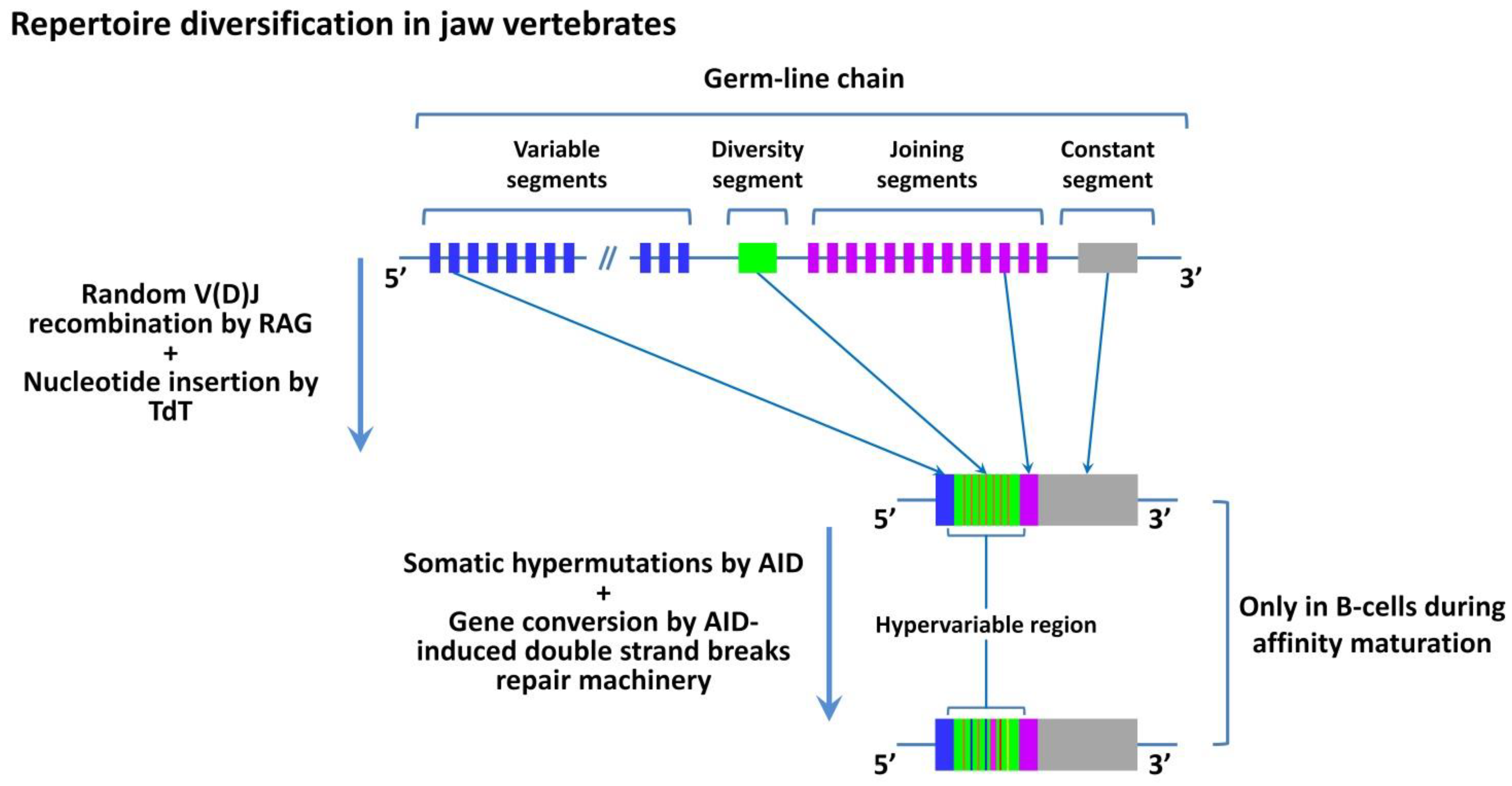
6. The Role of AIRE in the Formation of TCR and BCR Repertoires
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dumonde, D.C. Tissue-specific antigens. Adv. Immunol. 1966, 5, 245–412. [Google Scholar] [CrossRef]
- Steinmuller, D. Tissue-specific and tissue-restricted histocompatibility antigens. Immunol. Today 1984, 5, 234–240. [Google Scholar] [CrossRef]
- Geenen, V.; Legros, J.-J.; Franchimont, P.; Baudrihaye, M.; Defresne, M.-P.; Boniver, J. The neuroendocrine thymus: Coexistence of oxytocin and neurophysin in the human thymus. Science 1986, 232, 508–511. [Google Scholar] [CrossRef] [Green Version]
- Kirchner, T.; Hoppe, F.; Müller-Hermelink, H.K.; Schalke, B.; Tzartos, S. Acetylcholine receptor epitopes on epithelial cells of thymoma in myasthenia gravis. Lancet 1987, 1, 218. [Google Scholar] [CrossRef]
- Fuller, P.J.; Verity, K. Somatostatin gene expression in the thymus gland. J. Immunol. 1989, 143, 1015–1017. [Google Scholar] [PubMed]
- Kyewski, B.; Derbinski, J.; Gotter, J.; Klein, L. Promiscuous gene expression and central T-cell tolerance: More than meets the eye. Trends Immunol. 2002, 23, 364–371. [Google Scholar] [CrossRef]
- Pugliese, A.; Zeller, M.; Fernandez, A., Jr.; Zalcberg, L.J.; Bartlett, R.J.; Ricordi, C.; Pietropaolo, M.; Eisenbarth, G.S.; Bennett, S.T.; Patel, D.D. The insulin gene is transcribed in the human thymus and transcription levels correlate with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat. Genet. 1997, 15, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Vafiadis, P.; Bennett, S.T.; Todd, J.A.; Nadeau, J.; Grabs, R.; Goodyer, C.G.; Wickramasinghe, S.; Colle, E.; Polychronakos, C. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat. Genet. 1997, 15, 289–292. [Google Scholar] [CrossRef]
- Werdelin, O.; Cordes, U.; Jensen, T. Aberrant expression of tissue-specific proteins in the thymus: A hypothesis for the development of central Tolerance. Scand. J. Immunol. 1998, 47, 95–100. [Google Scholar] [CrossRef]
- Takase, H.; Yu, C.R.; Mahdi, R.M.; Douek, D.C.; Dirusso, G.B.; Midgley, F.M.; Dogra, R.; Allende, G.; Rosenkranz, E.; Pugliese, A.; et al. Thymic expression of peripheral tissue antigens in humans: A remarkable variability among individuals. Int. Immunol. 2005, 17, 1131–1140. [Google Scholar] [CrossRef] [Green Version]
- Ahonen, P. Autoimmune polyendocrinopathy-candidosis-ectodermal dystrophy (APECED): Autosomal recessive inheritance. Clin. Genet. 1985, 27, 535–542. [Google Scholar] [CrossRef]
- Björses, P.; Aaltonen, J.; Horelli-Kuitunen, N.; Yaspo, M.-L.; Peltonen, L. Gene defect behind APECED: A new clue to autoimmunity. Hum. Mol. Genet. 1998, 7, 1547–1553. [Google Scholar] [CrossRef] [Green Version]
- Aaltonen, J.; Björses, P.; Sandkuijl, L.; Perheentupa, J.; Peltonen, L.J. An autosomal locus causing autoimmune disease: Autoimmune polyglandular disease type I assigned to chromosome 21. Nat. Genet. 1994, 8, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Peterson, P.; Scott, H.S.; Kudoh, J.; Minoshima, S.; Heino, M.; Krohn, K.J.E.; Lalioti, M.D.; Mullis, P.E.; Antonarakis, S.; et al. Positional cloning of the APECED gene. Nat. Genet. 1997, 17, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, J.; Björses, P. Cloning of the APECED gene provides new insight into human autoimmunity. Ann. Med. 1999, 31, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Blechschmidt, K.; Schweiger, M.; Wertz, K.; Poulson, R.; Christensen, H.M.; Rosenthal, A.; Lehrach, H.; Yaspo, M.L. The mouse Aire gene: Comparative genomic sequencing, gene organization, and expression. Genome Res. 1999, 9, 158–166. [Google Scholar] [CrossRef]
- Derbinski, J.; Schulte, A.; Kyewski, B.; Klein, L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2001, 2, 1032–1039. [Google Scholar] [CrossRef]
- Saltis, M.; Criscitiello, M.F.; Ohta, Y.; Keefe, M.; Trede, N.S.; Goitsuka, R.; Flajnik, M.F. Evolutionarily conserved and divergent regions of the Autoimmune Regulator (Aire) gene: A comparative analysis. Immunogenetics 2008, 60, 105–114. [Google Scholar] [CrossRef]
- Sutoh, Y.; Kondo, M.; Ohta, Y.; Ota, T.; Tomaru, U.; Flajnik, M.F.; Kasahara, M. Comparative genomic analysis of the proteasome β5t subunit gene: Implications for the origin and evolution of thymoproteasomes. Immunogenetics 2012, 64, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Dehal, P.; Boore, J.L. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef] [Green Version]
- Holland, L.Z.; Daza, D.O. A new look at an old question: When did the second whole genome duplication occur in vertebrate evolution? Genome Biol. 2018, 19, 209. [Google Scholar] [CrossRef]
- Klein, J.; Figueroa, F. Evolution of the major histocompatibility complex. Crit. Rev. Immunol. 1986, 6, 295–386. [Google Scholar] [CrossRef]
- Poole, J.R.M.; Huang, S.F.; Xu, A.; Bayet, J.; Pontarotti, P. The RAG transposon is active through the deuterostome evolution and domesticated in jawed vertebrates. Immunogenetics 2017, 69, 391–400. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, T.C.; Huang, G.; Lu, Q.; Surleac, M.D.; Mandell, J.D.; Pontarotti, P.; Petrescu, A.-J.; Xu, A.; Xiong, Y.; et al. Transposon molecular domestication and the evolution of the RAG recombinase. Nature 2019, 569, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Yakovenko, I.; Agronin, J.; Smith, L.C.; Oren, M. Guardian of the Genome: An Alternative RAG/Transib Co-Evolution Hypothesis for the Origin of V(D)J Recombination. Front. Immunol. 2021, 12, 709165. [Google Scholar] [CrossRef]
- Kasahara, M.; Nakaya, J.; Satta, Y.; Takahata, N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997, 13, 90–92. [Google Scholar] [CrossRef]
- Kasahara, M.; Flajnik, M.F. Origin and evolution of the specialized forms of proteasomes involved in antigen presentation. Immunogenetics 2019, 71, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 2018, 18, 438–453. [Google Scholar] [CrossRef]
- Murata, S.; Takahama, Y.; Kasahara, M.; Tanaka, K. The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat. Immunol. 2018, 19, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Bajoghli, B.; Guo, P.; Aghaallaei, N.; Hirano, M.; Strohmeier, C.; McCurley, N.; Bockman, D.E.; Schorpp, M.; Cooper, M.D.; Boehm, T. A thymus candidate in lampreys. Nature 2011, 470, 90–94. [Google Scholar] [CrossRef]
- Alder, M.N.; Rogozin, I.B.; Iyer, L.M.; Glazko, G.V.; Cooper, M.D.; Pancer, Z. Diversity and Function of Adaptive Immune Receptors in a Jawless Vertebrate. Science 2005, 310, 1970–1973. [Google Scholar] [CrossRef] [Green Version]
- Herrin, B.R.; Cooper, M.D. Alternative Adaptive Immunity in Jawless Vertebrates. J. Immunol. 2010, 185, 1367–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagawa, F.; Kishishita, N.; Shimizu, K.; Hirose, S.; Miyoshi, M.; Nezu, J.; Nishimura, T.; Nishizumi, H.; Takahashi, Y.; Hashimoto, S.; et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat. Immunol. 2006, 8, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Holland, S.J.; Berghuis, L.M.; King, J.J.; Iyer, L.M.; Sikora, K.; Fifield, H.; Peter, S.; Quinlan, E.; Sugahara, F.; Shingate, P.; et al. Expansions, diversification, and interindividual copy number variations of AID/APOBEC family cytidine deaminase genes in lampreys. Proc. Natl. Acad. Sci. USA 2018, 115, E3211–E3220. [Google Scholar] [CrossRef] [Green Version]
- Maul, R.W.; Gearhart, P.J. AID and Somatic Hypermutation. Adv. Immunol. 2010, 105, 159–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajoghli, B.; Aghaallaei, N.; Hess, I.; Rode, I.; Netuschil, N.; Tay, B.-H.; Venkatesh, B.; Yu, J.-K.; Kaltenbach, S.L.; Holland, N.D.; et al. Evolution of Genetic Networks Underlying the Emergence of Thymopoiesis in Vertebrates. Cell 2009, 138, 186–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaidya, H.J.; Leon, A.B.; Blackburn, C.C. FOXN1 in thymus organogenesis and development. Eur. J. Immunol. 2016, 46, 1826–1837. [Google Scholar] [CrossRef] [Green Version]
- Holland, S.J.; Gao, M.; Hirano, M.; Iyer, L.M.; Luo, M.; Schorpp, M.; Cooper, M.D.; Aravind, L.; Mariuzza, R.A.; Boehm, T. Selection of the lamprey VLRC antigen receptor repertoire. Proc. Natl. Acad. Sci. USA 2014, 111, 14834–14839. [Google Scholar] [CrossRef] [Green Version]
- Flajnik, M.F. Re-evaluation of the Immunological Big Bang. Curr. Biol. 2014, 24, R1060–R1065. [Google Scholar] [CrossRef] [Green Version]
- Flajnik, M.F. A Convergent Immunological Holy Trinity of Adaptive Immunity in Lampreys: Discovery of the Variable Lymphocyte Receptors. J. Immunol. 2018, 201, 1331–1335. [Google Scholar] [CrossRef]
- Velikovsky, C.A.; Deng, L.; Tasumi, S.; Iyer, L.M.; Kerzic, M.C.; Aravind, L.; Pancer, Z.; Mariuzza, R.A. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat. Struct. Mol. Biol. 2009, 16, 725–730. [Google Scholar] [CrossRef]
- Collins, B.C.; Gunn, R.J.; McKitrick, T.R.; Cummings, R.D.; Cooper, M.D.; Herrin, B.R.; Wilson, I.A. Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates. Structure 2017, 25, 1667–1678.e4. [Google Scholar] [CrossRef] [Green Version]
- de Greef, P.C.; Oakes, T.; Gerritsen, B.; Ismail, M.; Heather, J.M.; Hermsen, R.; Chain, B.; de Boer, R.J. The naive T-cell receptor repertoire has an extremely broad distribution of clone sizes. eLife 2020, 9, e49900. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, Y.; Cheng, Y.; Glanville, J.; Zhang, D.; Lee, J.-Y.; Olshen, R.A.; Weyand, C.M.; Boyd, S.D.; Goronzy, J.J. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl. Acad. Sci. USA 2014, 111, 13139–13144. [Google Scholar] [CrossRef] [Green Version]
- Neuberger, M.S.; Ehrenstein, M.R.; Rada, C.; Sale, J.; Batista, F.D.; Williams, G.; Milstein, C. Memory in the B–cell compartment: Antibody affinity maturation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, X.; Li, Y.; Qiu, X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: Mechanism and regulation. Immunology 2020, 160, 233–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flajnik, M. Immunology: The Origin of Sweetbreads in Lampreys? Curr. Biol. 2011, 21, R218–R220. [Google Scholar] [CrossRef] [Green Version]
- Boehm, T. Design principles of adaptive immune systems. Nat. Rev. Immunol. 2011, 11, 307–317. [Google Scholar] [CrossRef]
- Trancoso, I.; Morimoto, R.; Boehm, T. Co-evolution of mutagenic genome editors and vertebrate adaptive immunity. Curr. Opin. Immunol. 2020, 65, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, A.; Quinlan, E.M.; Larijani, M. Evolutionary Comparative Analyses of DNA-Editing Enzymes of the Immune System: From 5-Dimensional Description of Protein Structures to Immunological Insights and Applications to Protein Engineering. Front. Immunol. 2021, 12, 642343. [Google Scholar] [CrossRef]
- Schwaiger, F.-W.; Epplen, J.T. Exonic MHC-DRB Polymorphisms and Intronic Simple Repeat Sequenees: Janus’Faces of DNA Sequence Evolution. Immunol. Rev. 1995, 143, 199–224, Erratum in: Immunol. Rev. 1995, 144, 314. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, L.B.; Kuchroo, V.K. T cell recognition of self and altered self antigens. Crit. Rev. Immunol. 1997, 17, 449–462. [Google Scholar] [PubMed]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef]
- Lutes, L.K.; Steier, Z.; McIntyre, L.L.; Pandey, S.; Kaminski, J.; Hoover, A.R.; Ariotti, S.; Streets, A.; Yosef, N.; Robey, E.A. T cell self-reactivity during thymic development dictates the timing of positive selection. eLife 2021, 10, e65435. [Google Scholar] [CrossRef]
- Koncz, B.; Balogh, G.M.; Papp, B.T.; Asztalos, L.; Kemény, L.; Manczinger, M. Self-mediated positive selection of T cells sets an obstacle to the recognition of nonself. Proc. Natl. Acad. Sci. USA 2021, 118, e2100542118. [Google Scholar] [CrossRef]
- Ohigashi, I.; Frantzeskakis, M.; Jacques, A.; Fujimori, S.; Ushio, A.; Yamashita, F.; Ishimaru, N.; Yin, D.; Cam, M.; Kelly, M.C.; et al. The thymoproteasome hardwires the TCR repertoire of CD8+ T cells in the cortex independent of negative selection. J. Exp. Med. 2021, 218, e20201904. [Google Scholar] [CrossRef]
- Chida, A.S.; Goyos, A.; Robert, J. Phylogenetic and developmental study of CD4, CD8 α and β T cell co-receptor homologs in two amphibian species, Xenopus tropicalis and Xenopus laevis. Dev. Comp. Immunol. 2011, 35, 366–377. [Google Scholar] [CrossRef] [Green Version]
- Swann, J.B.; Nusser, A.; Morimoto, R.; Nagakubo, D.; Boehm, T. Retracing the evolutionary emergence of thymopoiesis. Sci. Adv. 2020, 6, eabd9585. [Google Scholar] [CrossRef]
- Peterson, P.; Org, T.; Rebane, A. Transcriptional regulation by AIRE: Molecular mechanisms of central tolerance. Nat. Rev. Immunol. 2008, 8, 948–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passos, G.A.; Speck-Hernandez, C.A.; Assis, A.F.; Mendes-Da-Cruz, D.A. Update onAireand thymic negative selection. Immunology 2018, 153, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Perniola, R. Twenty Years of AIRE. Front. Immunol. 2018, 9, 98. [Google Scholar] [CrossRef]
- Abramson, J.; Husebye, E.S. Autoimmune regulator and self-tolerance-molecular and clinical aspects. Immunol. Rev. 2016, 271, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Alexander, C.; Rossi, S.W.; Liiv, I.; Rebane, A.; Worth, C.; Wong, J.; Laan, M.; Peterson, P.; Jenkinson, E.J.; et al. AIRE’s CARD Revealed, a New Structure for Central Tolerance Provokes Transcriptional Plasticity. J. Biol. Chem. 2008, 283, 1723–1731. [Google Scholar] [CrossRef] [Green Version]
- Pitkänen, J.; Vähämurto, P.; Krohn, K.; Peterson, P. Subcellular Localization of the Autoimmune Regulator Protein. Characterization of nuclear targeting and transcriptional activation domain. J. Biol. Chem. 2001, 276, 19597–19602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilmarinen, T.; Eskelin, P.; Halonen, M.; Rüppell, T.; Kilpikari, R.; Torres, G.D.; Kangas, H.; Ulmanen, I. Functional analysis of SAND mutations in AIRE supports dominant inheritance of the G228W mutation. Hum. Mutat. 2005, 26, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, C.; Bukrinsky, A.; Peltonen, L. Systematic mutagenesis of the functional domains of AIRE reveals their role in intracellular targeting. Hum. Mol. Genet. 2002, 11, 3299–3308. [Google Scholar] [CrossRef] [Green Version]
- Akiyoshi, H.; Hatakeyama, S.; Pitkänen, J.; Mouri, Y.; Doucas, V.; Kudoh, J.; Tsurugaya, K.; Uchida, D.; Matsushima, A.; Oshikawa, K.; et al. Subcellular Expression of Autoimmune Regulator Is Organized in a Spatiotemporal Manner. J. Biol. Chem. 2004, 279, 33984–33991. [Google Scholar] [CrossRef] [Green Version]
- Gardner, J.M.; Fletcher, A.L.; Anderson, M.S.; Turley, S.J. AIRE in the thymus and beyond. Curr. Opin. Immunol. 2009, 21, 582–589. [Google Scholar] [CrossRef]
- Takahama, Y.; Ohigashi, I.; Baik, S.; Anderson, G. Generation of diversity in thymic epithelial cells. Nat. Rev. Immunol. 2017, 17, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Takaba, H.; Takayanagi, H. The Mechanisms of T Cell Selection in the Thymus. Trends Immunol. 2017, 38, 805–816. [Google Scholar] [CrossRef]
- Kadouri, N.; Nevo, S.; Goldfarb, Y.; Abramson, J. Thymic epithelial cell heterogeneity: TEC by TEC. Nat. Rev. Immunol. 2020, 20, 239–253. [Google Scholar] [CrossRef]
- Bleul, C.C.; Corbeaux, T.; Reuter, A.; Fisch, P.; Mönting, J.S.; Boehm, T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 2006, 441, 992–996. [Google Scholar] [CrossRef]
- Li, J.; Gordon, J.; Chen, E.L.Y.; Xiao, S.; Wu, L.; Zúñiga-Pflücker, J.C.; Manley, N.R. NOTCH1 signaling establishes the medullary thymic epithelial cell progenitor pool during mouse fetal development. Development 2020, 147, dev178988. [Google Scholar] [CrossRef]
- Liu, D.; Kousa, A.I.; O’Neill, K.E.; Rouse, P.; Popis, M.; Farley, A.M.; Tomlinson, S.R.; Ulyanchenko, S.; Guillemot, F.; Seymour, P.A.; et al. Canonical Notch signaling controls the early thymic epithelial progenitor cell state and emergence of the medullary epithelial lineage in fetal thymus development. Development 2020, 147, dev178582. [Google Scholar] [CrossRef] [PubMed]
- Bocci, F.; Onuchic, J.N.; Jolly, M.K. Understanding the Principles of Pattern Formation Driven by Notch Signaling by Integrating Experiments and Theoretical Models. Front. Physiol. 2020, 11, 929. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.; Pongracz, J.; Parnell, S.; Jenkinson, E.J. Notch ligand-bearing thymic epithelial cells initiate and sustain Notch signaling in thymocytes independently of T cell receptor signaling. Eur. J. Immunol. 2001, 31, 3349–3354. [Google Scholar] [CrossRef]
- Sjöqvist, M.; Andersson, E.R. Do as I say, Not(ch) as I do: Lateral control of cell fate. Dev. Biol. 2019, 447, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.E.; Žuklys, S.; Zhanybekova, S.; Ohigashi, I.; Teh, H.-Y.; Sansom, S.N.; Shikama-Dorn, N.; Hafen, K.; Macaulay, I.C.; Deadman, M.E.; et al. Dynamic spatio-temporal contribution of single β5t+ cortical epithelial precursors to the thymus medulla. Eur. J. Immunol. 2016, 46, 846–856. [Google Scholar] [CrossRef] [Green Version]
- Ohigashi, I.; Zuklys, S.; Sakata, M.; Mayer, C.E.; Hamazaki, Y.; Minato, N.; Hollander, G.A.; Takahama, Y. Adult Thymic Medullary Epithelium Is Maintained and Regenerated by Lineage-Restricted Cells Rather Than Bipotent Progenitors. Cell Rep. 2015, 13, 1432–1443. [Google Scholar] [CrossRef] [Green Version]
- Alawam, A.S.; Anderson, G.; Lucas, B. Generation and Regeneration of Thymic Epithelial Cells. Front. Immunol. 2020, 11, 858. [Google Scholar] [CrossRef]
- Baik, S.; Sekai, M.; Hamazaki, Y.; Jenkinson, W.E.; Anderson, G. Relb acts downstream of medullary thymic epithelial stem cells and is essential for the emergence of RANK+ medullary epithelial progenitors. Eur. J. Immunol. 2016, 46, 857–862. [Google Scholar] [CrossRef] [Green Version]
- Burkly, L.; Hession, C.; Ogata, L.; Reilly, C.; Marconl, L.A.; Olson, D.; Tizard, R.; Gate, R.; Lo, D. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 1995, 373, 531–536. [Google Scholar] [CrossRef]
- Boehm, T.; Scheu, S.; Pfeffer, K.; Bleul, C.C. Thymic Medullary Epithelial Cell Differentiation, Thymocyte Emigration, and the Control of Autoimmunity Require Lympho–Epithelial Cross Talk via LTβR. J. Exp. Med. 2003, 198, 757–769. [Google Scholar] [CrossRef] [Green Version]
- Nowell, C.S.; Bredenkamp, N.; Tetélin, S.; Jin, X.; Tischner, C.; Vaidya, H.; Sheridan, J.M.; Stenhouse, F.H.; Heussen, R.; Smith, A.J.H.; et al. Foxn1 Regulates Lineage Progression in Cortical and Medullary Thymic Epithelial Cells But Is Dispensable for Medullary Sublineage Divergence. PLoS Genet. 2011, 7, e1002348. [Google Scholar] [CrossRef] [Green Version]
- Irla, M. RANK Signaling in the Differentiation and Regeneration of Thymic Epithelial Cells. Front. Immunol. 2021, 11, 623265. [Google Scholar] [CrossRef]
- Goldfarb, Y.; Kadouri, N.; Levi, B.; Sela, A.; Herzig, Y.; Cohen, R.N.; Hollenberg, A.N.; Abramson, J. HDAC3 Is a Master Regulator of mTEC Development. Cell Rep. 2016, 15, 651–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, N.; Sergé, A.; Ferrier, P.; Irla, M. Thymic Crosstalk Coordinates Medulla Organization and T-Cell Tolerance Induction. Front. Immunol. 2015, 6, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Laan, M.; Bichele, R.; Kisand, K.; Scott, H.S.; Peterson, P. Post-Aire Maturation of Thymic Medullary Epithelial Cells Involves Selective Expression of Keratinocyte-Specific Autoantigens. Front. Immunol. 2012, 3, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornstein, C.; Nevo, S.; Giladi, A.; Kadouri, N.; Pouzolles, M.; Gerbe, F.; David, E.; Machado, A.; Chuprin, A.; Tóth, B.; et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 2018, 559, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Ohigashi, I.; Nitta, T.; Sakata, M.; Tanaka, K.; Murata, S.; Kanagawa, O.; Takahama, Y. Thymic nurse cells provide microenvironment for secondary T cell receptor rearrangement in cortical thymocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 20572–20577. [Google Scholar] [CrossRef] [Green Version]
- Hendrix, T.M.; Chilukuri, R.V.; Martinez, M.; Olushoga, Z.; Blake, A.; Brohi, M.; Walker, C.; Samms, M.; Guyden, J.C. Thymic nurse cells exhibit epithelial progenitor phenotype and create unique extra-cytoplasmic membrane space for thymocyte selection. Cell. Immunol. 2010, 261, 81–92. [Google Scholar] [CrossRef] [Green Version]
- Gardner, J.M.; Metzger, T.C.; McMahon, E.J.; Au-Yeung, B.B.; Krawisz, A.K.; Lu, W.; Price, J.D.; Johannes, K.P.; Satpathy, A.T.; Murphy, K.M.; et al. Extrathymic Aire-Expressing Cells Are a Distinct Bone Marrow-Derived Population that Induce Functional Inactivation of CD4+ T Cells. Immunity 2013, 39, 560–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fergusson, J.R.; Morgan, M.D.; Bruchard, M.; Huitema, L.; Heesters, B.A.; Van Unen, V.; Van Hamburg, J.P.; van der Wel, N.; Picavet, D.; Koning, F.; et al. Maturing Human CD127+ CCR7+ PDL1+ Dendritic Cells Express AIRE in the Absence of Tissue Restricted Antigens. Front. Immunol. 2019, 9, 2902. [Google Scholar] [CrossRef]
- Zhao, B.; Chang, L.; Fu, H.; Sun, G.; Yang, W. The Role of Autoimmune Regulator (AIRE) in Peripheral Tolerance. J. Immunol. Res. 2018, 2018, 3930750. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, K.; Nitta, T.; Sugawa, R.; Shirai, M.; Schwartz, R.J.; Amagai, T.; Nitta, S.; Takahama, Y.; Morisaki, T. Differentiation of Pharyngeal Endoderm from Mouse Embryonic Stem Cell. Stem Cells Dev. 2010, 19, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Lambert, J.-P.; Cockburn, K.; Gingras, A.-C.; Rossant, J. AIRE is a critical spindle-associated protein in embryonic stem cells. eLife 2017, 6, e28131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillis-Buck, E.; Miller, H.; Sirota, M.; Sanders, S.J.; Ntranos, V.; Anderson, M.S.; Gardner, J.M.; MacKenzie, T.C. Extrathymic Aire -expressing cells support maternal-fetal tolerance. Sci. Immunol. 2021, 6, eabf1968. [Google Scholar] [CrossRef]
- Wang, J.; Lareau, C.A.; Bautista, J.L.; Gupta, A.R.; Sandor, K.; Germino, J.; Yin, Y.; Arvedson, M.P.; Reeder, G.C.; Cramer, N.T.; et al. Single-cell multiomics defines tolerogenic extrathymic Aire-expressing populations with unique homology to thymic epithelium. Sci. Immunol. 2021, 6, eabl5053. [Google Scholar] [CrossRef]
- Abramson, J.; Giraud, M.; Benoist, C.; Mathis, D. Aire’s Partners in the Molecular Control of Immunological Tolerance. Cell 2010, 140, 123–135. [Google Scholar] [CrossRef] [Green Version]
- Brennecke, P.; Reyes, A.; Pinto, S.; Rattay, K.; Nguyen, M.; Küchler, R.; Huber, W.; Kyewski, B.; Steinmetz, L.M. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat. Immunol. 2015, 16, 933–941. [Google Scholar] [CrossRef] [Green Version]
- Pinto, S.; Michel, C.; Schmidt-Glenewinkel, H.; Harder, N.; Rohr, K.; Wild, S.; Brors, B.; Kyewski, B. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc. Natl. Acad. Sci. USA 2013, 110, E3497–E3505. [Google Scholar] [CrossRef] [Green Version]
- Pitkänen, J.; Doucas, V.; Sternsdorf, T.; Nakajima, T.; Aratani, S.; Jensen, K.; Will, H.; Vähämurto, P.; Ollila, J.; Vihinen, M.; et al. The Autoimmune Regulator Protein Has Transcriptional Transactivating Properties and Interacts with the Common Coactivator CREB-binding Protein. J. Biol. Chem. 2000, 275, 16802–16809. [Google Scholar] [CrossRef] [Green Version]
- Pitkänen, J.; Rebane, A.; Rowell, J.; Murumägi, A.; Ströbel, P.; Möll, K.; Saare, M.; Heikkilä, J.; Doucas, V.; Marx, A.; et al. Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem. Biophys. Res. Commun. 2005, 333, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Chuprin, A.; Avin, A.; Goldfarb, Y.; Herzig, Y.; Levi, B.; Jacob, A.; Sela, A.; Katz, S.; Grossman, M.; Guyon, C.; et al. The deacetylase Sirt1 is an essential regulator of Aire-mediated induction of central immunological tolerance. Nat. Immunol. 2015, 16, 737–745. [Google Scholar] [CrossRef]
- Liiv, I.; Rebane, A.; Org, T.; Saare, M.; Maslovskaja, J.; Kisand, K.; Juronen, E.; Valmu, L.; Bottomley, M.J.; Kalkkinen, N.; et al. DNA-PK contributes to the phosphorylation of AIRE: Importance in transcriptional activity. Biochim. Biophys. Acta 2008, 1783, 74–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Li, Y.; Xiong, J.; Lan, B.; Wang, X.; Liu, J.; Lin, J.; Fei, Z.; Zheng, X.; Chen, C. Role of PRKDC in cancer initiation, progression, and treatment. Cancer Cell Int. 2021, 21, 563. [Google Scholar] [CrossRef]
- McClendon, A.K.; Rodriguez, A.C.; Osheroff, N. Human Topoisomerase IIα Rapidly Relaxes Positively Supercoiled DNA: Implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005, 280, 39337–39345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oven, I.; Brdičková, N.; Kohoutek, J.; Vaupotič, T.; Narat, M.; Peterlin, B.M. AIRE Recruits P-TEFb for Transcriptional Elongation of Target Genes in Medullary Thymic Epithelial Cells. Mol. Cell. Biol. 2007, 27, 8815–8823. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Inukai, N.; Narita, T.; Wada, T.; Handa, H. Evidence that Negative Elongation Factor Represses Transcription Elongation through Binding to a DRB Sensitivity-Inducing Factor/RNA Polymerase II Complex and RNA. Mol. Cell. Biol. 2002, 22, 2918–2927. [Google Scholar] [CrossRef] [Green Version]
- Giraud, M.; Jmari, N.; Du, L.; Carallis, F.; Nieland, T.J.F.; Perez-Campo, F.M.; Bensaude, O.; Root, D.E.; Hacohen, N.; Mathis, D.; et al. An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc. Natl. Acad. Sci. USA 2014, 111, 1491–1496. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Bansal, K.; Schaefer, U.; Chapman, T.; Rioja, I.; Proekt, I.; Anderson, M.S.; Prinjha, R.K.; Tarakhovsky, A.; Benoist, C.; et al. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc. Natl. Acad. Sci. USA 2015, 112, E4448–E4457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, K.; Yoshida, H.; Benoist, C.; Mathis, D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Bansal, K.; Michelson, D.A.; Ramirez, R.N.; Viny, A.D.; Levine, R.L.; Benoist, C.; Mathis, D. Aire regulates chromatin looping by evicting CTCF from domain boundaries and favoring accumulation of cohesin on superenhancers. Proc. Natl. Acad. Sci. USA 2021, 118, e2110991118. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, C.; Trofimov, A.; Brochu, S.; Lemieux, S.; Perreault, C. Differential Features of AIRE-Induced and AIRE-Independent Promiscuous Gene Expression in Thymic Epithelial Cells. J. Immunol. 2015, 195, 498–506. [Google Scholar] [CrossRef]
- Sansom, S.N.; Shikama-Dorn, N.; Zhanybekova, S.; Nusspaumer, G.; Macaulay, I.C.; Deadman, M.E.; Heger, A.; Ponting, C.P.; Holländer, G.A. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014, 24, 1918–1931. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Ortega, L.F.; Martínez, O. How Many Genes Are Expressed in a Transcriptome? Estimation and Results for RNA-Seq. PLoS ONE 2015, 10, e0130262. [Google Scholar] [CrossRef] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jonge, H.J.; Fehrmann, R.S.; De Bont, E.S.; Hofstra, R.; Gerbens, F.; Kamps, W.A.; de Vries, E.G.; Van Der Zee, A.G.J.; Meerman, G.J.; Ter Elst, A. Evidence Based Selection of Housekeeping Genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keane, P.; Ceredig, R.; Seoighe, C. Promiscuous mRNA splicing under the control of AIRE in medullary thymic epithelial cells. Bioinformatics 2015, 31, 986–990. [Google Scholar] [CrossRef] [Green Version]
- Danan-Gotthold, M.; Guyon, C.; Giraud, M.; Levanon, E.Y.; Abramson, J. Extensive RNA editing and splicing increase immune self-representation diversity in medullary thymic epithelial cells. Genome Biol. 2016, 17, 219. [Google Scholar] [CrossRef] [Green Version]
- Raposo, B.; Merky, P.; Lundqvist, C.; Yamada, H.; Urbonaviciute, V.; Niaudet, C.; Viljanen, J.; Kihlberg, J.; Kyewski, B.; Ekwall, O.; et al. T cells specific for post-translational modifications escape intrathymic tolerance induction. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Doyle, H.A.; Mamula, M.J. Autoantigenesis: The evolution of protein modifications in autoimmune disease. Curr. Opin. Immunol. 2012, 24, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Shevyrev, D.V.; Kozlov, V.A. The role of homeostatic proliferation and SNP mutations in MHC genes in the development of rheumatoid arthritis. Ann. Russ. Acad. Med. Sci. 2020, 75, 638–646. [Google Scholar] [CrossRef]
- Aichinger, M.; Wu, C.; Nedjic, J.; Klein, L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J. Exp. Med. 2013, 210, 287–300. [Google Scholar] [CrossRef] [Green Version]
- Schmid, D.; Pypaert, M.; Münz, C. Antigen-Loading Compartments for Major Histocompatibility Complex Class II Molecules Continuously Receive Input from Autophagosomes. Immunity 2007, 26, 79–92. [Google Scholar] [CrossRef] [Green Version]
- Nedjic, J.; Aichinger, M.; Emmerich, J.; Mizushima, N.; Klein, L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 2008, 455, 396–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, A.J.; Simon, A.K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 2019, 19, 170–183. [Google Scholar] [CrossRef]
- Shi, L.; Hu, L.-H.; Li, Y.-R. Autoimmune regulator regulates autophagy in THP-1 human monocytes. Front. Med. China 2010, 4, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Hubert, F.-X.; Kinkel, S.A.; Davey, G.M.; Phipson, B.; Mueller, S.N.; Liston, A.; Proietto, A.I.; Cannon, P.Z.; Forehan, S.; Smyth, G.; et al. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood 2011, 118, 2462–2472. [Google Scholar] [CrossRef]
- Skogberg, G.; Lundberg, V.; Berglund, M.; Gudmundsdottir, J.; Telemo, E.; Lindgren, S.; Ekwall, O. Human thymic epithelial primary cells produce exosomes carrying tissue-restricted antigens. Immunol. Cell Biol. 2015, 93, 727–734. [Google Scholar] [CrossRef] [Green Version]
- Perry, J.S.A.; Russler-Germain, E.V.; Zhou, Y.W.; Purtha, W.; Cooper, M.L.; Choi, J.; Schroeder, M.A.; Salazar, V.; Egawa, T.; Lee, B.-C.; et al. Transfer of Cell-Surface Antigens by Scavenger Receptor CD36 Promotes Thymic Regulatory T Cell Receptor Repertoire Development and Allo-tolerance. Immunity 2018, 48, 923–936.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takaba, H.; Morishita, Y.; Tomofuji, Y.; Danks, L.; Nitta, T.; Komatsu, N.; Kodama, T.; Takayanagi, H. Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell 2015, 163, 975–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabrielsen, I.S.M.; Helgeland, H.; Akselsen, H.D.; Aass, H.C.D.; Sundaram, A.; Snowhite, I.V.; Pugliese, A.; Flåm, S.T.; Lie, B.A. Transcriptomes of antigen presenting cells in human thymus. PLoS ONE 2019, 14, e0218858. [Google Scholar] [CrossRef] [PubMed]
- Tomofuji, Y.; Takaba, H.; Suzuki, H.I.; Benlaribi, R.; Martinez, C.D.P.; Abe, Y.; Morishita, Y.; Okamura, T.; Taguchi, A.; Kodama, T.; et al. Chd4 choreographs self-antigen expression for central immune tolerance. Nat. Immunol. 2020, 21, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Derbinski, J.; Gäbler, J.; Brors, B.; Tierling, S.; Jonnakuty, S.; Hergenhahn, M.; Peltonen, L.; Walter, J.; Kyewski, B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 2005, 202, 33–45. [Google Scholar] [CrossRef]
- Baba, T.; Nakamoto, Y.; Mukaida, N. Crucial Contribution of Thymic Sirpα+ Conventional Dendritic Cells to Central Tolerance against Blood-Borne Antigens in a CCR2-Dependent Manner. J. Immunol. 2009, 183, 3053–3063. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Shin, J.-S. The Role of Dendritic Cells in Central Tolerance. Immune Netw. 2015, 15, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, H.; Matsumoto, T. Mechanisms of Tolerance Induction by Dendritic Cells In Vivo. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Perry, J.S.A.; Lio, C.J.; Kau, A.L.; Nutsch, K.; Yang, Z.; Gordon, J.I.; Murphy, K.M.; Hsieh, C.-S. Distinct Contributions of Aire and Antigen-Presenting-Cell Subsets to the Generation of Self-Tolerance in the Thymus. Immunity 2014, 41, 414–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starr, T.K.; Jameson, S.C.; Hogquist, K.A. Positive and Negative Selection of T Cells. Annu. Rev. Immunol. 2003, 21, 139–176. [Google Scholar] [CrossRef]
- Shevyrev, D.; Tereshchenko, V.; Kozlov, V. Immune Equilibrium Depends on the Interaction between Recognition and Presentation Landscapes. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Malchow, S.; Leventhal, D.S.; Lee, V.; Nishi, S.; Socci, N.D.; Savage, P.A. Aire Enforces Immune Tolerance by Directing Autoreactive T Cells into the Regulatory T Cell Lineage. Immunity 2016, 44, 1102–1113. [Google Scholar] [CrossRef] [Green Version]
- Daniely, D.; Kern, J.; Cebula, A.; Ignatowicz, L. Diversity of TCRs on Natural Foxp3+ T Cells in Mice Lacking Aire Expression. J. Immunol. 2010, 184, 6865–6873. [Google Scholar] [CrossRef] [Green Version]
- Ogishi, M.; Yotsuyanagi, H. Quantitative Prediction of the Landscape of T Cell Epitope Immunogenicity in Sequence Space. Front. Immunol. 2019, 10, 827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calis, J.J.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; De Silva, A.D.; Sette, A.; Keşmir, C.; Peters, B. Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef] [Green Version]
- Baccala, R.; Theofilopoulos, A.N. The new paradigm of T-cell homeostatic proliferation-induced autoimmunity. Trends Immunol. 2005, 26, 5–8. [Google Scholar] [CrossRef]
- Shevyrev, D.; Tereshchenko, V.; Blinova, E.; Knauer, N.; Pashkina, E.; Sizikov, A.; Kozlov, V. Regulatory T Cells Fail to Suppress Fast Homeostatic Proliferation In Vitro. Life 2021, 11, 245. [Google Scholar] [CrossRef]
- Shevyrev, D.; Tereshchenko, V.; Manova, O.; Kozlov, V. Homeostatic proliferation as a physiological process and a risk factor for autoimmune pathology. AIMS Allergy Immunol. 2021, 5, 18–32. [Google Scholar] [CrossRef]
- Goronzy, J.J.; Weyand, C.M. Immune aging and autoimmunity. Cell. Mol. Life Sci. 2012, 69, 1615–1623. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.-J.; Lee, J.K.; Shin, O.S. Aging and the Immune System: The Impact of Immunosenescence on Viral Infection, Immunity and Vaccine Immunogenicity. Immune Netw. 2019, 19, e37. [Google Scholar] [CrossRef]
- Izraelson, M.; Metsger, M.; Davydov, A.N.; Shagina, I.A.; Dronina, M.A.; Obraztsova, A.S.; Miskevich, D.A.; Mamedov, I.Z.; Volchkova, L.N.; Nakonechnaya, T.O.; et al. Distinct organization of adaptive immunity in the long-lived rodent Spalax galili. Nat. Aging 2021, 1, 179–189. [Google Scholar] [CrossRef]
- Shevyrev, D.; Blinova, E.A.; Kozlov, V.A. The influence of humoral factors of homeostatistic proliferation on t-regulatory cells in vitro. Bull. Sib. Med. 2019, 18, 286–293. [Google Scholar] [CrossRef]
- Wortel, I.M.N.; Keşmir, C.; de Boer, R.J.; Mandl, J.N.; Textor, J. Is T Cell Negative Selection a Learning Algorithm? Cells 2020, 9, 690. [Google Scholar] [CrossRef] [Green Version]
- Sng, J.; Ayoglu, B.; Chen, J.W.; Schickel, J.-N.; Ferre, E.M.N.; Glauzy, S.; Romberg, N.; Hoenig, M.; Cunningham-Rundles, C.; Utz, P.J.; et al. AIRE expression controls the peripheral selection of autoreactive B cells. Sci. Immunol. 2019, 4, eaav6778. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.S.B.; Braun, S.; Husebye, E.S.; Oftedal, B.E. B Cells and Autoantibodies in AIRE Deficiency. Biomedicines 2021, 9, 1274. [Google Scholar] [CrossRef]
- Koivula, T.-T.; Laakso, S.M.; Niemi, H.J.; Kekäläinen, E.; Laine, P.; Paulin, L.; Auvinen, P.; Arstila, T.P. Clonal Analysis of Regulatory T Cell Defect in Patients with Autoimmune Polyendocrine Syndrome Type 1 Suggests Intrathymic Impairment. Scand. J. Immunol. 2017, 86, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Bacchetta, R.; Weinberg, K. Thymic origins of autoimmunity—lessons from inborn errors of immunity. Semin. Immunopathol. 2021, 43, 65–83. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shevyrev, D.; Tereshchenko, V.; Kozlov, V.; Sennikov, S. Phylogeny, Structure, Functions, and Role of AIRE in the Formation of T-Cell Subsets. Cells 2022, 11, 194. https://doi.org/10.3390/cells11020194
Shevyrev D, Tereshchenko V, Kozlov V, Sennikov S. Phylogeny, Structure, Functions, and Role of AIRE in the Formation of T-Cell Subsets. Cells. 2022; 11(2):194. https://doi.org/10.3390/cells11020194
Chicago/Turabian StyleShevyrev, Daniil, Valeriy Tereshchenko, Vladimir Kozlov, and Sergey Sennikov. 2022. "Phylogeny, Structure, Functions, and Role of AIRE in the Formation of T-Cell Subsets" Cells 11, no. 2: 194. https://doi.org/10.3390/cells11020194






