Super-Enhancers Dysregulations in Hematological Malignancies
Abstract
1. Introduction
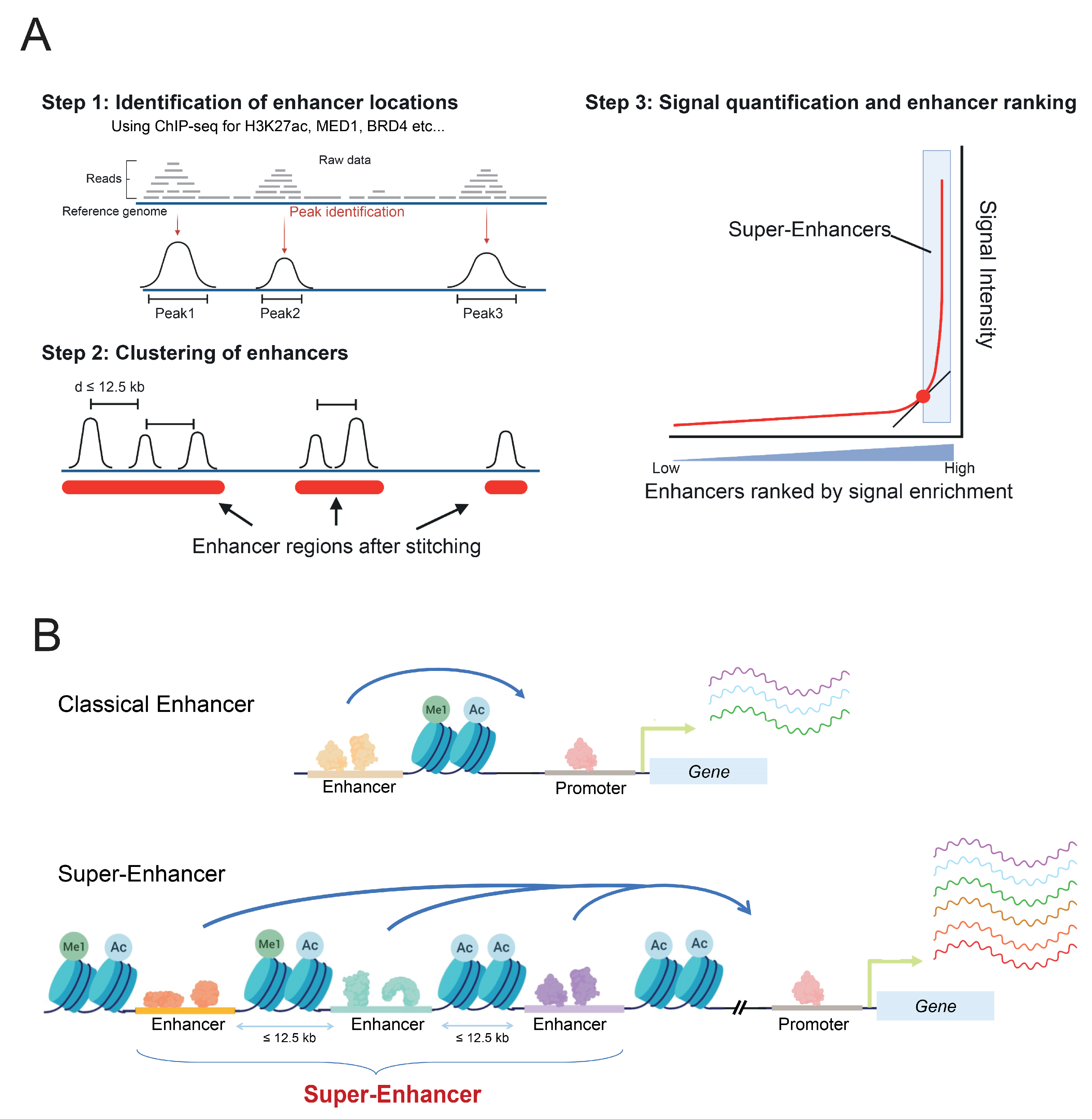
2. Enhancers and Super-Enhancers
2.1. Characteristics
2.2. Modes of Action in Gene Regulation
3. Enhancers and Super-Enhancers in Hematological Malignancies: Enhancerophathies
3.1. Point Mutations and Indels
3.2. Focal Amplification of Super-Enhancers
3.3. Translocations Involving Super-Enhancers
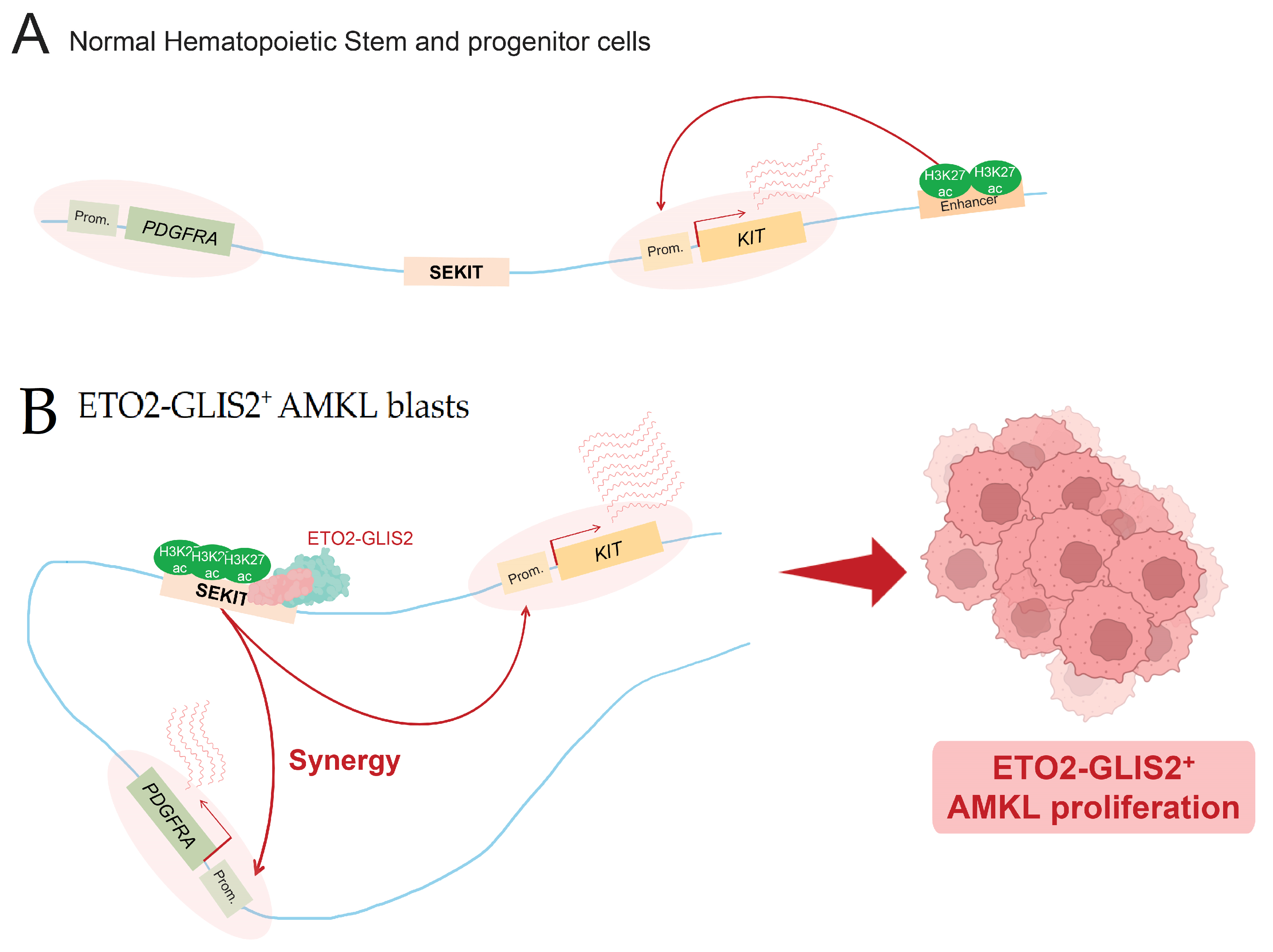
3.4. Super-Enhancer Hijacking
3.5. Viral Oncogenes Activation of SE
4. Targeting Oncogenic Super-Enhancers in Cancer Therapy: An Effective Approach?
5. Limitations of Super-Enhancer Targeting
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Longo, D.L.; Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar]
- Shah, M.Y.; Licht, J. DNMT3A mutations in acute myeloid leukemia. Nat. Genet. 2011, 43, 289–290. [Google Scholar] [CrossRef]
- Nakao, M.; Yokota, S.; Iwai, T.; Kaneko, H.; Horiike, S.; Kashima, K.; Sonoda, Y.; Fujimoto, T.; Misawa, S. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia 1996, 10, 1911–1918. [Google Scholar] [PubMed]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R.; Ding, L.; Dooling, D.J.; Larson, D.E.; McLellan, M.D.; Chen, K.; Koboldt, D.C.; Fulton, R.S.; Delehaunty, K.D.; McGrath, S.D.; et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009, 361, 1058–1066. [Google Scholar] [CrossRef]
- Ward, P.S.; Patel, J.; Wise, D.R.; Abdel-Wahab, O.; Bennett, B.D.; Coller, H.A.; Cross, J.R.; Fantin, V.R.; Hedvat, C.V.; Perl, A.E.; et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010, 17, 225–234. [Google Scholar] [CrossRef] [PubMed]
- King-Underwood, L.; Renshaw, J.; Pritchard-Jones, K. Mutations in the Wilms’ tumor gene WT1 in leukemias. Blood 1996, 87, 2171–2179. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, Q.; Zi, J.; Ma, J.; Song, H.; Tian, Y.; McGrath, M.; Song, C.; Ge, Z. Mutations in EZH2 are associated with poor prognosis for patients with myeloid neoplasms. Genes Dis. 2019, 6, 276–281. [Google Scholar] [CrossRef]
- Lund, K.; Adams, P.D.; Copland, M. EZH2 in normal and malignant hematopoiesis. Leukemia 2014, 28, 44–49. [Google Scholar] [CrossRef]
- Kirito, K.; Sakoe, K.; Shinoda, D.; Takiyama, Y.; Kaushansky, K.; Komatsu, N. A novel RUNX1 mutation in familial platelet disorder with propensity to develop myeloid malignancies. Haematologica 2008, 93, 155–156. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Mardis, E.R.; Wilson, R.K. DNA sequencing of a cytogenetically normal acute myeloid leukemia genome. Nature 2008, 456, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Vlierberghe, P.V.; Patel, J.; Abdel-Wahab, O.; Hedvat, C.V.; Balbin, M.; Nicolas, C.; Payer, A.R.; Fernandez, H.F.; Tallman, M.S.; Paietta, E. PHF6 mutations in adult acute myeloid leukemia. Leukemia 2011, 25, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn-Khosrovani, S.B.; Spensberger, D.; de Knegt, Y.; Tang, M.; Lowenberg, B.; Delwel, R. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene 2005, 24, 4129–4137. [Google Scholar]
- Patel, J.P.; Gönen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef]
- Delhommeau, F.; Dupont, S.; Della Valle, V.; James, C.; Trannoy, S.; Massé, A.; Kosmider, O.; Le Couedic, J.-P.; Robert, F.; Alberdi, A.; et al. Mutation inTET2in myeloid cancers. N. Engl. J. Med. 2009, 360, 2289–2301. [Google Scholar] [CrossRef]
- Elhefni, A.M.; Ahmad, E.I.; Gawish, H.H.; Al Azizi, N.M. The prognostic impact of K-RAS mutations in adult acute myeloid leukemia patients treated with high-dose cytarabine. OncoTargets Ther. 2011, 4, 115–121. [Google Scholar] [CrossRef][Green Version]
- Ding, L.; Lew, T.J.; DiPersio, J.F. Clonal evolution in relapsed acute myeloid leukemia revealed by whole genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef]
- Martens, J.; Stunnenberg, H.G. The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS Lett. 2010, 584, 2662–2669. [Google Scholar] [CrossRef]
- Alcalay, M.; Meani, N.; Gelmetti, V.; Fantozzi, A.; Fagioli, M.; Orleth, A.; Riganelli, D.; Sebastiani, C.; Cappelli, E.; Casciari, C.; et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 2003, 112, 1751–1761. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.G.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [PubMed]
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef]
- Mora, A.; Sandve, G.K.; Gabrielsen, O.S.; Eskeland, R. In the loop: Promoter–enhancer interactions and bioinformatics. Brief. Bioinform. 2016, 17, 980–995. [Google Scholar] [CrossRef] [PubMed]
- Barolo, S. Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 2012, 34, 135–141. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Witte, S.; Bradley, A.; Enright, A.J.; Muljo, S.A. High-density P300 enhancers control cell state transitions. BMC Genom. 2015, 16, 903. [Google Scholar] [CrossRef]
- Pott, S.; Lieb, J.D. What are super-enhancers? Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef]
- Wu, M.; Shen, J. From super-enhancer non-coding RNA to immune checkpoint: Frameworks to functions. Front. Oncol. 2019, 9, 1307. [Google Scholar] [CrossRef]
- Bahr, C.; Von Paleske, L.; Uslu, V.V.; Remeseiro, S.; Takayama, N.; Ng, S.W.; Murison, A.; Langenfeld, K.; Petretich, M.; Scognamiglio, R.; et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 2018, 553, 515–520. [Google Scholar] [CrossRef]
- Yassin, M.; Aqaqe, N.; Yassin, A.A.; van Galen, P.; Kugler, E.; Bernstein, B.E.; Koren-Michowitz, M.; Canaani, J.; Nagler, A.; Lechman, E.; et al. A novel method for detecting the cellular stemness state in normal and leukemic human hematopoietic cells can predict disease outcome and drug sensitivity. Leukemia 2019, 33, 2061–2077. [Google Scholar] [CrossRef] [PubMed]
- Thirant, C.; Ignacimouttou, C.; Lopez, C.K.; Diop, M.; Le Mouël, L.; Thiollier, C.; Siret, A.; Dessen, P.; Aid, Z.; Rivière, J.; et al. ETO2-GLIS2 hijacks transcriptional complexes to drive cellular identity and self-renewal in pediatric acute megakaryoblastic leukemia. Cancer Cell 2017, 31, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Orgilles, B.; Saldaña-Meyer, R.; Wang, E.; Trompouki, E.; Fassl, A.; Lau, S.; Mullenders, J.; Rocha, P.P.; Raviram, R.; Guillamot, M.; et al. MED12 regulates HSC-specific enhancers independently of mediator kinase activity to control hematopoiesis. Cell Stem Cell 2016, 19, 784–799. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Schmidt, S.C.; Jiang, S.; Willox, B.; Bernhardt, K.; Liang, J.; Johannsen, E.C.; Kharchenko, P.; Gewurz, B.E.; Kieff, E.; et al. Epstein-barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 2015, 17, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.R.; Abraham, B.J.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.D.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014, 346, 1373–1377. [Google Scholar] [CrossRef]
- de Laat, W.; Duboule, D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 2013, 502, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Pombo, A.; Dillon, N. Three-dimensional genome architecture: Players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015, 16, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Bulger, M.; Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 2011, 144, 327–339. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Dekker, J. The hierarchy of the 3D genome. Mol. Cell 2013, 49, 773–782. [Google Scholar] [CrossRef]
- Uhlmann, F. SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 2016, 17, 399–412. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 2017, 169, 930–944.e22. [Google Scholar] [CrossRef]
- Alipour, E.; Marko, J.F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012, 40, 11202–11212. [Google Scholar] [CrossRef]
- Kim, Y.; Shi, Z.; Zhang, H.; Finkelstein, I.J.; Yu, H. Human cohesin compacts DNA by loop extrusion. Science 2019, 366, 1345–1349. [Google Scholar] [CrossRef]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J.-M. DNA loop extrusion by human cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A.; Zhang, X. Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers. Epigenetics 2018, 13, 910–922. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.Y.; Willi, M.; Yoo, K.H.; Zeng, X.; Wang, C.; Metser, G.; Hennighausen, L. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016, 48, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Dukler, N.; Gulko, B.; Huang, Y.-F.; Siepel, A. Is a super-enhancer greater than the sum of its parts? Nat. Genet. 2017, 49, 2–3. [Google Scholar] [CrossRef]
- Hay, D.; Hughes, J.R.; Babbs, C.; Davies, J.; Graham, B.J.; Hanssen, L.; Kassouf, M.T.; Oudelaar, A.M.; Sharpe, J.A.; Suciu, M.; et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet. 2016, 48, 895–903. [Google Scholar] [CrossRef]
- Abraham, B.J.; Hnisz, D.; Wientraub, A.S.; Kwiatkowski, N.; Li, C.H.; Li, Z.; Weichert-Leahey, N.; Rahman, S.; Liu, Y.; Etchin, J.; et al. Small genomic insertions form enhancers that misregulate oncogenes. Nat. Commun. 2017, 8, 14385. [Google Scholar] [CrossRef]
- Hnisz, D.; Weintraub, A.S.; Day, D.S.; Valton, A.-L.; Bak, R.O.; Li, C.H.; Goldmann, J.; Lajoie, B.R.; Fan, Z.P.; Sigova, A.A.; et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 2016, 351, 1454–1458. [Google Scholar] [CrossRef] [PubMed]
- Flavahan, W.A.; Drier, Y.; Johnstone, S.E.; Hemming, M.L.; Tarjan, D.R.; Hegazi, E.; Shareef, S.J.; Javed, N.M.; Raut, C.P.; Eschle, B.K.; et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 2019, 575, 229–233. [Google Scholar] [CrossRef]
- Yoshida, K.; Toki, T.; Okuno, Y.; Kanezaki, R.; Shiraishi, Y.; Sato-Otsubo, A.; Sanada, M.; Park, M.-J.; Terui, K.; Suzuki, H.; et al. The landscape of somatic mutations in Down syndrome–related myeloid disorders. Nat. Genet. 2013, 45, 1293–1299. [Google Scholar] [CrossRef]
- Zhang, X.; Choi, P.; Francis, J.M.; Imielinski, M.; Watanabe, H.; Cherniack, A.D.; Meyerson, M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2015, 48, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Herranz, D.; Ambesi-Impiombato, A.; Palomero, T.; Schnell, S.A.; Belver, L.; Wendorff, A.A.; Xu, L.; Castillo-Martin, M.; Llobet-Navas, D.; Cardo, C.C.; et al. N-Me, a long range oncogenic enhancer in T-cell acute lymphoblastic leukemia. Nat. Med. 2014, 20, 1130–1137. [Google Scholar] [CrossRef]
- Shi, J.; Whyte, W.A.; Zepeda-Mendoza, C.J.; Milazzo, J.P.; Shen, C.; Roe, J.-S.; Minder, J.L.; Mercan, F.; Wang, E.; Eckersley-Maslin, M.A.; et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013, 27, 2648–2662. [Google Scholar] [CrossRef] [PubMed]
- Kubota, S.; Tokunaga, K.; Umezu, T.; Yokomizo-Nakano, T.; Sun, Y.; Oshima, M.; Tan, K.T.; Yang, H.; Kanai, A.; Iwanaga, E.; et al. Lineage-specific RUNX2 super-enhancer activates MYC and promotes the development of blastic plasmacytoid dendritic cell neoplasm. Nat. Commun. 2019, 10, 1653. [Google Scholar] [CrossRef]
- Gröschel, S.; Sanders, M.A.; Hoogenboezem, R.; de Wit, E.; Bouwman, B.A.M.; Erpelinck, C.; van der Velden, V.H.J.; Havermans, M.; Avellino, R.; van Lom, K.; et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014, 157, 369–381. [Google Scholar] [CrossRef]
- Walker, B.; Wardell, C.; Brioli, A.; Boyle, E.M.; Kaiser, M.; Begum, D.B.; Dahir, N.B.; Johnson, D.C.; Ross, F.M.; Davies, F.; et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014, 4, e191. [Google Scholar] [CrossRef]
- Affer, M.; Chesi, M.; Chen, W.-D.G.; Keats, J.J.; Demchenko, Y.N.; Roschke, A.V.; Van Wier, S.; Fonseca, R.; Bergsagel, P.L.; Kuehl, W.M. Promiscuous rearrangements of the MYC locus hijack enhancers and super-enhancers to dysregulate MYC Expression in multiple myeloma. Leukemia 2014, 28, 1725–1735. [Google Scholar] [CrossRef]
- Drier, Y.; Cotton, M.J.; Williamson, K.E.; Gillespie, S.; Ryan, R.; Kluk, M.J.; Carey, C.D.; Rodig, S.J.; Sholl, L.M.; Afrogheh, A.H.; et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat. Genet. 2016, 48, 265–272. [Google Scholar] [CrossRef]
- Katerndahl, C.D.; Heltemes-Harris, L.M.; Willette, M.J.; Henzler, C.M.; Frietze, S.; Yang, R.; Schjerven, H.; Silverstein, K.A.; Ramsey, L.B.; Hubbard, G.; et al. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat. Immunol. 2017, 18, 694–704. [Google Scholar] [CrossRef]
- Ottema, S.; Mulet-Lazaro, R.; Erpelinck-Verschueren, C.; van Herk, S.; Havermans, M.; Varea, A.A.; Vermeulen, M.; Beverloo, H.B.; Gröschel, S.; Haferlach, T.; et al. The leukemic oncogene EVI1 hijacks a MYC super-enhancer by CTCF-facilitated loops. Nat. Commun. 2021, 12, 5679. [Google Scholar] [CrossRef] [PubMed]
- Benbarche, S. Screening of ETO2-GLIS2 induced Super Enhancers identifies targetable cooperative dependencies in acute megakaryoblastic leukemia. Sci Adv. 2022, 8, eabg9455. [Google Scholar]
- Tian, Y.; Wang, G.; Hu, Q.; Xiao, X.; Chen, S. AML1/ETO trans-activates c-KIT expression through the long range interaction between promoter and intronic enhancer. J. Cell. Biochem. 2018, 119, 3706–3715. [Google Scholar] [CrossRef] [PubMed]
- Smeenk, L.; Ottema, S.; Mulet-Lazaro, R.; Ebert, A.; Havermans, M.; Varea, A.A.; Fellner, M.; Pastoors, D.; van Herk, S.; Erpelinck-Verschueren, C.; et al. Selective requirement of MYB for oncogenic hyperactivation of a translocated enhancer in leukemia. Cancer Discov. 2021, 11, 2868–2883. [Google Scholar] [CrossRef] [PubMed]
- Tulstrup, M.; Soerensen, M.; Hansen, J.W.; Gillberg, L.; Needhamsen, M.; Kaastrup, K.; Helin, K.; Christensen, K.; Weischenfeldt, J.; Grønbæk, K. TET2 mutations are associated with hypermethylation at key regulatory enhancers in normal and malignant hematopoiesis. Nat. Commun. 2021, 12, 6061. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; Berest, I.; Keβler, S.; Nishimura, K.; Simón-Carrasco, L.; Vassiliou, G.S.; Pedersen, M.T.; Christensen, J.; Zaugg, J.B.; Helin, K. TET2 binding to enhancers facilitates transcription factor recruitment in hematopoietic cells. Genome Res. 2019, 29, 564–575. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Jia, G.; Johansen, J.V.; Pedersen, M.T.; Rapin, N.; Bagger, F.O.; Porse, B.T.; Bernard, O.A.; Christensen, J.; Helin, K. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015, 29, 910–922. [Google Scholar] [CrossRef]
- Dooley, K.E.; Warburton, A.; McBride, A.A. Tandemly integrated HPV16 can form a Brd4-dependent super-enhancer-like element that drives transcription of viral oncogenes. mBio 2016, 7, e01446. [Google Scholar] [CrossRef]
- Weinstein, I.B.; Joe, A.K. Mechanisms of Disease: Oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat. Clin. Pr. Oncol. 2006, 3, 448–457. [Google Scholar] [CrossRef]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef]
- Chapuy, B.; McKeown, M.R.; Lin, C.Y.; Monti, S.; Roemer, M.G.M.; Qi, J.; Rahl, P.B.; Sun, H.H.; Yeda, K.T.; Doench, J.G.; et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 2013, 24, 777–790. [Google Scholar] [CrossRef]
- Hajmirza, A.; Emadali, A.; Gauthier, A.; Casasnovas, O.; Gressin, R.; Callanan, M.B. BET family protein BRD4: An emerging actor in NFκB signaling in inflammation and cancer. Biomedicines 2018, 6, 16. [Google Scholar] [CrossRef]
- Zuber, J.; Shi, J.; Wang, E.; Rappaport, A.R.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef] [PubMed]
- Mertz, J.A.; Conery, A.R.; Bryant, B.; Sandy, P.; Balasubramanian, S.; Mele, D.A.; Bergeron, L.; Sims, R.J. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 2011, 108, 16669–16674. [Google Scholar] [CrossRef] [PubMed]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 2018, 17, 164. [Google Scholar] [CrossRef]
- Nilson, K.A.; Guo, J.; Turek, M.E.; Brogie, J.E.; Delaney, E.; Luse, D.S.; Price, D.H. THZ1 reveals roles for Cdk7 in co-transcriptional capping and pausing. Mol. Cell 2015, 59, 576–587. [Google Scholar] [CrossRef] [PubMed]
- Sampathi, S.; Acharya, P.; Zhao, Y.; Wang, J.; Stengerl, K.R.; Liu, Q.; Savona, M.R.; Hiebert, S.W. The CDK7 inhibitor THZ1 alters RNA polymerase dynamics at the 5′ and 3′ ends of genes. Nucleic Acids Res. 2019, 47, 3921–3936. [Google Scholar] [CrossRef]
- Chipumuro, E.; Marco, E.; Christensen, C.L.; Kwiatkowski, N.; Zhang, T.; Hatheway, C.M.; Abraham, B.J.; Sharma, B.; Yeung, C.; Altabef, A.; et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 2014, 159, 1126–1139. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Elyada, E.; Pribluda, A.; Goldstein, R.E.; Morgenstern, Y.; Brachya, G.; Cojocaru, G.; Snir-Alkalay, I.; Burstain, I.; Haffner-Krausz, R.; Jung, S.; et al. CKIα ablation highlights a critical role for p53 in invasiveness control. Nature 2011, 470, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Minzel, W.; Venkatachalam, A.; Fink, A.; Hung, E.; Brachya, G.; Burstain, I.; Shaham, M.; Rivlin, A.; Omer, I.; Zinger, A.; et al. Small molecules co-targeting CKIα and the transcriptional kinases CDK7/9 control AML in preclinical models. Cell 2018, 175, 171–185.e25. [Google Scholar] [CrossRef] [PubMed]
- Rathert, P.; Roth, M.; Neumann, T.; Muerdter, F.; Roe, J.-S.; Muhar, M.; Deswal, S.; Cerny-Reiterer, S.; Peter, B.; Jude, J.; et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 2015, 525, 543–547. [Google Scholar] [CrossRef]
- Guo, L.; Li, J.; Zeng, H.; Guzman, A.G.; Li, T.; Lee, M.; Zhou, Y.; Goodell, M.A.; Stephan, C.; Davies, P.J.A.; et al. A combination strategy targeting enhancer plasticity exerts synergistic lethality against BETi-resistant leukemia cells. Nat. Commun. 2020, 11, 740. [Google Scholar] [CrossRef]
- Call, S.G.; Duren, R.P.; Panigrahi, A.K.; Nguyen, L.; Freire, P.R.; Grimm, S.L.; Coarfa, C.; Conneely, O.M. Targeting oncogenic super enhancers in MYC-dependent AML using a small molecule activator of NR4A nuclear receptors. Sci. Rep. 2020, 10, 2851. [Google Scholar] [CrossRef]
- Bakshi, S.; McKee, C.; Walker, K.; Brown, C.; Chaudhry, G.R. Toxicity of JQ1 in neuronal derivatives of human umbilical cord mesenchymal stem cells. Oncotarget 2018, 9, 33853–33864. [Google Scholar] [CrossRef]
- Alghamdi, S.; Khan, I.; Beeravolu, N.; McKee, C.; Thibodeau, B.; Wilson, G.; Chaudhry, G.R. BET protein inhibitor JQ1 inhibits growth and modulates WNT signaling in mesenchymal stem cells. Stem Cell Res. Ther. 2016, 7, 22. [Google Scholar] [CrossRef]
- Matzuk, M.M.; McKeown, M.R.; Filippakopoulos, P.; Li, Q.; Ma, L.; Agno, J.E.; Lemieux, M.E.; Picaud, S.; Yu, R.N.; Qi, J.; et al. Small-molecule inhibition of BRDT for male contraception. Cell 2012, 150, 673–684. [Google Scholar] [CrossRef]
- Berthon, C.; Raffoux, E.; Thomas, X.; Vey, N.; Gomez-Roca, C.; Yee, K.; Taussig, D.C.; Rezai, K.; Roumier, C.; Herait, P.; et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016, 3, e186–e195. [Google Scholar] [CrossRef]
- Sengupta, S.; George, R.E. Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer 2017, 3, 269–281. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belloucif, Y.; Lobry, C. Super-Enhancers Dysregulations in Hematological Malignancies. Cells 2022, 11, 196. https://doi.org/10.3390/cells11020196
Belloucif Y, Lobry C. Super-Enhancers Dysregulations in Hematological Malignancies. Cells. 2022; 11(2):196. https://doi.org/10.3390/cells11020196
Chicago/Turabian StyleBelloucif, Yannis, and Camille Lobry. 2022. "Super-Enhancers Dysregulations in Hematological Malignancies" Cells 11, no. 2: 196. https://doi.org/10.3390/cells11020196
APA StyleBelloucif, Y., & Lobry, C. (2022). Super-Enhancers Dysregulations in Hematological Malignancies. Cells, 11(2), 196. https://doi.org/10.3390/cells11020196






