Microtubular TRIM36 E3 Ubiquitin Ligase in Embryonic Development and Spermatogenesis
Abstract
:1. Introduction
2. Tripartite Motif 36 (TRIM36)
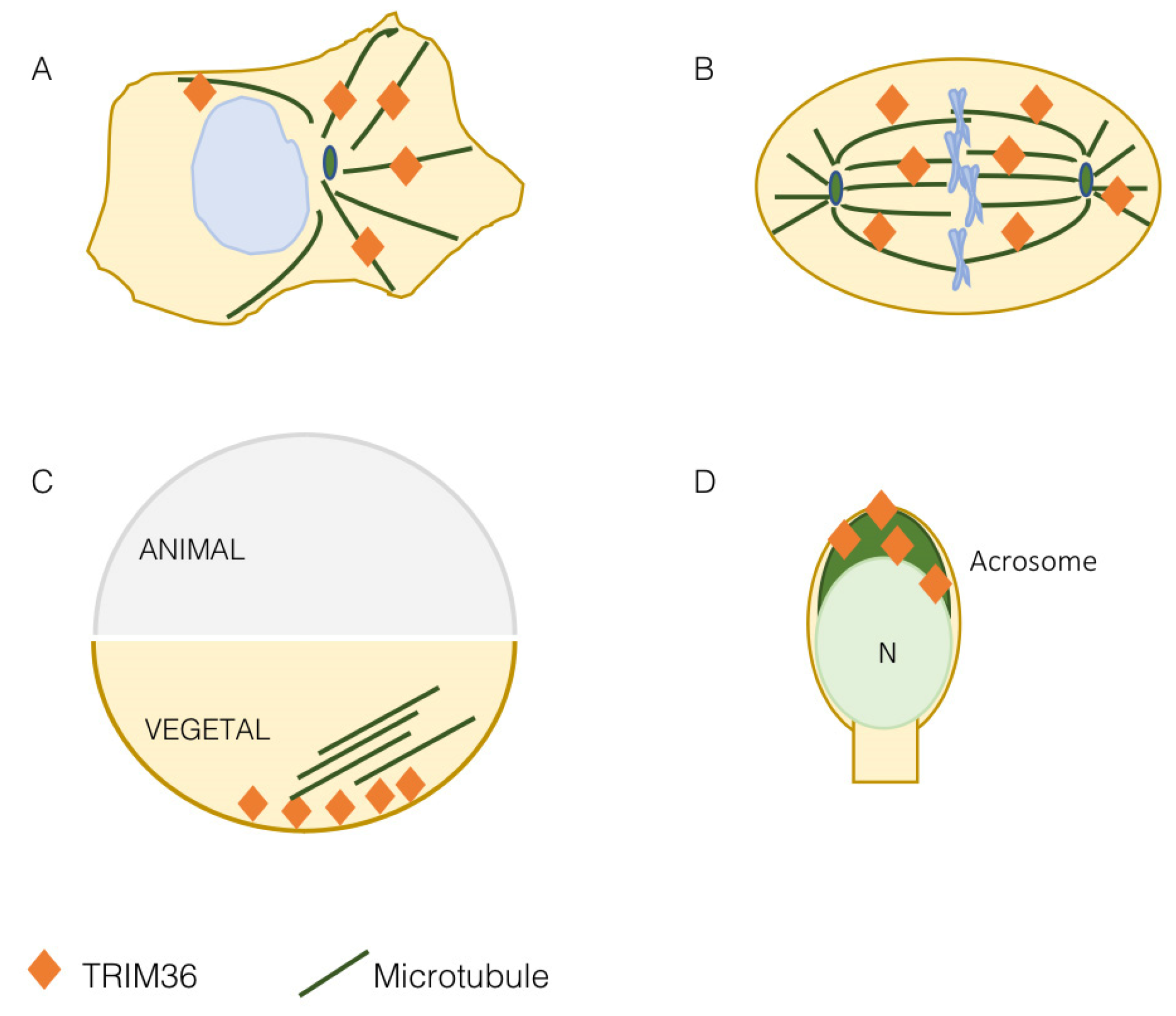
3. TRIM36 Associates with the Microtubules
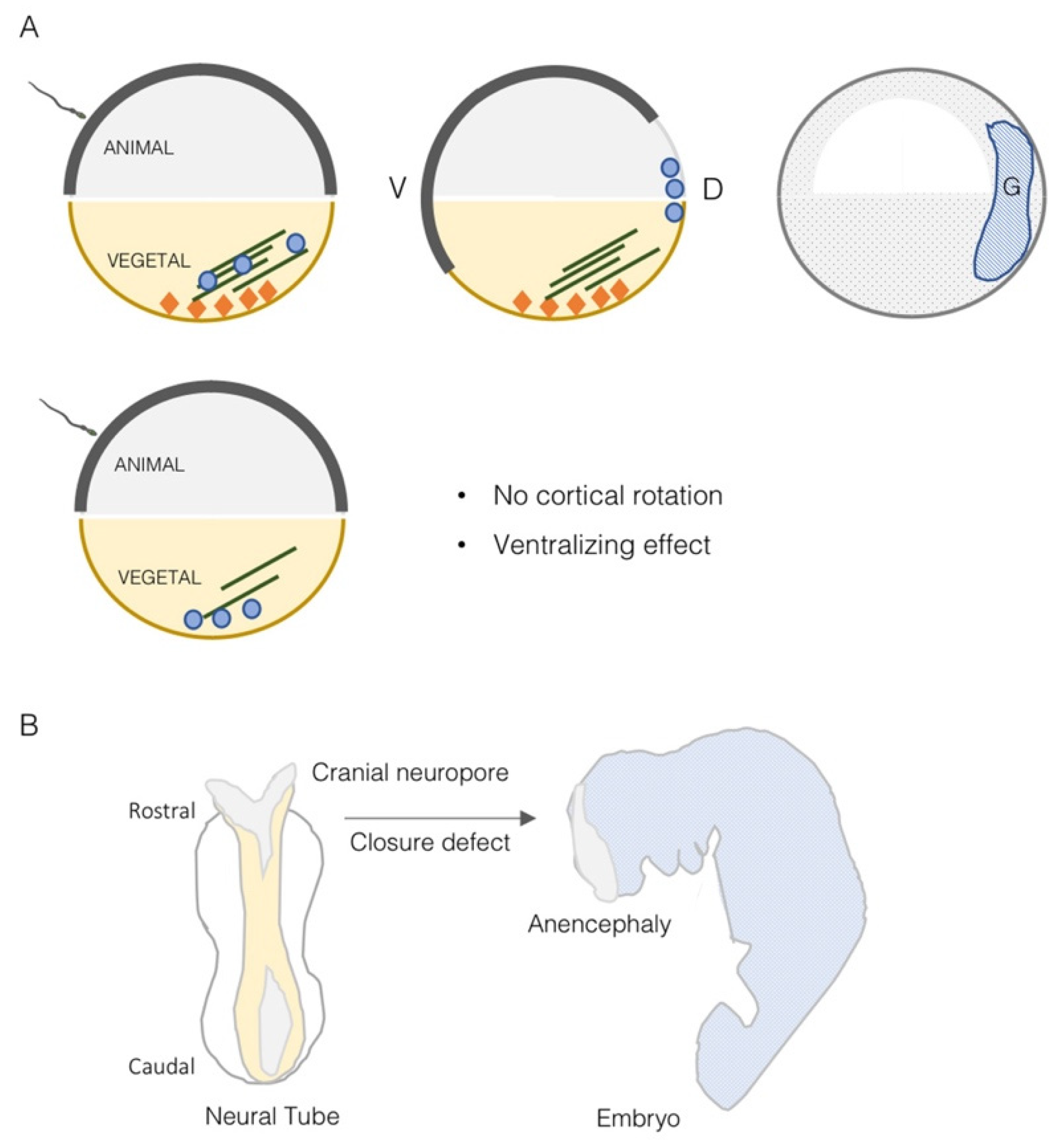
4. TRIM36 Function in Embryonic Development
5. TRIM36 Is Involved in Spermatogenesis
6. Involvement of TRIM36 in Cancer
7. TRIM36 Biochemical Roles
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, G.; Diez-Roux, G. TRIM/RBCC, a novel class of ’single protein RING finger’ E3 ubiquitin ligases. Bioessays 2005, 27, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [Green Version]
- Williams, F.P.; Haubrich, K.; Perez-Borrajero, C.; Hennig, J. Emerging RNA-binding roles in the TRIM family of ubiquitin ligases. Biol. Chem. 2019, 400, 1443–1464. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S. TRIM proteins and cancer. Nat. Rev. Cancer 2011, 11, 792–804. [Google Scholar] [CrossRef]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef]
- Di Rienzo, M.; Romagnoli, A.; Antonioli, M.; Piacentini, M.; Fimia, G.M. TRIM proteins in autophagy: Selective sensors in cell damage and innate immune responses. Cell Death Differ. 2020, 27, 887–902. [Google Scholar] [CrossRef]
- Mohammadi, A.; Abbasi, M.S.P.; Khorrami, S.; Khodamoradi, S.; Goldar, Z.M.; Ebrahimzadeh, F. The TRIM proteins in cancer: From expression to emerging regulatory mechanisms. Clin. Transl. Oncol. 2021; online ahead of print. [Google Scholar]
- Yang, W.; Gu, Z.; Zhang, H.; Hu, H. To TRIM the Immunity: From Innate to Adaptive Immunity. Front. Immunol. 2020, 11, 02157. [Google Scholar] [CrossRef]
- Ozato, K.; Shin, D.M.; Chang, T.H.; Morse, H.C., 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 2008, 8, 849–860. [Google Scholar] [CrossRef] [Green Version]
- Short, K.M.; Cox, T.C. Sub-classification of the rbcc/trim superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 2006, 281, 8970–8980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cainarca, S.; Messali, S.; Ballabio, A.; Meroni, G. Functional characterization of the Opitz syndrome gene product (midin): Evidence for homodimerization and association with microtubules throughout the cell cycle. Hum. Mol. Genet. 1999, 8, 1387–1396. [Google Scholar] [CrossRef] [Green Version]
- Schweiger, S.; Foerster, J.; Lehmann, T.; Suckow, V.; Muller, Y.A.; Walter, G.; Davies, T.; Porter, H.; van Bokhoven, H.; Lunt, P.W.; et al. The Opitz syndrome gene product, MID1, associates with microtubules. Proc. Natl. Acad. Sci. USA 1999, 96, 2794–2799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, G. TRIM E3 Ubiquitin Ligases in Rare Genetic Disorders. Adv. Exp. Med. Biol. 2020, 1233, 311–325. [Google Scholar] [PubMed]
- Sardiello, M.; Cairo, S.; Fontanella, B.; Ballabio, A.; Meroni, G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008, 8, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, K.; Tanaka, H.; Nishimune, Y. Haprin, a novel haploid germ cell-specific RING finger protein involved in the acrosome reaction. J. Biol. Chem. 2003, 278, 44417–44423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balint, I.; Muller, A.; Nagy, A.; Kovacs, G. Cloning and characterisation of the RBCC728/TRIM36 zinc-binding protein from the tumor suppressor gene region at chromosome 5q22.3. Gene 2004, 332, 45–50. [Google Scholar] [CrossRef]
- Kitamura, K.; Nishimura, H.; Nishimune, Y.; Tanaka, H. Identification of human HAPRIN potentially involved in the acrosome reaction. J. Androl. 2005, 26, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Bhat, V.K.; Tiwari, A.; Kodaganur, S.G.; Tontanahal, S.J.; Sarda, A.; Malini, K.V.; Kumar, A. A homozygous mutation in TRIM36 causes autosomal recessive anencephaly in an Indian family. Hum. Mol. Genet. 2017, 26, 1104–1114. [Google Scholar] [CrossRef] [Green Version]
- Cuykendall, T.N.; Houston, D.W. Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development 2009, 136, 3057–3065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyajima, N.; Maruyama, S.; Nonomura, K.; Hatakeyama, S. TRIM36 interacts with the kinetochore protein CENP-H and delays cell cycle progression. Biochem. Biophys. Res. Commun. 2009, 381, 383–387. [Google Scholar] [CrossRef]
- Gudimchuk, N.B.; McIntosh, J.R. Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 2021, 22, 777–795. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, T.; Matsushita, K.; Ishibashi, M.; Nezu, M.; Shimada, H.; Ochiai, T.; Yoda, K.; Nomura, F. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy. Cancer Res. 2005, 65, 4683–4689. [Google Scholar] [CrossRef] [Green Version]
- Olson, D.J.; Oh, D.; Houston, D.W. The dynamics of plus end polarization and microtubule assembly during Xenopus cortical rotation. Dev. Biol. 2015, 401, 249–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, W.; Jin, Z.; Lai, F.; Schwend, T.; Houston, D.W.; King, M.L.; Yang, J. Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development 2013, 140, 2334–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houston, D.W. Cortical rotation and messenger RNA localization in Xenopus axis formation. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 371–388. [Google Scholar] [CrossRef]
- Aoki, Y.; Tsujimura, A.; Kaseda, K.; Okabe, M.; Tokuhiro, K.; Ohta, T.; O’Bryan, M.K.; Okuda, H.; Kitamura, K.; Ogawa, Y.; et al. Haprin-deficient spermatozoa are incapable of in vitro fertilization. Mol. Reprod. Dev. 2020, 87, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Quaderi, N.A.; Schweiger, S.; Gaudenz, K.; Franco, B.; Rugarli, E.I.; Berger, W.; Feldman, G.J.; Volta, M.; Andolfi, G.; Gilgenkrantz, S.; et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat. Genet. 1997, 17, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Geetha, T.S.; Michealraj, K.A.; Kabra, M.; Kaur, G.; Juyal, R.C.; Thelma, B.K. Targeted deep resequencing identifies MID2 mutation for X-linked intellectual disability with varied disease severity in a large kindred from India. Hum. Mutat. 2014, 35, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Bhide, P.; Sagoo, G.S.; Moorthie, S.; Burton, H.; Kar, A. Systematic review of birth prevalence of neural tube defects in India. Birth Defects Res. A Clin. Mol. Teratol. 2013, 97, 437–443. [Google Scholar] [CrossRef]
- Copp, A.J.; Stanier, P.; Greene, N.D. Neural tube defects: Recent advances, unsolved questions, and controversies. Lancet Neurol. 2013, 12, 799–810. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Hara, Y.; Takagi, C.; Yamamoto, T.S.; Ueno, N. MID1 and MID2 are required for Xenopus neural tube closure through the regulation of microtubule organization. Development 2010, 137, 2329–2339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancioni, A.; Pizzo, M.; Fontanella, B.; Ferrentino, R.; Napolitano, L.M.; de Leonibus, E.; Meroni, G. Lack of Mid1, the mouse ortholog of the Opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J. Neurosci. 2010, 30, 2880–2887. [Google Scholar] [CrossRef]
- Griswold, M.D. Spermatogenesis: The Commitment to Meiosis. Physiol. Rev. 2016, 96, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramalho-Santos, J.; Schatten, G.; Moreno, R.D. Control of membrane fusion during spermiogenesis and the acrosome reaction. Biol. Reprod. 2002, 67, 1043–1051. [Google Scholar] [CrossRef] [Green Version]
- Plooster, M.; Menon, S.; Winkle, C.C.; Urbina, F.L.; Monkiewicz, C.; Phend, K.D.; Weinberg, R.J.; Gupton, S.L. TRIM9-dependent ubiquitination of DCC constrains kinase signaling, exocytosis, and axon branching. Mol. Biol. Cell 2017, 28, 2374–2385. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Li, D.; Xu, C. Downregulation of Col1a1 induces differentiation in mouse spermatogonia. Asian J. Androl. 2012, 14, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.M.; Saini, N.; Singh, M.K.; Manik, R.; Singla, S.K.; Palta, P.; Chauhan, M.S. Testicular cell-conditioned medium supports embryonic stem cell differentiation toward germ lineage and to spermatocyte- and oocyte-like cells. Theriogenology 2016, 86, 715–729. [Google Scholar] [CrossRef]
- Shah, S.M.; Saini, N.; Ashraf, S.; Singh, M.K.; Manik, R.S.; Singla, S.K.; Palta, P.; Chauhan, M.S. Cumulus cell-conditioned medium supports embryonic stem cell differentiation to germ cell-like cells. Reprod. Fertil. Dev. 2017, 29, 679–693. [Google Scholar] [CrossRef] [Green Version]
- Endo, T.; Mikedis, M.M.; Nicholls, P.K.; Page, D.C.; de Rooij, D.G. Retinoic Acid and Germ Cell Development in the Ovary and Testis. Biomolecules 2019, 9, 775. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.M.; Singla, S.K.; Palta, P.; Manik, R.S.; Chauhan, M.S. Retinoic acid induces differentiation of buffalo (Bubalus bubalis) embryonic stem cells into germ cells. Gene 2017, 631, 54–67. [Google Scholar] [CrossRef]
- Huang, Y.L.; Zhang, P.F.; Fu, Q.; He, W.T.; Xiao, K.; Zhang, M. Novel targets identified by integrated proteomic and phosphoproteomic analysis in spermatogenesis of swamp buffalo (Bubalus bubalis). Sci. Rep. 2020, 10, 15659. [Google Scholar] [CrossRef] [PubMed]
- Schlatt, S.; Ehmcke, J. Regulation of spermatogenesis: An evolutionary biologist’s perspective. Semin. Cell Dev. Biol. 2014, 29, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.Y.; Hirano, T.; Shibata, S.; Seki, N.M.; Kitajima, R.; Sedohara, A.; Siomi, M.C.; Sasaki, E.; Siomi, H.; Imamura, M.; et al. Gene expression ontogeny of spermatogenesis in the marmoset uncovers primate characteristics during testicular development. Dev. Biol. 2015, 400, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, S.; Qin, C.; Bao, M.; Cheng, G.; Liu, B.; Shao, P.; Lv, Q.; Song, N.; Hua, L.; et al. TRIM36, a novel androgen-responsive gene, enhances anti-androgen efficacy against prostate cancer by inhibiting MAPK/ERK signaling pathways. Cell Death Dis. 2018, 9, 155. [Google Scholar] [CrossRef]
- Fujimura, T.; Takahashi, S.; Urano, T.; Takayama, K.; Sugihara, T.; Obinata, D.; Yamada, Y.; Kumagai, J.; Kume, H.; Ouchi, Y.; et al. Expression of androgen and estrogen signaling components and stem cell markers to predict cancer progression and cancer-specific survival in patients with metastatic prostate cancer. Clin. Cancer Res. 2014, 20, 4625–4635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, N.; Yamada, Y.; Takayama, K.I.; Fujimura, T.; Takahashi, S.; Kume, H.; Inoue, S. Androgen-responsive tripartite motif 36 enhances tumor-suppressive effect by regulating apoptosis-related pathway in prostate cancer. Cancer Sci. 2018, 109, 3840–3852. [Google Scholar] [CrossRef] [Green Version]
- Ren, F.; Wang, D.B.; Li, T.; Chen, Y.H.; Li, Y. Identification of differentially methylated genes in the malignant transformation of ovarian endometriosis. J. Ovarian Res. 2014, 7, 73. [Google Scholar] [CrossRef] [Green Version]
- Zhan, W.; Han, T.; Zhang, C.; Xie, C.; Gan, M.; Deng, K.; Fu, M.; Wang, J.B. TRIM59 Promotes the Proliferation and Migration of Non-Small Cell Lung Cancer Cells by Upregulating Cell Cycle Related Proteins. PLoS ONE 2015, 10, e0142596. [Google Scholar] [CrossRef]
- Olsson, M.; Beck, S.; Kogner, P.; Martinsson, T.; Caren, H. Genome-wide methylation profiling identifies novel methylated genes in neuroblastoma tumors. Epigenetics 2016, 11, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Wang, J.; Ling, L.; Xue, D.; Wang, S.; Zhao, Y. Screening and analysis of breast cancer genes regulated by the human mammary microenvironment in a humanized mouse model. Oncol. Lett. 2016, 12, 5261–5268. [Google Scholar] [CrossRef] [Green Version]
- Man, Z.; Chen, T.; Zhu, Z.; Zhang, H.; Ao, L.; Xi, L.; Zhou, J.; Tang, Z. High expression of TRIM36 is associated with radiosensitivity in gastric cancer. Oncol. Lett. 2019, 17, 4401–4408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Sun, W.; Qiao, G.; Zhao, B.; Liu, X.; Zhu, F. The Expression of Tripartite Motif Protein 36 and beta-Catenin Correlates with the Prognosis of Esophageal Cancer. Gastroenterol. Res. Pract. 2020, 2020, 7641761. [Google Scholar] [CrossRef] [PubMed]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015, 16, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [Green Version]

| Type of Cancer | TRIM36 Role | Ref. |
|---|---|---|
| Prostate cancer | TRIM36 mRNA expression is upregulated in patients’ tissues; no mutations gene detected. | [17] |
| TRIM36 protein expression is negatively associated with patients’ poor prognosis. In the prostate cancer cell line PCa TRIM36 reduces cell growth. Smaller tumors generated in TRIM36 overexpressing PC3 cells mouse xenograft. TRIM36 is an androgen-responsive gene and enhances the efficacy of anti-androgen drugs in PCa cell lines. | [45] | |
| Increased TRIM36 mRNA expression is associated with no PSA recurrence patients. | [46] | |
| Patients with low TRIM36 have poor progression-free survival. TRIM36 overexpression in LNCaP cells suppresses cell proliferation and migration and induces apoptosis by increasing Bax and TNFSF10. | [47] | |
| Endometriosis | TRIM36 is hypermethylated in patients’ tissues. | [48] |
| Non-Small Cell Lung | TRIM36 mRNA is downregulated in NSCLC cell line. | [49] |
| Neuroblastoma | TRIM36 is hypermethylated in patients’ tissues. | [50] |
| Breast cancer | TRIM36 mRNA is downregulated in humanized breast and conventional subcutaneous mouse models. | [51] |
| Gastric cancer | TRIM36 mRNA expression is increased in patients receiving radiotherapy. | [52] |
| Esophageal cancer | Patients with low levels of TRIM36 (mRNA and protein) display larger tumor size, advanced stage, and lymph node metastasis. | [53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mascaro, M.; Lages, I.; Meroni, G. Microtubular TRIM36 E3 Ubiquitin Ligase in Embryonic Development and Spermatogenesis. Cells 2022, 11, 246. https://doi.org/10.3390/cells11020246
Mascaro M, Lages I, Meroni G. Microtubular TRIM36 E3 Ubiquitin Ligase in Embryonic Development and Spermatogenesis. Cells. 2022; 11(2):246. https://doi.org/10.3390/cells11020246
Chicago/Turabian StyleMascaro, Martina, Inês Lages, and Germana Meroni. 2022. "Microtubular TRIM36 E3 Ubiquitin Ligase in Embryonic Development and Spermatogenesis" Cells 11, no. 2: 246. https://doi.org/10.3390/cells11020246







