PFN1 Inhibits Myogenesis of Bovine Myoblast Cells via Cdc42-PAK/JNK
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. Antibody and Inhibitors
2.3. qRT-PCR Analysis
2.4. Western Blot Analysis
2.5. Plasmids, siRNAs and Transfection
2.6. Co-IP
2.7. Cdc42 Activity Assay
2.8. Statistical Analysis
3. Results
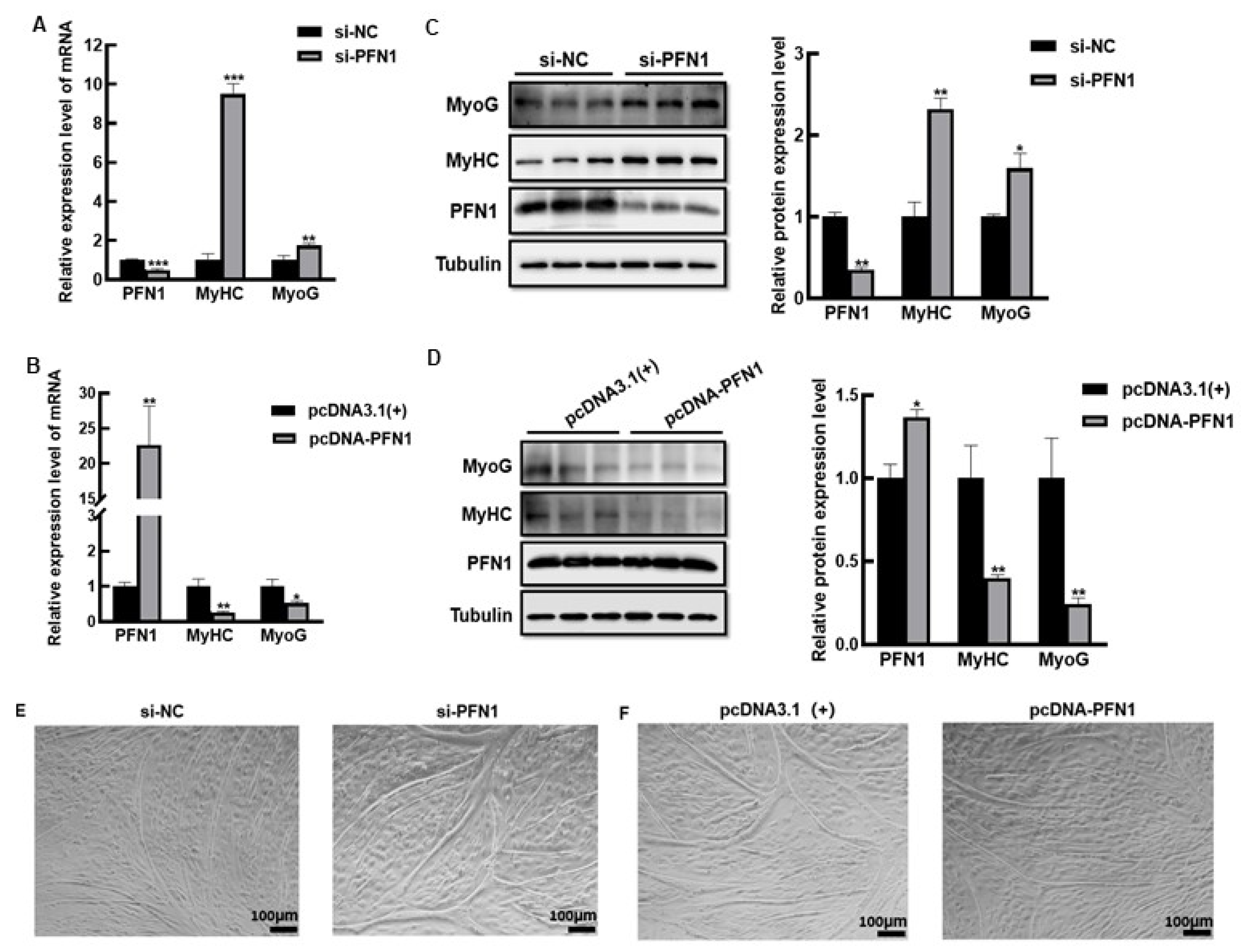
3.1. PFN1 Inhibits Bovine Myoblasts Differentiation
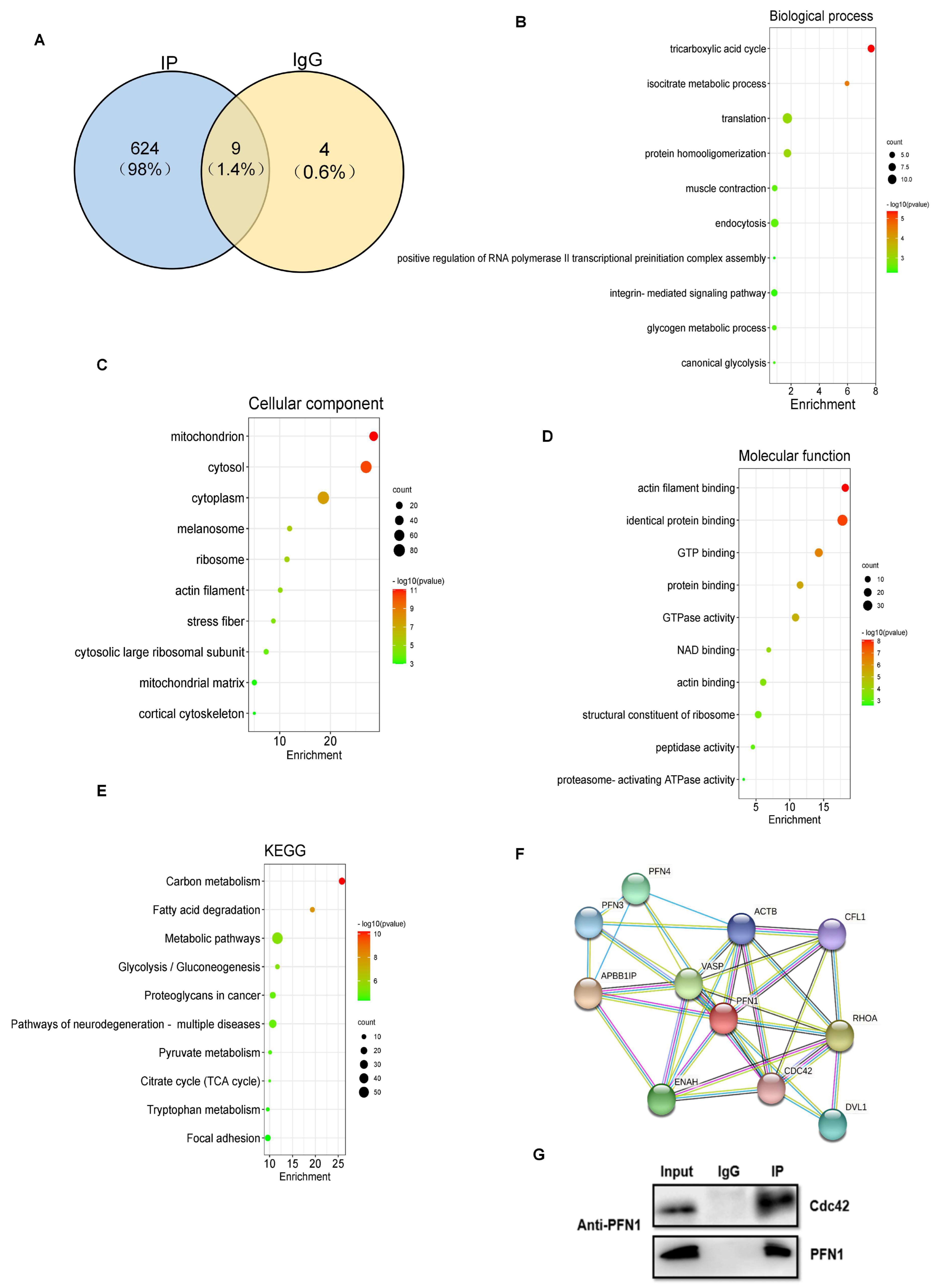
3.2. Analysis of PFN1 Potentially Binding Proteins
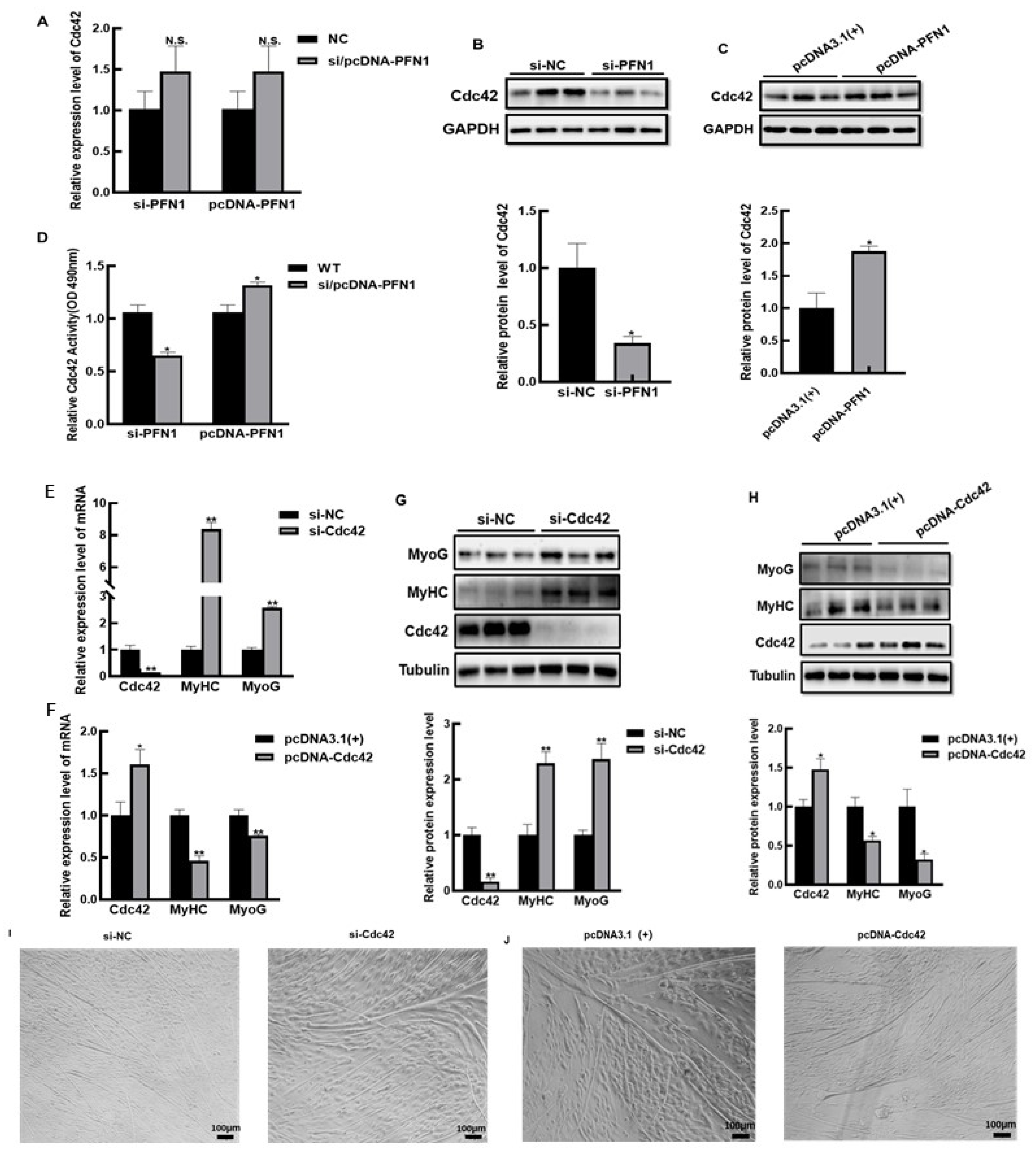
3.3. PFN1 Inhibits the Differentiation of Bovine Myoblasts via Cdc42
3.4. PFN1-Cdc42 Negatively Regulates the Differentiation of Myoblasts via Activating PAK/JNK Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sousa-Victor, P.; García-Prat, L.; Muñoz-Cánoves, P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2021, 23, 204–226. [Google Scholar] [CrossRef]
- Singh, K.; Cassano, M.; Planet, E.; Sebastian, S.; Jang, S.M.; Sohi, G.; Faralli, H.; Choi, J.; Youn, H.D.; Dilworth, F.J.; et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes Dev. 2015, 29, 513–525. [Google Scholar] [CrossRef] [Green Version]
- Wagers, A.J.; Conboy, I.M. Cellular and molecular signatures of muscle regeneration: Current concepts and controversies in adult myo-genesis. Cell 2005, 122, 659–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Zhang, L.; Guo, Y.; Liu, X.; Song, Y.; Li, X.; Ding, X.; Guo, H. A novel lncRNA promotes myogenesis of bovine skeletal muscle satellite cells via PFN1-RhoA/Rac1. J. Cell. Mol. Med. 2021, 25, 5988–6005. [Google Scholar] [CrossRef]
- Metzler, W.J.; Farmer, B.T.; Constantine, K.L.; Friedrichs, M.S.; Lavoie, T.; Mueller, L. Refined solution structure of human pro-filin I. Protein Sci. 1995, 4, 450–459. [Google Scholar] [CrossRef] [Green Version]
- Carlsson, L.; Nyström, L.-E.; Sundkvist, I.; Markey, F.; Lindberg, U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 1977, 115, 465–483. [Google Scholar] [CrossRef]
- Zou, L.; Ding, Z.; Roy, P. Profilin-1 overexpression inhibits proliferation of MDA-MB-231 breast cancer cells partly through p27kip1upregulation. J. Cell. Physiol. 2010, 223, 623–629. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Bae, Y.H.; Wells, A.; Roy, P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast can-cer cells. J. Cell Physiol. 2009, 218, 436–443. [Google Scholar] [CrossRef] [Green Version]
- Moustafa-Bayoumi, M.; Alhaj, M.A.; El-Sayed, O.; Wisel, S.; Chotani, M.A.; Abouelnaga, Z.A.; Hassona, M.D.H.; Rigatto, K.; Morris, M.; Nuovo, G.; et al. Vascular Hypertrophy and Hypertension Caused by Transgenic Overexpression of Profilin 1. J. Biol. Chem. 2007, 282, 37632–37639. [Google Scholar] [CrossRef] [Green Version]
- Freischmidt, A.; Schöpflin, M.; Feiler, M.S.; Fleck, A.K.; Ludolph, A.C.; Weishaupt, J.H. Profilin 1 with the amyotrophic lateral scle-rosis associated mutation T109M displays unaltered actin binding and does not affect the actin cytoskeleton. BMC Neurosci. 2015, 16, 77. [Google Scholar] [CrossRef]
- Del Poggetto, E.; Bemporad, F.; Tatini, F.; Chiti, F. Mutations of Profilin-1 Associated with Amyotrophic Lateral Sclerosis Pro-mote Aggregation Due to Structural Changes of Its Native State. ACS Chem. Biol. 2015, 10, 2553–2563. [Google Scholar] [CrossRef]
- Del Poggetto, E.; Gori, L.; Chiti, F. Biophysical analysis of three novel profilin-1 variants associated with amyotrophic lateral sclerosis indicates a correlation between their aggregation propensity and the structural features of their globular state. Biol. Chem. 2016, 397, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Fallini, C.; Ticozzi, N.; Keagle, P.J.; Sapp, P.C.; Piotrowska, K.; Lowe, P.; Koppers, M.; McKenna-Yasek, D.; Baron, D.M.; et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 2012, 488, 499–503. [Google Scholar] [CrossRef] [Green Version]
- Melendez, J.; Grogg, M.; Zheng, Y. Signaling Role of Cdc42 in Regulating Mammalian Physiology. J. Biol. Chem. 2011, 286, 2375–2381. [Google Scholar] [CrossRef] [Green Version]
- Vasyutina, E.; Martarelli, B.; Brakebusch, C.; Wende, H.; Birchmeier, C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc. Natl. Acad. Sci. USA 2009, 106, 8935–8940. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.S.; Bae, G.U.; Yi, M.J.; Yang, Y.J.; Oh, J.E.; Takaesu, G.; Zhou, Y.T.; Low, B.C.; Krauss, R.S. A Cdo-Bnip-2-Cdc42 signaling pathway regulates p38alpha/beta MAPK activity and myogen-ic differentiation. J. Cell Biol. 2008, 182, 497–507. [Google Scholar] [CrossRef] [Green Version]
- Meriane, M.; Roux, P.; Primig, M.; Fort, P.; Gauthier-Rouvière, C. Critical Activities of Rac1 and Cdc42Hs in Skeletal Myogenesis: Antagonistic Effects of JNK and p38 Pathways. Mol. Biol. Cell 2000, 11, 2513–2528. [Google Scholar] [CrossRef] [Green Version]
- Sanders, L.C.; Matsumura, F.; Bokoch, G.M.; de Lanerolle, P. Inhibition of Myosin Light Chain Kinase by p21-Activated Kinase. Science 1999, 283, 2083–2085. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, Y.M.; Zhang, W.R.; Liu, X.F.; Li, X.; Ding, X.B.; Guo, H. The role of microRNA-1 and microRNA-206 in the prolifera-tion and differentiation of bovine skeletal muscle satellite cells. In Vitro Cell Dev. Biol. Anim. 2016, 52, 27–34. [Google Scholar] [CrossRef]
- Ding, Z.; Bae, Y.H.; Roy, P. Molecular insights on context-specific role of profilin-1 in cell migration. Cell Adhes. Migr. 2012, 6, 442–534. [Google Scholar] [CrossRef]
- Wang, Z.; Shi, Z.; Zhang, L.; Zhang, H.; Zhang, Y. Profilin 1, negatively regulated by microRNA-19a-3p, serves as a tumor suppressor in hu-man hepatocellular carcinoma. Pathol. Res. Pract. 2019, 215, 499–505. [Google Scholar] [CrossRef]
- Janke, J.; Schlüter, K.; Jandrig, B.; Theile, M.; Kölble, K.; Arnold, W.; Grinstein, E.; Schwartz, A.; Estevéz-Schwarz, L.; Schlag, P.M.; et al. Suppression of Tumorigenicity in Breast Cancer Cells by the Microfilament Protein Profilin 1. J. Exp. Med. 2000, 191, 1675–1686. [Google Scholar] [CrossRef] [Green Version]
- Zou, L.; Jaramillo, M.; Whaley, D.; Wells, A.; Panchapakesa, V.; Das, T.; Roy, P. Profilin-1 is a negative regulator of mammary car-cinoma aggressiveness. Br. J. Cancer 2007, 97, 1361–1371. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.; Lambrechts, A.; Parepally, M.; Roy, P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J. Cell Sci. 2006, 119, 4127–4137. [Google Scholar] [CrossRef] [Green Version]
- Witke, W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004, 14, 461–469. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Cdc42—The centre of polarity. J. Cell Sci. 2004, 117, 1291–1300. [Google Scholar] [CrossRef] [Green Version]
- Cerione, R.A.; Zheng, Y. The Dbl family of oncogenes. Curr. Opin. Cell Biol. 1996, 8, 216–222. [Google Scholar] [CrossRef]
- Cerione, R.A. Cdc42: New roads to travel. Trends Cell Biol. 2004, 14, 127–132. [Google Scholar] [CrossRef]
- Rodríguez-Fdez, S.; Bustelo, X.R. Rho GTPases in Skeletal Muscle Development and Homeostasis. Cells 2021, 10, 2984. [Google Scholar] [CrossRef]
- Simionescu-Bankston, A.; Leoni, G.; Wang, Y.; Pham, P.P.; Ramalingam, A.; DuHadaway, J.B.; Faundez, V.; Nusrat, A.; Prendergast, G.C.; Pavlath, G.K. The N-BAR domain protein, Bin3, regulates Rac1- and Cdc42-dependent processes in myogenesis. Dev. Biol. 2013, 382, 160–171. [Google Scholar] [CrossRef]
- Gallo, R.; Serafini, M.; Castellani, L.; Falcone, G.; Alemà, S. Distinct effects of Rac1 on differentiation of primary avian my-oblasts. Mol. Biol. Cell 1999, 10, 3137–3150. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Yang, G.; Chen, Y.; Zhou, X.; Chen, H.; Li, M.; Yu, K.; Zhang, X.; Xie, S.; Zhang, Y.; et al. CEP2 attenuates myoblast differenti-ation but does not affect proliferation. Int. J. Biol. Sci. 2015, 11, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Bagci, H.; Sriskandarajah, N.; Robert, A.; Boulais, J.; Elkholi, I.E.; Tran, V.; Lin, Z.Y.; Thibault, M.P.; Dubé, N.; Faubert, D.; et al. Map-ping the proximity interaction network of the Rho-family GTPases reveals signalling pathways and regulatory mecha-nisms. Nat. Cell Biol. 2020, 22, 120–134. [Google Scholar] [CrossRef]
- Hill, C.S.; Wynne, J.; Treisman, R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 1995, 81, 1159–1170. [Google Scholar] [CrossRef] [Green Version]
- Perona, R.; Montaner, S.; Saniger, L.; Sánchez-Pérez, I.; Bravo, R.; Lacal, J.C. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997, 11, 463–475. [Google Scholar] [CrossRef] [Green Version]
- Miralles, F.; Posern, G.; Zaromytidou, A.-I.; Treisman, R. Actin Dynamics Control SRF Activity by Regulation of Its Coactivator MAL. Cell 2003, 113, 329–342. [Google Scholar] [CrossRef] [Green Version]
- Teramoto, H.; Coso, O.A.; Miyata, H.; Igishi, T.; Miki, T.; Gutkind, J.S. Signaling from the Small GTP-binding Proteins Rac1 and Cdc42 to the c-Jun N-terminal Kinase/Stress-activated Protein Kinase Pathway. J. Biol. Chem. 1996, 271, 27225–27228. [Google Scholar] [CrossRef] [Green Version]
- Heller, H.; Gredinger, E.; Bengal, E. Rac1 inhibits myogenic differentiation by preventing the complete withdrawal of my-oblasts from the cell cycle. J. Biol. Chem. 2001, 276, 37307–37316. [Google Scholar] [CrossRef] [Green Version]
- van Leeuwen, F.N.; van Delft, S.; Kain, H.E.; van der Kammen, R.A.; Collard, J.G. Rac regulates phosphorylation of the myosin-II heavy chain, actinomyosin disassembly and cell spreading. Nat. Cell Biol. 1999, 1, 242–248. [Google Scholar] [CrossRef]
- Rudel, T.; Zenke, F.T.; Chuang, T.H.; Bokoch, G.M. p21-activated kinase (PAK) is required for Fas-induced JNK activation in Jurkat cells. J. Immunol. 1998, 160. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zi, J.; Xu, J.; Luo, J.; Yang, X.; Zhen, Z.; Li, X.; Hu, D.; Guo, Y.; Guo, H.; Ding, X.; et al. PFN1 Inhibits Myogenesis of Bovine Myoblast Cells via Cdc42-PAK/JNK. Cells 2022, 11, 3188. https://doi.org/10.3390/cells11203188
Zi J, Xu J, Luo J, Yang X, Zhen Z, Li X, Hu D, Guo Y, Guo H, Ding X, et al. PFN1 Inhibits Myogenesis of Bovine Myoblast Cells via Cdc42-PAK/JNK. Cells. 2022; 11(20):3188. https://doi.org/10.3390/cells11203188
Chicago/Turabian StyleZi, Jingjing, Jing Xu, Jintang Luo, Xu Yang, Zhen Zhen, Xin Li, Debao Hu, Yiwen Guo, Hong Guo, Xiangbin Ding, and et al. 2022. "PFN1 Inhibits Myogenesis of Bovine Myoblast Cells via Cdc42-PAK/JNK" Cells 11, no. 20: 3188. https://doi.org/10.3390/cells11203188
APA StyleZi, J., Xu, J., Luo, J., Yang, X., Zhen, Z., Li, X., Hu, D., Guo, Y., Guo, H., Ding, X., & Zhang, L. (2022). PFN1 Inhibits Myogenesis of Bovine Myoblast Cells via Cdc42-PAK/JNK. Cells, 11(20), 3188. https://doi.org/10.3390/cells11203188





