The Role of N6-Methyladenosine Modification in Microvascular Dysfunction
Abstract
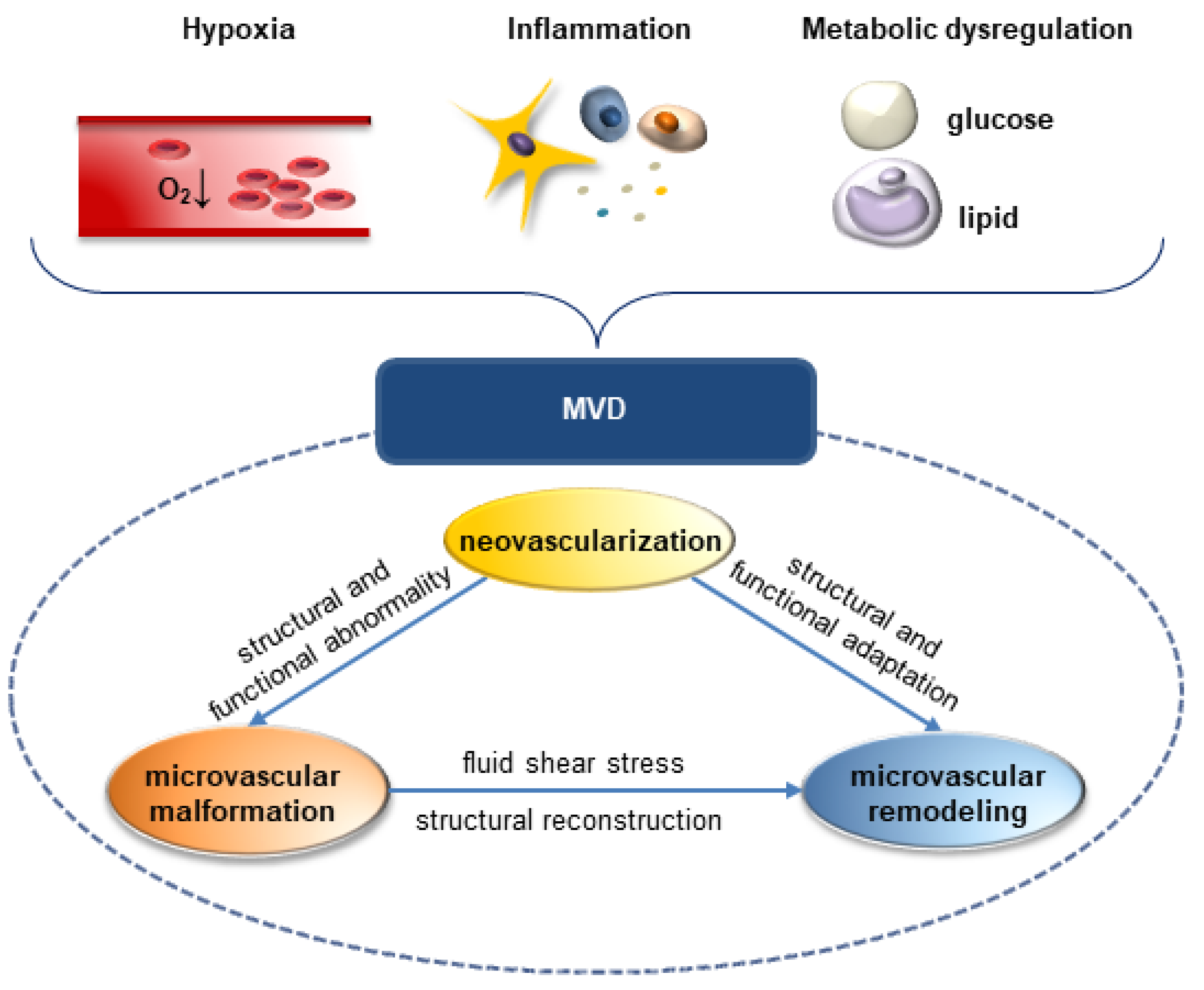
:1. Introduction
2. RNA m6A Methylation
3. M6A Modifications in Pathological Neovascularization
3.1. M6A Modifications in Hypoxia-Related Neovascularization
3.2. M6A Modifications in Inflammation-Related Pathological Neovascularization
3.3. Others
4. M6A Modifications in Microvascular Malformation
4.1. M6A Modifications in Hypoxia-Related Microvascular Malformation
4.2. M6A Modifications in Inflammation-Related Microvascular Malformation
4.3. Others
5. M6A Modifications in Microvascular Remodeling
5.1. M6A Modifications in Hypoxia-Related Microvascular Remodeling
5.2. M6A Modifications in Inflammation-Related Microvascular Remodeling
5.3. M6A Modifications in Metabolism-Related Microvascular Remodeling
6. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Horton, W.B.; Barrett, E.J. Microvascular Dysfunction in Diabetes Mellitus and Cardiometabolic Disease. Endocr. Rev. 2021, 42, 29–55. [Google Scholar] [CrossRef] [PubMed]
- Marseglia, A.; Fratiglioni, L.; Kalpouzos, G.; Wang, R.; Bäckman, L.; Xu, W. Prediabetes and diabetes accelerate cognitive decline and predict microvascular lesions: A population-based cohort study. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2019, 15, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Suhrs, H.E.; Schroder, J.; Bové, K.B.; Mygind, N.D.; Frestad, D.; Michelsen, M.M.; Lange, T.; Gustafsson, I.; Kastrup, J.; Prescott, E. Inflammation, non-endothelial dependent coronary microvascular function and diastolic function-Are they linked? PLoS ONE 2020, 15, e0236035. [Google Scholar] [CrossRef] [PubMed]
- Virdis, A.; Colucci, R.; Bernardini, N.; Blandizzi, C.; Taddei, S.; Masi, S. Microvascular Endothelial Dysfunction in Human Obesity: Role of TNF-α. J. Clin. Endocrinol. Metab. 2019, 104, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, B.W.; Marsch, E.; Treps, L.; Baes, M.; Carmeliet, P. Endothelial cell metabolism in health and disease: Impact of hypoxia. EMBO J. 2017, 36, 2187–2203. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m(6)A modification in physiology and disease. Cell Death Dis. 2020, 11, 960. [Google Scholar] [CrossRef] [PubMed]
- Garbo, S.; Zwergel, C.; Battistelli, C. m6A RNA methylation and beyond—The epigenetic machinery and potential treatment options. Drug Discov. Today 2021, 26, 2559–2574. [Google Scholar] [CrossRef]
- Wang, X.; Ma, R.; Zhang, X.; Cui, L.; Ding, Y.; Shi, W.; Guo, C.; Shi, Y. Crosstalk between N6-methyladenosine modification and circular RNAs: Current understanding and future directions. Mol. Cancer 2021, 20, 121. [Google Scholar] [CrossRef]
- Gu, Y.; Wu, X.; Zhang, J.; Fang, Y.; Pan, Y.; Shu, Y.; Ma, P. The evolving landscape of N(6)-methyladenosine modification in the tumor microenvironment. Mol. Ther. 2021, 29, 1703–1715. [Google Scholar] [CrossRef]
- Hu, B.B.; Wang, X.Y.; Gu, X.Y.; Zou, C.; Gao, Z.J.; Zhang, H.; Fan, Y. N(6)-methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: Roles, mechanisms, and applications. Mol. Cancer 2019, 18, 178. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 49, 7239–7255. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Li, L.; Luo, E.; Hou, J.; Yan, G.; Wang, D.; Qiao, Y.; Tang, C. Role of m6A RNA methylation in cardiovascular disease (Review). Int. J. Mol. Med. 2020, 46, 1958–1972. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.B.; Wang, Z.; Yang, C. The m(6)A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 2021, 20, 61. [Google Scholar] [CrossRef]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Chai, P.; Jia, R.; Fan, X. Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol. Cancer 2020, 19, 78. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, T.; Wu, D.; Min, Z.; Tan, J.; Yu, B. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J. Exp. Clin. Cancer Res. 2020, 39, 203. [Google Scholar] [CrossRef]
- Zimna, A.; Kurpisz, M. Hypoxia-Inducible Factor-1 in Physiological and Pathophysiological Angiogenesis: Applications and Therapies. BioMed Res. Int. 2015, 2015, 549412. [Google Scholar] [CrossRef] [Green Version]
- Cho, W.C.; Jour, G.; Aung, P.P. Role of angiogenesis in melanoma progression: Update on key angiogenic mechanisms and other associated components. Semin. Cancer Biol. 2019, 59, 175–186. [Google Scholar] [CrossRef]
- Shmakova, A.; Frost, M.; Batie, M.; Kenneth, N.S.; Rocha, S. PBRM1 Cooperates with YTHDF2 to Control HIF-1α Protein Translation. Cells 2021, 10, 1425. [Google Scholar] [CrossRef]
- Jiang, L.; Li, Y.; He, Y.; Wei, D.; Yan, L.; Wen, H. Knockdown of m6A Reader IGF2BP3 Inhibited Hypoxia-Induced Cell Migration and Angiogenesis by Regulating Hypoxia Inducible Factor-1α in Stomach Cancer. Front. Oncol. 2021, 11, 711207. [Google Scholar] [CrossRef]
- Panneerdoss, S.; Eedunuri, V.K.; Yadav, P.; Timilsina, S.; Rajamanickam, S.; Viswanadhapalli, S.; Abdelfattah, N.; Onyeagucha, B.C.; Cui, X.; Lai, Z.; et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 2018, 4, eaar8263. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M.; et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 2019, 18, 163. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Niu, Y.; Wan, A.; Chen, D.; Liang, H.; Chen, X.; Sun, L.; Zhan, S.; Chen, L.; Cheng, C.; et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020, 39, e103181. [Google Scholar] [CrossRef]
- Yao, M.D.; Jiang, Q.; Ma, Y.; Liu, C.; Zhu, C.Y.; Sun, Y.N.; Shan, K.; Ge, H.M.; Zhang, Q.Y.; Zhang, H.Y.; et al. Role of METTL3-Dependent N(6)-Methyladenosine mRNA Modification in the Promotion of Angiogenesis. Mol. Ther. 2020, 28, 2191–2202. [Google Scholar] [CrossRef]
- Shan, K.; Zhou, R.M.; Xiang, J.; Sun, Y.N.; Liu, C.; Lv, M.W.; Xu, J.J. FTO regulates ocular angiogenesis via m(6)A-YTHDF2-dependent mechanism. Exp. Eye Res. 2020, 197, 108107. [Google Scholar] [CrossRef]
- Qi, Y.; Yao, R.; Zhang, W.; Cui, Q. KAT1 triggers YTHDF2-mediated ITGB1 mRNA instability to alleviate the progression of diabetic retinopathy. Pharmacol. Res. 2021, 170, 105713. [Google Scholar] [CrossRef]
- Chang, G.; Shi, L.; Ye, Y.; Shi, H.; Zeng, L.; Tiwary, S.; Huse, J.T.; Huo, L.; Ma, L.; Ma, Y.; et al. YTHDF3 Induces the Translation of m(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020, 38, 857–871.e7. [Google Scholar] [CrossRef]
- Wang, H.; Deng, Q.; Lv, Z.; Ling, Y.; Hou, X.; Chen, Z.; Dinglin, X.; Ma, S.; Li, D.; Wu, Y.; et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol. Cancer 2019, 18, 181. [Google Scholar] [CrossRef]
- Rong, Z.X.; Li, Z.; He, J.J.; Liu, L.Y.; Ren, X.X.; Gao, J.; Mu, Y.; Guan, Y.D.; Duan, Y.M.; Zhang, X.P.; et al. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Front. Oncol. 2019, 9, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Kang, Y.; Wang, L.; Huff, S.; Tang, R.; Hui, H.; Agrawal, K.; Gonzalez, G.M.; Wang, Y.; Patel, S.P.; et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA 2020, 117, 20159–20170. [Google Scholar] [CrossRef]
- Tirpe, A.A.; Gulei, D.; Ciortea, S.M.; Crivii, C.; Berindan-Neagoe, I. Hypoxia: Overview on Hypoxia-Mediated Mechanisms with a Focus on the Role of HIF Genes. Int J. Mol. Sci. 2019, 20, 6140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serocki, M.; Bartoszewska, S.; Janaszak-Jasiecka, A.; Ochocka, R.J.; Collawn, J.F.; Bartoszewski, R. miRNAs regulate the HIF switch during hypoxia: A novel therapeutic target. Angiogenesis 2018, 21, 183–202. [Google Scholar] [CrossRef] [Green Version]
- Taylor, C.T.; Colgan, S.P. Regulation of immunity and inflammation by hypoxia in immunological niches. Nat. Rev. Immunol. 2017, 17, 774–785. [Google Scholar] [CrossRef]
- Lee, J.W.; Ko, J.; Ju, C.; Eltzschig, H.K. Hypoxia signaling in human diseases and therapeutic targets. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Watts, E.R.; Walmsley, S.R. Inflammation and Hypoxia: HIF and PHD Isoform Selectivity. Trends Mol. Med. 2019, 25, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Knutson, A.K.; Williams, A.L.; Boisvert, W.A.; Shohet, R.V. HIF in the heart: Development, metabolism, ischemia, and atherosclerosis. J. Clin. Investig. 2021, 131, e137557. [Google Scholar] [CrossRef]
- Maynard, M.A.; Evans, A.J.; Hosomi, T.; Hara, S.; Jewett, M.A.; Ohh, M. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005, 19, 1396–1406. [Google Scholar] [CrossRef]
- Wang, Y.J.; Yang, B.; Lai, Q.; Shi, J.F.; Peng, J.Y.; Zhang, Y.; Hu, K.S.; Li, Y.Q.; Peng, J.W.; Yang, Z.Z.; et al. Reprogramming of m(6)A epitranscriptome is crucial for shaping of transcriptome and proteome in response to hypoxia. RNA Biol. 2021, 18, 131–143. [Google Scholar] [CrossRef]
- Jiang, L.; Yin, M.; Wei, X.; Liu, J.; Wang, X.; Niu, C.; Kang, X.; Xu, J.; Zhou, Z.; Sun, S.; et al. Bach1 Represses Wnt/β-Catenin Signaling and Angiogenesis. Circ. Res. 2015, 117, 364–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Liu, C.H.; Huang, S.; Chen, J. Wnt Signaling in vascular eye diseases. Prog. Retin. Eye Res. 2019, 70, 110–133. [Google Scholar] [CrossRef]
- Yang, N.; Wang, T.; Li, Q.; Han, F.; Wang, Z.; Zhu, R.; Zhou, J. HBXIP drives metabolic reprogramming in hepatocellular carcinoma cells via METTL3-mediated m6A modification of HIF-1α. J. Cell Physiol. 2021, 236, 3863–3880. [Google Scholar] [CrossRef]
- Zhang, Q.; Cao, X. Epigenetic Remodeling in Innate Immunity and Inflammation. Annu. Rev. Immunol. 2021, 39, 279–311. [Google Scholar] [CrossRef]
- Ma, Y.S.; Shi, B.W.; Guo, J.H.; Liu, J.B.; Yang, X.L.; Xin, R.; Shi, Y.; Zhang, D.D.; Lu, G.X.; Jia, C.Y.; et al. microRNA-320b suppresses HNF4G and IGF2BP2 expression to inhibit angiogenesis and tumor growth of lung cancer. Carcinogenesis 2021, 42, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Lawton, M.T.; Rutledge, W.C.; Kim, H.; Stapf, C.; Whitehead, K.J.; Li, D.Y.; Krings, T.; terBrugge, K.; Kondziolka, D.; Morgan, M.K.; et al. Brain arteriovenous malformations. Nat. Rev. Dis. Primers 2015, 1, 15008. [Google Scholar] [CrossRef] [PubMed]
- Sadick, M.; Müller-Wille, R.; Wildgruber, M.; Wohlgemuth, W.A. Vascular Anomalies (Part I): Classification and Diagnostics of Vascular Anomalies. Rofo Fortschr. Auf Dem Geb. der Rontgenstrahlen der Nukl. 2018, 190, 825–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darden, J.; Payne, L.B.; Zhao, H.; Chappell, J.C. Excess vascular endothelial growth factor-A disrupts pericyte recruitment during blood vessel formation. Angiogenesis 2019, 22, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, Y.; Jiang, X.; Peng, M.; Liu, Y.; Mo, Y.; Ren, D.; Hua, Y.; Yu, B.; Zhou, Y.; et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol. Cancer 2021, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Liu, Y.; Xu, Z.; Zhang, H.; Zhang, H.; Zhang, C.; Chang, Z.; Lu, X.; Li, Z.; Luo, C.; et al. RNA m6A methylation promotes the formation of vasculogenic mimicry in hepatocellular carcinoma via Hippo pathway. Angiogenesis 2021, 24, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Suo, L.; Liu, C.; Zhang, Q.Y.; Yao, M.D.; Ma, Y.; Yao, J.; Jiang, Q.; Yan, B. METTL3-mediated N (6)-methyladenosine modification governs pericyte dysfunction during diabetes-induced retinal vascular complication. Theranostics 2022, 12, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Deng, L.; Xu, G. METTL14 promotes glomerular endothelial cell injury and diabetic nephropathy via m6A modification of α-klotho. Mol. Med. 2021, 27, 106. [Google Scholar] [CrossRef]
- Tu, J.; Li, Y.; Hu, Z.; Chen, Z. Radiosurgery inhibition of the Notch signaling pathway in a rat model of arteriovenous malformations. J. Neurosurg. 2014, 120, 1385–1396. [Google Scholar] [CrossRef] [Green Version]
- ZhuGe, Q.; Zhong, M.; Zheng, W.; Yang, G.Y.; Mao, X.; Xie, L.; Chen, G.; Chen, Y.; Lawton, M.T.; Young, W.L.; et al. Notch-1 signalling is activated in brain arteriovenous malformations in humans. Brain 2009, 132, 3231–3241. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.J.; Xue, Y.; Huo, R.; Yan, Z.; Xu, H.; Li, H.; Wang, J.; Zhang, Q.; Cao, Y.; Zhao, J.Z. N6-methyladenosine methyltransferase METTL3 affects the phenotype of cerebral arteriovenous malformation via modulating Notch signaling pathway. J. Biomed. Sci. 2020, 27, 62. [Google Scholar] [CrossRef]
- Wang, L.J.; Xue, Y.; Li, H.; Huo, R.; Yan, Z.; Wang, J.; Xu, H.; Wang, J.; Cao, Y.; Zhao, J.Z. Wilms’ tumour 1-associating protein inhibits endothelial cell angiogenesis by m6A-dependent epigenetic silencing of desmoplakin in brain arteriovenous malformation. J. Cell Mol. Med. 2020, 24, 4981–4991. [Google Scholar] [CrossRef] [Green Version]
- Karar, J.; Maity, A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Parial, R.; Li, H.; Li, J.; Archacki, S.; Yang, Z.; Wang, I.Z.; Chen, Q.; Xu, C.; Wang, Q.K. Role of epigenetic m(6) A RNA methylation in vascular development: Mettl3 regulates vascular development through PHLPP2/mTOR-AKT signaling. FASEB J. 2021, 35, e21465. [Google Scholar] [CrossRef]
- Tian, C.; Huang, Y.; Li, Q.; Feng, Z.; Xu, Q. Mettl3 Regulates Osteogenic Differentiation and Alternative Splicing of Vegfa in Bone Marrow Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 551. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Hua, W.; Huang, X.; Chen, Y.; Zhang, J.; Li, G. Regulatory Role of RNA N(6)-Methyladenosine Modification in Bone Biology and Osteoporosis. Front. Endocrinol. 2019, 10, 911. [Google Scholar] [CrossRef]
- Jiang, W.; Zhu, P.; Huang, F.; Zhao, Z.; Zhang, T.; An, X.; Liao, F.; Guo, L.; Liu, Y.; Zhou, N.; et al. The RNA Methyltransferase METTL3 Promotes Endothelial Progenitor Cell Angiogenesis in Mandibular Distraction Osteogenesis via the PI3K/AKT Pathway. Front. Cell Dev. Biol. 2021, 9, 720925. [Google Scholar] [CrossRef] [PubMed]
- Rizzoni, D.; De Ciuceis, C.; Szczepaniak, P.; Paradis, P.; Schiffrin, E.L.; Guzik, T.J. Immune System and Microvascular Remodeling in Humans. Hypertension 2022, 79, 691–705. [Google Scholar] [CrossRef]
- Koh, M.Y.; Powis, G. Passing the baton: The HIF switch. Trends Biochem. Sci. 2012, 37, 364–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.A.R.; Lawrie, A. Targeting Vascular Remodeling to Treat Pulmonary Arterial Hypertension. Trends Mol. Med. 2017, 23, 31–45. [Google Scholar] [CrossRef]
- Zhou, X.L.; Huang, F.J.; Li, Y.; Huang, H.; Wu, Q.C. SEDT2/METTL14-mediated m6A methylation awakening contributes to hypoxia-induced pulmonary arterial hypertension in mice. Aging 2021, 13, 7538–7548. [Google Scholar] [CrossRef]
- Mourani, P.M.; Garl, P.J.; Wenzlau, J.M.; Carpenter, T.C.; Stenmark, K.R.; Weiser-Evans, M.C. Unique, highly proliferative growth phenotype expressed by embryonic and neointimal smooth muscle cells is driven by constitutive Akt, mTOR, and p70S6K signaling and is actively repressed by PTEN. Circulation 2004, 109, 1299–1306. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Qiao, Y.; Li, L.; Luo, E.; Wang, D.; Yao, Y.; Tang, C.; Yan, G. The m(6)A methyltransferase METTL3 promotes hypoxic pulmonary arterial hypertension. Life Sci. 2021, 274, 119366. [Google Scholar] [CrossRef]
- Chien, C.S.; Li, J.Y.; Chien, Y.; Wang, M.L.; Yarmishyn, A.A.; Tsai, P.H.; Juan, C.C.; Nguyen, P.; Cheng, H.M.; Huo, T.I.; et al. METTL3-dependent N(6)-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc. Natl. Acad. Sci. USA 2021, 118, e2025070118. [Google Scholar] [CrossRef]
- Jian, D.; Wang, Y.; Jian, L.; Tang, H.; Rao, L.; Chen, K.; Jia, Z.; Zhang, W.; Liu, Y.; Chen, X.; et al. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics 2020, 10, 8939–8956. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging role of m(6) A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2020, 21, e12942. [Google Scholar] [CrossRef]
- Zhang, B.F.; Wu, Z.H.; Deng, J.; Jin, H.J.; Chen, W.B.; Zhang, S.; Liu, X.J.; Wang, W.T.; Zheng, X.T. M(6)A methylation-mediated elevation of SM22α inhibits the proliferation and migration of vascular smooth muscle cells and ameliorates intimal hyperplasia in type 2 diabetes mellitus. Biol. Chem. 2021, 403, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Liu, Y.; Zhou, L.; Xue, Y.; Lu, Z.; Gan, J. YTHDC2-Mediated circYTHDC2 N6-Methyladenosine Modification Promotes Vascular Smooth Muscle Cells Dysfunction Through Inhibiting Ten-Eleven Translocation 2. Front. Cardiovasc. Med. 2021, 8, 686293. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jin, Y.; Tang, W.H.; Qin, L.; Zhang, X.; Tellides, G.; Hwa, J.; Yu, J.; Martin, K.A. Ten-eleven translocation-2 (TET2) is a master regulator of smooth muscle cell plasticity. Circulation 2013, 128, 2047–2057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messner, B.; Bernhard, D. Smoking and cardiovascular disease: Mechanisms of endothelial dysfunction and early atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 509–515. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Fan, Y.; Liu, J. METTL14 mediated m6A modification to LncRNA ZFAS1/ RAB22A: A novel therapeutic target for atherosclerosis. Int. J. Cardiol. 2021, 328, 177. [Google Scholar] [CrossRef]
- Mo, C.; Yang, M.; Han, X.; Li, J.; Gao, G.; Tai, H.; Huang, N.; Xiao, H. Fat mass and obesity-associated protein attenuates lipid accumulation in macrophage foam cells and alleviates atherosclerosis in apolipoprotein E-deficient mice. J. Hypertens. 2017, 35, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Y.; Han, L.; Tang, Y.F.; Zhang, G.X.; Fan, X.L.; Zhang, J.J.; Xue, Q.; Xu, Z.Y. METTL14 regulates M6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7015–7023. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, S.; Shim, J. YTHDF2 Suppresses Notch Signaling through Post-transcriptional Regulation on Notch1. Int. J. Biol. Sci. 2021, 17, 3776–3785. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Pollyea, D.A.; Pratz, K.; Letai, A.; Jonas, B.A.; Wei, A.H.; Pullarkat, V.; Konopleva, M.; Thirman, M.J.; Arellano, M.; Becker, P.S.; et al. Venetoclax with azacitidine or decitabine in patients with newly diagnosed acute myeloid leukemia: Long term follow-up from a phase 1b study. Am. J. Hematol. 2021, 96, 208–217. [Google Scholar] [CrossRef]
- Scott, L.J. Azacitidine: A Review in Myelodysplastic Syndromes and Acute Myeloid Leukaemia. Drugs 2016, 76, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Yang, Y.; Kang, M.; Wang, Y.; Wang, Y.; Bi, Y.; He, S.; Shimamoto, F. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 2020, 19, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef] [PubMed]

| Pathological Process | Disease | M6A Regulators | Model System | Mechanism | Reference | ||
|---|---|---|---|---|---|---|---|
| Human Tissue | Animal Model | Cell Line | |||||
| hypoxia | lung cancer | YTHDF2↑ | √ | √ | promote HIF-1 expression | [21] | |
| stomach cancer | IGF2BP3↑ | √ | √ | promote HIF-1 expression | [22] | ||
| breast cancer | METTL14 /ALKBH5↑ | √ | √ | √ | increase TGFβ1 expression | [23] | |
| HCC | YTHDF2↓ | √ | √ | √ | stabilize IL-11 and SERPINE2 mRNA | [24] | |
| METTL3↓ | √ | √ | √ | increase PDGF and VEGF expression | [25] | ||
| oxygen-induced retinopathy | METTL3↑ | √ | √ | activate the Wnt pathway | [26] | ||
| inflammation | HCC | YTHDF2↓ | √ | √ | √ | stabilize IL-11 and SERPINE2 mRNA | [24] |
| corneal neovascularization | FTO↑ | √ | √ | increase FAK expression | [27] | ||
| METTL3↑ | √ | √ | activate the Wnt signaling pathway | [26] | |||
| diabetic retinopathy | YTHDF2↓ | √ | √ | activate FAK/PI3K/AKT pathway | [28] | ||
| others | breast cancer | YTHDF3↑ | √ | √ | √ | enhance translation of VEGF | [29] |
| lung cancer | METTL3↑ | √ | √ | √ | increase VEGFA expression | [30] | |
| intrahepatic cholangiocarcinoma | FTO↓ | √ | √ | √ | increase CCL19 expression | [31] | |
| colorectal cancer/melanoma | ALKBH5↑ | √ | √ | √ | promote VEGF expression | [32] | |
| Pathological Process | Disease | M6A Regulators | Model System | Mechanism | Reference | ||
|---|---|---|---|---|---|---|---|
| Human Tissue | Animal Model | Cell Line | |||||
| hypoxia | HCC | YTHDF2↓ | √ | √ | √ | stabilize IL-11 and SERPINE2 mRNA | [24] |
| METTL3↑ | √ | √ | √ | activate Hippo pathway | [50] | ||
| inflammation | diabetic nephropathy | METTL14↑ | √ | √ | √ | decrease α-klotho expression | [52] |
| diabetic retinopathy | METTL3↑ | √ | √ | suppress PKC/FAT4/PDGFRA pathway | [51] | ||
| others | arteriovenous malformation | METTL3↓ | √ | √ | activate the Notch pathway | [55] | |
| WTAP↓ | √ | √ | block the Wnt pathway | [56] | |||
| model system (endothelial cells) | METTL3↓ | √ | √ | inhibit the PI3K/AKT pathway | [58] | ||
| model system (bone mesenchymal stem cells) | METTL3↓ | √ | inhibit the PI3K/AKT pathway | [59,60] | |||
| Pathological Process | Disease | M6A Regulators | Model System | Mechanism | Reference | ||
|---|---|---|---|---|---|---|---|
| Human Tissue | Animal Model | Cell Line | |||||
| hypoxia | HCC | YTHDF2↓ | √ | √ | √ | stabilize IL-11 and SERPINE2 mRNA | [24] |
| pulmonary arterial hypertension | METTL3↑ | √ | √ | degrade PETN mRNAs | [67] | ||
| METTL14↑ | √ | cooperate with SETD2 | [65] | ||||
| inflammation | atherosclerosis | METTL3↑ | √ | √ | increase NLRP1 and decrease KLF4 expression | [68] | |
| METTL14↑ | √ | √ | increase VCAM-A and ICAM-1 expression | [69,77] | |||
| metabolism | type 2 diabetes mellitus | FTO↑ | √ | √ | destabilize SM22α mRNAs | [71] | |
| YTHDC2↑ | √ | √ | inhibit TET2 expression | [72] | |||
| atherosclerosis | FTO↑ | √ | √ | √ | reduce CD36 and PPARγ level | [76] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.-R.; Ji, J.-D.; Wang, J.-N.; Wang, Y.; Zhu, H.-J.; Sun, R.-X.; Liu, Q.-H.; Chen, X. The Role of N6-Methyladenosine Modification in Microvascular Dysfunction. Cells 2022, 11, 3193. https://doi.org/10.3390/cells11203193
Zhang Y-R, Ji J-D, Wang J-N, Wang Y, Zhu H-J, Sun R-X, Liu Q-H, Chen X. The Role of N6-Methyladenosine Modification in Microvascular Dysfunction. Cells. 2022; 11(20):3193. https://doi.org/10.3390/cells11203193
Chicago/Turabian StyleZhang, Ye-Ran, Jiang-Dong Ji, Jia-Nan Wang, Ying Wang, Hong-Jing Zhu, Ru-Xu Sun, Qing-Huai Liu, and Xue Chen. 2022. "The Role of N6-Methyladenosine Modification in Microvascular Dysfunction" Cells 11, no. 20: 3193. https://doi.org/10.3390/cells11203193
APA StyleZhang, Y.-R., Ji, J.-D., Wang, J.-N., Wang, Y., Zhu, H.-J., Sun, R.-X., Liu, Q.-H., & Chen, X. (2022). The Role of N6-Methyladenosine Modification in Microvascular Dysfunction. Cells, 11(20), 3193. https://doi.org/10.3390/cells11203193






