The Relevance of DNA Methylation and Histone Modification in Periodontitis: A Scoping Review
Abstract
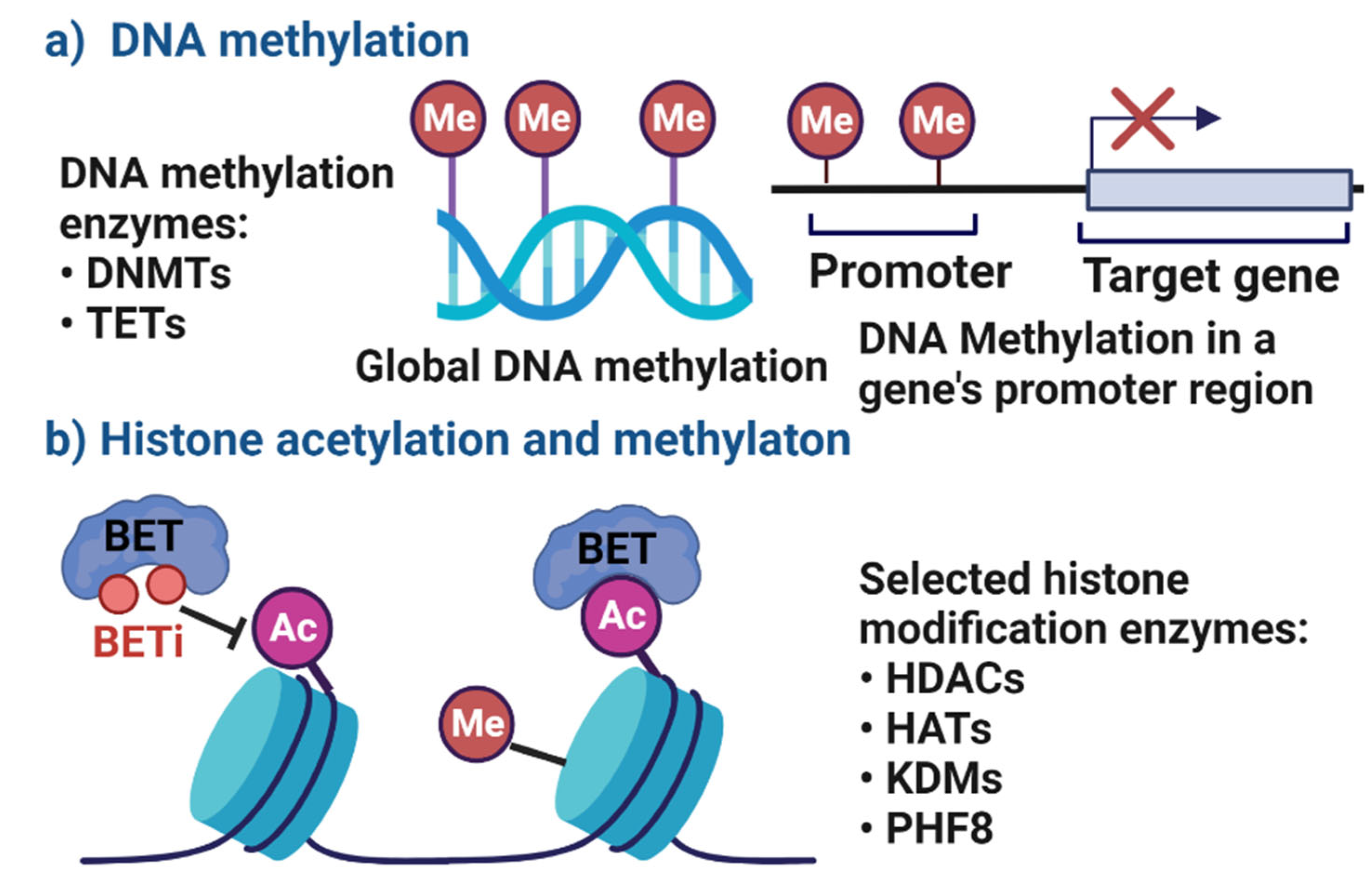
:1. Introduction
2. Materials and Methods
2.1. Study Eligibility Criteria
- Patients with periodontitis;
- Gingival biopsies, gingival crevicular fluid or saliva samples;
- Retrospective and prospective studies of the observational, case–control, cohort, or randomised controlled trial design;
- English language.
- In vitro and in vivo studies (for DNA methylation—refer to 2.3);
- Case reports;
- Literature reviews;
- Diagnoses of aggressive periodontitis;
- Human blood or mouth rinse samples.
2.2. Search Strategy
2.3. Study Selection Strategy
3. Results
3.1. Literature Search and Screening Process
3.2. Study Characteristics
3.3. Changes in DNA Methylation Patterns and Gene Expression Profiles in Periodontitis Patients
3.3.1. Changes in DNA Methylation Patterns of Cytokine-Encoding Genes
3.3.2. Changes in DNA Methylation Patterns of Cellular Signalling Genes
3.3.3. Changes in DNA Methylation Patterns following Periodontal Therapy
3.3.4. Changes of sEVs-Associated Epigenetic Markers in Periodontitis
3.3.5. Conclusions from the Included Studies
3.4. Histone Modification and Its Relevance in Periodontal Inflammation
3.4.1. The Role of Histone Deacetylases, Methylases and Demethylases in Periodontal Inflammation
3.4.2. Changes in Histone Deacetylase Expression in Periodontitis versus Health
3.4.3. The Effects of Histone Deacetylase Inhibitors on the Periodontal Inflammatory Response
3.4.4. The Effect of Bromodomain and Extra-Terminal Proteins Inhibitors in Periodontitis
3.4.5. Conclusions from the Included Studies
4. Discussion and Conclusions
4.1. Implications of the Findings for Research
4.2. Implications of the Findings for Clinical Practice
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bartold, P.M.; Van Dyke, T.E. An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis. J. Clin. Periodontol. 2019, 46, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Meyle, J.; Chapple, I. Molecular aspects of the pathogenesis of periodontitis. Periodontology 2000, 69, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 2014, 35, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, P.; Liu, T.; Vaquette, C.; Frazer, D.; Anderson, G.; Ivanovski, S. Iron accumulation is associated with periodontal destruction in a mouse model of HFE-related haemochromatosis. J. Periodontal Res. 2022, 57, 294–304. [Google Scholar] [CrossRef]
- Pan, W.; Wang, Q.; Chen, Q. The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 2019, 11, 30. [Google Scholar] [CrossRef] [Green Version]
- Bartold, P.M.; Van Dyke, T.E. Periodontitis: A host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontology 2000 2013, 62, 203–217. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Mealey, B.L.; Mariotti, A.; Chapple, I.L. Dental plaque-induced gingival conditions. J. Periodontol. 2018, 89, S17–S27. [Google Scholar] [CrossRef] [Green Version]
- Ivanovski, S. Periodontal regeneration. Aust. Dent. J. 2009, 54 (Suppl. 1), S118–S128. [Google Scholar] [CrossRef]
- Bartold, P.M. Lifestyle and periodontitis: The emergence of personalized periodontics. Periodontology 2000 2018, 78, 7–11. [Google Scholar] [CrossRef]
- Larsson, L. Current Concepts of Epigenetics and Its Role in Periodontitis. Curr. Oral Health Rep. 2017, 4, 286–293. [Google Scholar] [CrossRef]
- Bartold, P.M.; Ivanovski, S. P4 Medicine as a model for precision periodontal care. Clin. Oral Investig. 2022, 26, 5517–5533. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A Landscape Takes Shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, P.; Gomez, G.A.; Duda, G.N.; Ivanovski, S.; Poh, P.S. Scaffold geometry modulation of mechanotransduction and its influence on epigenetics. Acta Biomater. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Barros, S.P.; Offenbacher, S. Modifiable risk factors in periodontal disease: Epigenetic regulation of gene expression in the inflammatory response. Periodontol 2000 2014, 64, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Johnson, I.T.; Belshaw, N.J. Environment, diet and CpG island methylation: Epigenetic signals in gastrointestinal neoplasia. Food Chem. Toxicol. 2008, 46, 1346–1359. [Google Scholar] [CrossRef]
- Cantley, M.D.; Zannettino, A.C.W.; Bartold, P.M.; Fairlie, D.P.; Haynes, D.R. Histone deacetylases (HDAC) in physiological and pathological bone remodelling. Bone 2017, 95, 162–174. [Google Scholar] [CrossRef] [Green Version]
- Han, P.; Bartold, P.M.; Ivanovski, S. The emerging role of small extracellular vesicles in saliva and gingival crevicular fluid as diagnostics for periodontitis. J. Periodontal Res. 2021, 57, 219–231. [Google Scholar] [CrossRef]
- Han, P.; Bartold, P.M.; Salomon, C.; Ivanovski, S. Salivary Small Extracellular Vesicles Associated miRNAs in Periodontal Status—A Pilot Study. Int. J. Mol. Sci. 2020, 21, 2809. [Google Scholar] [CrossRef] [Green Version]
- Han, P.; Bartold, P.; Salomon, C.; Ivanovski, S. Salivary Outer Membrane Vesicles and DNA Methylation of Small Extracellular Vesicles as Biomarkers for Periodontal Status: A Pilot Study. Int. J. Mol. Sci. 2021, 22, 2423. [Google Scholar] [CrossRef]
- Han, P.; Ivanovski, S. Effect of Saliva Collection Methods on the Detection of Periodontium-Related Genetic and Epigenetic Biomarkers—A Pilot Study. Int. J. Mol. Sci. 2019, 20, 4729. [Google Scholar] [CrossRef]
- Hua, S.; Bartold, P.; Gulati, K.; Moran, C.; Ivanovski, S.; Han, P. Periodontal and Dental Pulp Cell-Derived Small Extracellular Vesicles: A Review of the Current Status. Nanomaterials 2021, 11, 1858. [Google Scholar] [CrossRef] [PubMed]
- Jiao, K.; Walsh, L.; Ivanovski, S.; Han, P. The Emerging Regulatory Role of Circular RNAs in Periodontal Tissues and Cells. Int. J. Mol. Sci. 2021, 22, 4636. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Lai, A.; Salomon, C.; Ivanovski, S. Detection of Salivary Small Extracellular Vesicles Associated Inflammatory Cytokines Gene Methylation in Gingivitis. Int. J. Mol. Sci. 2020, 21, 5273. [Google Scholar] [CrossRef]
- Benakanakere, M.R.; Finoti, L.; Palioto, D.B.; Teixeira, H.S.; Kinane, D.F. Epigenetics, Inflammation, and Periodontal Disease. Curr. Oral Health Rep. 2019, 6, 37–46. [Google Scholar] [CrossRef]
- Emfietzoglou, R.; Pachymanolis, E.; Piperi, C. Impact of Epigenetic Alterations in the Development of Oral Diseases. Curr Med Chem 2021, 28, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, Y. Emerging role of epigenetic regulations in periodontitis: A literature review. Am. J. Transl. Res. 2022, 14, 2162–2183. [Google Scholar]
- Khouly, I.; Braun, R.S.; Ordway, M.; Aouizerat, B.E.; Ghassib, I.; Larsson, L.; Asa’Ad, F. The Role of DNA Methylation and Histone Modification in Periodontal Disease: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 6217. [Google Scholar] [CrossRef]
- Suzuki, S.; Yamada, S. Epigenetics in susceptibility, progression, and diagnosis of periodontitis. Jpn. Dent. Sci. Rev. 2022, 58, 183–192. [Google Scholar] [CrossRef]
- Asa’Ad, F.; Bollati, V.; Pagni, G.; Castilho, R.M.; Rossi, E.; Pomingi, F.; Tarantini, L.; Consonni, D.; Giannobile, W.V.; Rasperini, G. Evaluation of DNA methylation of inflammatory genes following treatment of chronic periodontitis: A pilot case-control study. J. Clin. Periodontol. 2017, 44, 905–914. [Google Scholar] [CrossRef]
- Andia, D.; Planello, A.C.; Portinho, D.; da Silva, R.A.; Salmon, C.R.; Sallum, E.A.; Junior, F.H.N.; De Souza, A.P. DNA methylation analysis of SOCS1, SOCS3, and LINE-1 in microdissected gingival tissue. Clin. Oral Investig. 2015, 19, 2337–2344. [Google Scholar] [CrossRef]
- Zhang, S.; Barros, S.P.; Moretti, A.J.; Yu, N.; Zhou, J.; Preisser, J.S.; Niculescu, M.D.; Offenbacher, S. Epigenetic regulation of TNFA expression in periodontal disease. J. Periodontol. 2013, 84, 1606–1616. [Google Scholar] [PubMed]
- Zhang, S.; Barros, S.; Niculescu, M.; Moretti, A.; Preisser, J.; Offenbacher, S. Alteration of PTGS2 Promoter Methylation in Chronic Periodontitis. J. Dent. Res. 2010, 89, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Crivello, A.; Offenbacher, S.; Moretti, A.; Paquette, D.; Barros, S.P. Interferon-gamma promoter hypomethylation and increased expression in chronic periodontitis. J. Clin. Periodontol. 2010, 37, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Stefani, F.A.; Viana, M.B.; Dupim, A.C.; Brito, J.A.R.; Gomez, R.S.; da Costa, J.E.; Moreira, P.R. Expression, polymorphism and methylation pattern of interleukin-6 in periodontal tissues. Immunobiology 2013, 218, 1012–1017. [Google Scholar] [CrossRef]
- Li, X.; Lu, J.; Teng, W.; Zhao, C.; Ye, X. Quantitative Evaluation of MMP-9 and TIMP-1 Promoter Methylation in Chronic Periodontitis. DNA Cell Biol. 2018, 37, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Thorbert-Mros, S.; Lopez-Lago, A.; Kalm, J.; Shikhan, A.; Berglundh, T. Expression of TET2 enzyme indicates enhanced epigenetic modification of cells in periodontitis. Eur. J. Oral Sci. 2016, 124, 329–333. [Google Scholar] [CrossRef]
- De Souza, A.P.; Planello, A.C.; Marques, M.R.; De Carvalho, D.D.; Line, S.R.P. High-throughput DNA analysis shows the importance of methylation in the control of immune inflammatory gene transcription in chronic periodontitis. Clin. Epigenetics 2014, 6, 15. [Google Scholar] [CrossRef]
- De Oliveira, N.F.P.; Andia, D.C.; Planello, A.C.; Pasetto, S.; Marques, M.R.; Nociti, F.H., Jr.; Line, S.R.P.; De Souza, A.P. TLR2 and TLR4 gene promoter methylation status during chronic periodontitis. J. Clin. Periodontol. 2011, 38, 975–983. [Google Scholar] [CrossRef]
- Amormino, S.A.D.F.; Arão, T.C.; Saraiva, A.M.; Gomez, R.S.; Dutra, W.O.; da Costa, J.E.; Silva, J.D.F.C.; Moreira, P. Hypermethylation and low transcription of TLR2 gene in chronic periodontitis. Hum. Immunol. 2013, 74, 1231–1236. [Google Scholar] [CrossRef]
- Azevedo, A.M.; Rocha, L.P.C.; Amormino, S.A.D.F.; Gomes, C.C.; Dutra, W.O.; Gomez, R.S.; Da Costa, J.E.; Moreira, P.R. DNA methylation profile of genes related to immune response in generalized periodontitis. J. Periodontal Res. 2020, 55, 426–431. [Google Scholar] [CrossRef]
- Kim, H.; Momen-Heravi, F.; Chen, S.; Hoffmann, P.; Kebschull, M.; Papapanou, P.N. Differential DNA methylation and mRNA transcription in gingival tissues in periodontal health and disease. J. Clin. Periodontol. 2021, 48, 1152–1164. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.B.; Cardoso, F.P.; Diniz, M.G.; Costa, F.O.; da Costa, J.E.; Gomez, R.S.; Moreira, P.R. Methylation pattern of IFN-γ and IL-10 genes in periodontal tissues. Immunobiology 2011, 216, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Parachuru, V.P.B.; Coates, D.E.; Milne, T.J.; Rich, A.M.; Seymour, G.J. FoxP3+ regulatory T cells, interleukin 17 and mast cells in chronic inflammatory periodontal disease. J. Periodontal Res. 2018, 53, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hill, A.F. Therapeutically harnessing extracellular vesicles. Nat. Rev. Drug Discov. 2022, 21, 379–399. [Google Scholar] [CrossRef] [PubMed]
- Cantley, M.D.; Dharmapatni, A.A.S.S.K.; Algate, K.; Crotti, T.N.; Bartold, P.; Haynes, D.R. Class I and II histone deacetylase expression in human chronic periodontitis gingival tissue. J. Periodontal Res. 2016, 51, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Cantley, M.D.; Bartold, P.M.; Marino, V.; Fairlie, D.P.; Le, G.T.; Lucke, A.J.; Haynes, D.R. Histone deacetylase inhibitors and periodontal bone loss. J. Periodontal Res. 2011, 46, 697–703. [Google Scholar] [CrossRef]
- Huynh, N.C.-N.; Everts, V.; Nifuji, A.; Pavasant, P.; Ampornaramveth, R.S. Histone deacetylase inhibition enhances in-vivo bone regeneration induced by human periodontal ligament cells. Bone 2017, 95, 76–84. [Google Scholar] [CrossRef]
- Yin, L.; Chung, W.O. Epigenetic regulation of human beta-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 2011, 4, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Sun, J.; Dong, Z.; Xue, P.; He, X.; Liao, L.; Yuan, L.; Jin, Y. GCN5 modulates osteogenic differentiation of periodontal ligament stem cells through DKK1 acetylation in inflammatory microenvironment. Sci. Rep. 2016, 6, 26542. [Google Scholar] [CrossRef] [Green Version]
- Martins, M.D.; Jiao, Y.; Larsson, L.; Almeida, L.O.; Garaicoa-Pazmino, C.; Le, J.; Squarize, C.; Inohara, N.; Giannobile, W.; Castilho, R.M. Epigenetic Modifications of Histones in Periodontal Disease. J. Dent. Res. 2016, 95, 215–222. [Google Scholar] [CrossRef]
- Li, L.; Liu, W.; Wang, H.; Yang, Q.; Zhang, L.; Jin, F.; Jin, Y. Mutual inhibition between HDAC9 and miR-17 regulates osteogenesis of human periodontal ligament stem cells in inflammatory conditions. Cell Death Dis. 2018, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, J.E.; Kirkwood, K.L.; Woster, P.M. Inhibition of the histone demethylase KDM4B leads to activation of KDM1A, attenuates bacterial-induced pro-inflammatory cytokine release, and reduces osteoclastogenesis. Epigenetics 2018, 13, 557–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, M.; Pandya, M.; Gopinathan, G.; Lyu, H.; Ma, W.; Foyle, D.; Nares, S.; Luan, X.; Nares, S. Histone Methylation Mechanisms Modulate the Inflammatory Response of Periodontal Ligament Progenitors. Stem Cells Dev. 2019, 28, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Mehrazarin, S.; Alshaikh, A.; Kim, S.; Chen, W.; Lux, R.; Gwack, Y.; Kim, R.H.; Kang, M.K. Histone Lys demethylase KDM3C demonstrates anti-inflammatory effects by suppressing NF-κB signaling and osteoclastogenesis. FASEB J. 2019, 33, 10515–10527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; He, Y.; Xu, C.; Li, J.; Zeng, S.; Yang, X.; Han, Q. The role of PHF8 and TLR4 in osteogenic differentiation of periodontal ligament cells in inflammatory environment. J. Periodontol. 2021, 92, 1049–1059. [Google Scholar] [CrossRef]
- Cantley, M.; Fairlie, D.; Bartold, P.; Rainsford, K.; Le, G.; Lucke, A.; Holding, C.; Haynes, D. Inhibitors of histone deacetylases in class I and class II suppress human osteoclasts in vitro. J. Cell. Physiol. 2011, 226, 3233–3241. [Google Scholar] [CrossRef]
- Huynh, N.C.; Everts, V.; Pavasant, P.; Ampornaramveth, R.S. Inhibition of Histone Deacetylases Enhances the Osteogenic Differentiation of Human Periodontal Ligament Cells. J. Cell. Biochem. 2016, 117, 1384–1395. [Google Scholar] [CrossRef]
- Lagosz, K.; Bysiek, A.; Macina, J.; Bereta, G.; Kantorowicz, M.; Lipska, W.; Sochalska, M.; Gawron, K.; Kaczmarzyk, T.; Chomyszyn-Gajewska, M.; et al. HDAC3 Regulates Gingival Fibroblast Inflammatory Responses in Periodontitis. J. Dent. Res. 2020, 99, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Algate, K.; Haynes, D.; Fitzsimmons, T.; Romeo, O.; Wagner, F.; Holson, E.; Reid, R.; Fairlie, D.; Bartold, P.; Cantley, M. Histone deacetylases 1 and 2 inhibition suppresses cytokine production and osteoclast bone resorption in vitro. J. Cell. Biochem. 2020, 121, 244–258. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [Green Version]
- Maksylewicz, A.; Bysiek, A.; Lagosz, K.B.; Macina, J.M.; Kantorowicz, M.; Bereta, G.; Sochalska, M.; Gawron, K.; Chomyszyn-Gajewska, M.; Potempa, J.; et al. BET Bromodomain Inhibitors Suppress Inflammatory Activation of Gingival Fibroblasts and Epithelial Cells From Periodontitis Patients. Front. Immunol. 2019, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Zhang, L.; Tang, Y.; Tu, Q.; Zheng, L.; Yu, L.; Murray, D.; Cheng, J.; Kim, S.H.; Zhou, X. BET Inhibitor JQ1 Blocks Inflammation and Bone Destruction. J. Dent. Res. 2014, 93, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, H.; Chen, X.; Chen, Y.; Bian, T. BET protein inhibitor apabetalone represses Porphyromonas gingivalis LPS-induced macrophage M1 polarization via regulating miR-130a/STAT3 axis. BIOCELL 2022, 46, 2281–2289. [Google Scholar] [CrossRef]
- Goodson, J.M.; Tanner, A.C.R.; Haffajee, A.D.; Sornberger, G.C.; Socransky, S.S. Patterns of progression and regression of advanced destructive periodontal disease. J. Clin. Periodontol. 1982, 9, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Alminana-Pastor, P.; Boronat-Catala, M.; Mico-Martinez, P.; Bellot-Arcis, C.; Lopez-Rolda, A.; Alpiste-Illueca, F. Epigenetics and periodontics: A systematic review. Med. Oral Patol. Oral Cir. Bucal. 2019, 24, e659–e672. [Google Scholar] [CrossRef]
- Nishitani, S.; Parets, S.E.; Haas, B.W.; Smith, A.K. DNA methylation analysis from saliva samples for epidemiological studies. Epigenetics 2018, 13, 352–362. [Google Scholar] [CrossRef]


| Author/Year | Global or Target Genes | Tissue/Cell Type | Sample Size | Case Definitions | Key Epigenetic Events and Conclusions |
|---|---|---|---|---|---|
| Kim et al., 2021 |
| Gingival biopsies | 12 patients with Stage III periodontitis (n = 12), gingivitis (n = 12), periodontal health (n = 12) | Stage III periodontitis: PD ≥ 5 mm with CAL ≥ 3 mm, radiographic bone loss beyond the coronal third of the root, and BoP. Gingivitis: PD ≤ 3 mm, no CAL, and presence of BoP. Healthy: PD ≤3 mm, no CAL, and no BoP. |
|
| Han et al., 2021 |
| Unstimulated whole saliva (salivary extracellular vesicles) | Periodontal health (n = 7), gingivitis (n = 7) and periodontitis (n = 8) | Healthy: no periodontal disease history; PPD < 3 mm; BOP < 15 % sites. Gingivitis: no periodontal pocket, PPD < 3 mm; BOP > 30 % sites. Stage III/IV periodontitis: > 30% of the sites with PPD ≥ 3 mm and BOP, at least five sites with PPD ≥ 5mm on at least three non-adjacent teeth. |
|
| Azevedo et al., 2020 |
| Gingival biopsies | Periodontitis (n = 20) and healthy individuals (n = 20). | Periodontitis group: >30% of teeth with interproximal CAL ≥ 5 mm, radiographic bone loss extending the apical third of the roots, loss ≥ 5 teeth due to periodontitis, PD ≥ 7 mm. Control group: PD < 4 mm during tooth extraction for orthodontic treatment or of third molar and crown lengthening |
|
| Li et al., 2018 |
| Gingival crevicular fluid | Chronic periodontitis patients (n = 88) and healthy controls (n = 15) | Severe periodontitis group (n = 27) had PD (6.88 ± 0.83 mm) and CAL > 7 mm with bleeding upon probing each tooth. Moderate periodontitis (n = 29) had PD (5.78 ± 1.09 mm) and CAL <6 and 4 mm with bleeding upon probing. Mild periodontitis group (n = 32) had PD (2.45 ± 0.21 mm) and CAL<3 mm with bleeding upon probing. Subjects in the control group (n = 15) PD < 2 mm and no CAL, bleeding on probing, or radiographic bone loss. |
|
| Asa’ad et al., 2017 |
| Gingival biopsies | Periodontal health (n = 10) moderate chronic periodontitis (n = 10) | Healthy group: PPD ≤ 3 mm, without any signs of tooth mobility, BOP or CAL. Periodontitis group: Patients in this group displayed a minimum of a single “healthy site” (PPD < 4 mm without signs of bleeding on probing, mobility or inflammation) and a periodontitis site (PPD ≥ 5 mm with bleeding on probing at baseline). |
|
| Larsson et al., 2016 |
| Gingival biopsies | Generalized severe chronic periodontitis (n = 21) and gingivitis (n = 17) | Periodontitis: bone loss ≥ 50%, PPD) ≥ 6 mm and bleeding on probing (BoP) Gingivitis: no signs of bone loss, PPD ≤ 4 mm and BoP |
|
| Andia et al., 2015 |
| Gingival biopsies | Healthy (n = 10), periodontitis (n = 10) | Healthy: no BOP, no CAL, FMB S ≤ 25%, and absence of tooth mobility. Periodontitis: CAL ≥ 5 mm, and radiographic bone loss in at least eight teeth and bleeding following pocket probing |
|
| De Souza et al., 2014 |
| Gingival biopsies | Chronic periodontitis (n = 12) and healthy age- and gender-matched control individuals (n = 11) | Periodontitis: more than 3 CAL≥ 5 mm and bleeding on probing. Control group: no bleeding on probing, and all teeth with CAL ≤ 3.5 mm |
|
| Stefani et al., 2013 |
| Gingival biopsies | Periodontitis (n = 21) and control group (n = 21). | Periodontitis: PD ≥ 4 mm, gingival bleeding, CAL ≥ 3 mm Control group: absence of signs of periodontal disease, PD<4mm |
|
| De Faria Amormino et al., 2013 |
| Gingival biopsies | Periodontitis (n = 20) and control samples (n = 20) | Periodontitis group: a PD of ≥4 mm and CAL ≥ 3 mm and lesions distributed on more than two quadrants were included in this group. Control: absence of CAL, no sites with PD > 3 mm and absence of bleeding. |
|
| Zhang et al., 2013 |
| Gingival biopsies from healthy or periodontitis patient | Chronic periodontitis (n = 17) (CP) and periodontally healthy individuals (n = 18). Another 11 individuals participated in an experimentally induced gingivitis study. | Periodontitis: at least 5 mm PD and radiographic evidence of alveolar bone loss (ABL). Health: a PD measurement of ≤4 mm, no BOP, and no evidence of radiographic ABL. |
|
| Viana et al., 2011 |
| Gingival biopsies | Periodontitis group (n = 18), control group (n = 16). | Control group: no CAL, no bleeding on probing and PD ≤ 3 mm. Periodontitis: PD ≥ 4 mm, bleeding on probing and with extensive radiographic bone loss. |
|
| De Oliveira et al., 2011 |
| Gingival biopsies | Healthy controls (n = 11), periodontitis smokers (n = 11), periodontitis non-smokers (n = 12). | Healthy controls: absence of CAL and no sites with PD >3 mm. Periodontitis smokers (n = 11): Subjects with at least three teeth exhibiting sites ≥5 mm CAL in at least two different quadrants and had consumed five cigarettes per day for at least 5 years. Periodontitis non-smokers (n = 12): Subjects with at least three teeth exhibiting sites ≥5 mm CAL in at least two different quadrants. |
|
| Zhang et al., 2010 |
| Gingival biopsies | 16 participants, aged between 18 and 65 yrs. Inflamed (n = 10), non-inflamed (n = 6). | Periodontitis: PD>5 mm, bleeding on probing, and radiographic evidence of localized bone loss. Health: probing depth measurements of ≤4 mm at all 4 inter-proximal probing sites and no bleeding on probing. |
|
| Zhang et al., 2010 |
| Gingival biopsies | Periodontally healthy sites (n = 23), Experimentally induced gingivitis sites (n = 12) and chronic periodontitis sites (n = 12). | Health: PD<4mm, no BOP, no evidence of radiographic bone loss Periodontitis: Those biopsied tissues were from sites exhibiting PDs of ≥5 mm, BOP and radiographic evidence of localized bone loss. |
|
| Author/Year | Target Genes | Tissue/Cell Type | Sample Size | Case Definitions | Epigenetic Events and Conclusions |
|---|---|---|---|---|---|
| Liu et al. 2021 | Histone demethylase PHF8 (Real-time PCR for mRNA expression analysis) | PDL stem cells | Not reported | Not reported | mRNA expression levels of PHF8 were significantly upregulated during the osteogenic differentiation of PDL stem cells, and exposure to P. gingvalis-induced lipopolysaccharide (LPS) inhibited PHF8 expression. |
| Algate et al. 2020 | HDAC1, HDAC2, HDAC5 (2−ΔCT method) | Monocytic cells | Not reported | Not reported | Stimulation of monocytes with TNFα for 24 h resulted in increased expression of HDAC1 and HDAC2. |
| Lee et al. 2019 | KDM3C (RT-qPCR) | Monocytic cells | Not reported | Not reported | Cells challenged by P. gingivalis LPS showed suppression of KDM3C expression Knockout of KDM3C in cells exposed to LPS by P. gingivalis led to an elevated level of TNF-α, IL-1β, IL-6 and NF-κB signalling. |
| Francis et al. 2019 | H3K4me3, H3K9me3 and H3K27me3 (Chip-polymerase chain reaction) | PDL stem cells | Not reported | Not reported | In PDL stem cells exposed to P. gingivalis LPS, the expression of H3K27me3 increased on extracellular matrix and osteogenesis lineage gene promoters, and H3K4me3 expression increased on the promoters of inflammatory response genes. |
| Kirkpatrick et al., 2018 | KDM4B and KDM4E (RT-qPCR) | Connective tissue | Not reported | Periodontitis: at least 1 site with PD > 4 mm, GI 1–3, and PI 1–3. Healthy: PD ≤ 4 mm, GI ≤ 1, and PI ≤ 2. | A significant increase in KDM4B and KDM4E was observed in periodontally diseased vs. healthy tissue. |
| Li et al., 2018 | HDAC9 (RT-qPCR) | PDL stem cells | Twenty teeth of periodontitis patients were collected from the teeth extracted due to clinical diagnosed chronic periodontitis. | Not reported | HDAC9 RNA expression levels were significantly increased in human periodontitis PDL stem cells, and TNF-α stimulated healthy PDL stem cells, and their osteogenic capacity was impaired in inflammatory conditions. |
| Cantley et al., 2016 | HDACs1 to 10 (qPCR, Immunohistochemical detection of HDAC1, 5, 8 and 9, Semiquantitative analysis of immunohistochemistry) | Gingival biopsies | For mRNA analysis using real-time PCR, nine samples of periodontitis and eight samples of non-periodontitis tissues were analysed. For immunohistochemical staining, 15 samples of periodontitis tissues and seven samples of non-periodontitis tissues were analysed for both HDAC 1 and 9 staining. For HDAC 5 and 8 staining, eight samples of periodontitis tissues and six non-periodontitis tissues were analysed. | Periodontitis: histologically and radiographic evidence of bone loss Healthy: no or only mild levels of inflammation and had no evidence of bone loss. | mRNA expression of Class I HDAC1 and 8 and Class II HDAC5 and 9 were significantly higher in chronic periodontitis samples compared with non-periodontitis samples. There was a twofold increase in HDAC1, 8 and 5 expression and a threefold increase in HDAC9 expression in chronic periodontitis samples compared with their counterpart. HDAC1 was expressed by CD3 and TNF-α positive inflammatory cells in periodontitis tissues. |
| Martins et al., 2016 | Histone H3 | Oral epithelial cells | Not reported | Not reported | Abrupt but short-lived increased acetylation of H3 induced by LPS from P. gingivalis and heated inactivated F. nucleatum. E. coli LPS resulted in a delayed but powerful induction of histone acetylation compared with P. gingivalis and F. nucleatum. |
| Li et al., 2016 | Histone acetyltransferase GCN5 (RT-PCR) | PDL stem cells | Healthy (n = 5), periodontitis (n = 5) | Healthy: absence of BoP, PD < 4 mm and CAL< 3 mm. Periodontitis: alveolar bone loss (2/3) and more than 1 pocket (depth 5 mm) | Inflammation in the microenvironment resulted in downregulation of GCN5 expression, leading to decreased osteogenic differentiation of PDL stem cells. |
| Yin et al., 2011 | HDAC1 and HDAC2 | Gingival epithelial cells | Not reported | Not reported | The gene expression of both HDAC1 and HDAC2 significantly decreased in cells treated with P. gingivalis and F. nucleatum, compared with unstimulated controls after 24 h. |
| Histone H3K4me3 | Gingival epithelial cells | The protein amount was significantly decreased when cells were stimulated with P. gingivalis compared with unstimulated controls, but not with F. nucleatum. | |||
| HDAC1 and HDAC2 (qRT-PCR) | Immortalised keratinocyte cell line | The gene expression of HDAC1 was significantly decreased in cells treated with P. gingivalis after 24 h, but not with F. nucleatum. The gene expression of HDAC2 slightly increased with F. nucleatum and P. gingivalis initially, then decreased after 24 h. |
| Author/Year | Type of HDAC Inhibition | Tissue/Cell Type | Case Definitions | Epigenetic Events and Conclusions |
|---|---|---|---|---|
| Algate et al., 2020 | Selective Class I and II HDACs | Monocytic cells Osteoclast progenitors | Not reported | When monocytes were stimulated with TNF, HDAC-1i and HDAC-2i both reduced the mRNA expression of pro-inflammatory cytokines IL-1β and TNF and chemokines MIP-1α and MCP-1 in a dose-dependent manner. TNF-stimulated osteoclasts were sensitive to HDAC-1i, reducing the no. and size of osteoclastic cells in vitro. Both HDAC-1i and HDAC-2i diminished osteoclast activity. Both HDAC-1i and HDAC-2i reduced the mRNA expression of the osteoclast signalling factors TRAF-6, TRAP, Cath K and DC-STAMP. No effect was found when inhibiting Class II HDACs (HDAC5 and HDAC7). |
| Lagosz et al., 2020 | Combined and selective Class I and II HDACs | Gingival fibroblasts | Not reported | Treatment of healthy gingival fibroblasts with the pan-HDACi or HDAC3/6i prior to P. gingivalis infection significantly reduced the induction of several inflammatory mediators (CCL2, CCL5, CXCL10, IL1B, COX2 and MMP3) that contribute to periodontitis pathogenesis. Treatment of periodontally diseased gingival fibroblasts with HDAC3/6i suppressed the induction of CCL2, CCL5, CXCL10, IL1B, MMP3 and MMP1 to a similar degree as cells from healthy individuals. |
| Li et al., 2018 | HDAC9 (RT-PCR) | PDL stem cells | Not reported | Downregulation of HDAC9 by HDAC inhibitors rescued the osteogenic differentiation capacity of inflammatory PDL stem cells to a similar level with the healthy PDL stem cells. |
| Huynh et al., 2017 | Combined Class I and II HDACs | PDL stem cells | Not reported | Trichostatin A induced acetylated RUNX2, enhancing mineral deposition and the osteogenic potential of PDL stem cells. In vivo bone regeneration of mouse calvaria defects was also significantly enhanced by Trichostatin A pre-treated PDL stem cells |
| Huynh et al., 2016 | Combined Class I and II HDACs (RT-PCR) | PDL stem cells | Not reported | In the presence of the HDAC inhibitor Trichostatin A osteogenic differentiation was induced and osteoblast-related gene expression was increased significantly (without any adipogenic differentiation). Alkaline phosphatase activity and mineral nodule formation were also enhanced. |
| Cantley et al., 2011 | Combined Class I and II HDACs (Real-time PCR analysis) | Alveolar bone and soft tissue | Not reported | Periodontitis-induced mice treated with HDACi 1179.4b exhibited a significant reduction in osteoclast numbers and suppression of bone loss, although no suppression of inflammation. Periodontitis-induced mice treated with HDACi MS-275 had little effect on bone loss but had a non-significant reduction in inflammation. |
| Cantley et al., 2011 | Combined Class I and II HDACs (Real-time PCR analysis) | Monocytic cells Osteoclast progenitors | Not reported | During the late stages of osteoclast formation, 1179.4b significantly reduced the expression of osteoclast transcription factors NFATc1 and OSCAR. Furthermore, 1179.4b almost completely inhibited TRAP expression and osteoclast resorption, reducing the no. of pits formed in a dose-dependent manner. This was confirmed with a 13-fold reduction in CTR gene expression (osteoclast gene associated with ability to resorb bone) corresponding with a marked inhibition of resorption. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liaw, A.; Liu, C.; Ivanovski, S.; Han, P. The Relevance of DNA Methylation and Histone Modification in Periodontitis: A Scoping Review. Cells 2022, 11, 3211. https://doi.org/10.3390/cells11203211
Liaw A, Liu C, Ivanovski S, Han P. The Relevance of DNA Methylation and Histone Modification in Periodontitis: A Scoping Review. Cells. 2022; 11(20):3211. https://doi.org/10.3390/cells11203211
Chicago/Turabian StyleLiaw, Andrew, Chun Liu, Sašo Ivanovski, and Pingping Han. 2022. "The Relevance of DNA Methylation and Histone Modification in Periodontitis: A Scoping Review" Cells 11, no. 20: 3211. https://doi.org/10.3390/cells11203211
APA StyleLiaw, A., Liu, C., Ivanovski, S., & Han, P. (2022). The Relevance of DNA Methylation and Histone Modification in Periodontitis: A Scoping Review. Cells, 11(20), 3211. https://doi.org/10.3390/cells11203211








