Mapping Proteome and Lipidome Changes in Early-Onset Non-Alcoholic Fatty Liver Disease Using Hepatic 3D Spheroids
Abstract
1. Introduction
2. Materials and Methods
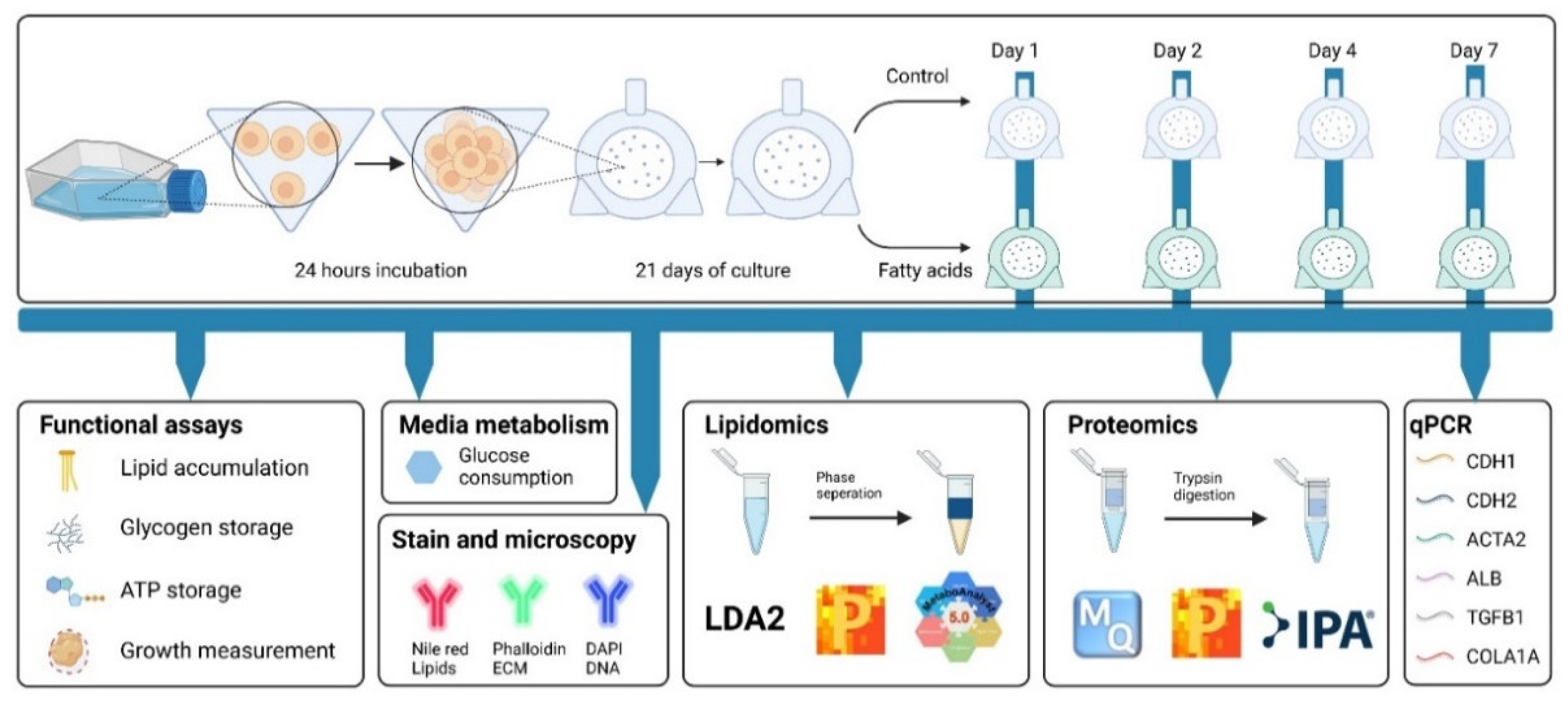
2.1. Monolayer Cell Culture
2.2. Formation and Maintenance of Spheroids
2.3. Spheroids Treatment (Palmitic and Oleic Acid or TGF-β1)
2.4. Monitoring Spheroid Size
2.5. ATP Assay
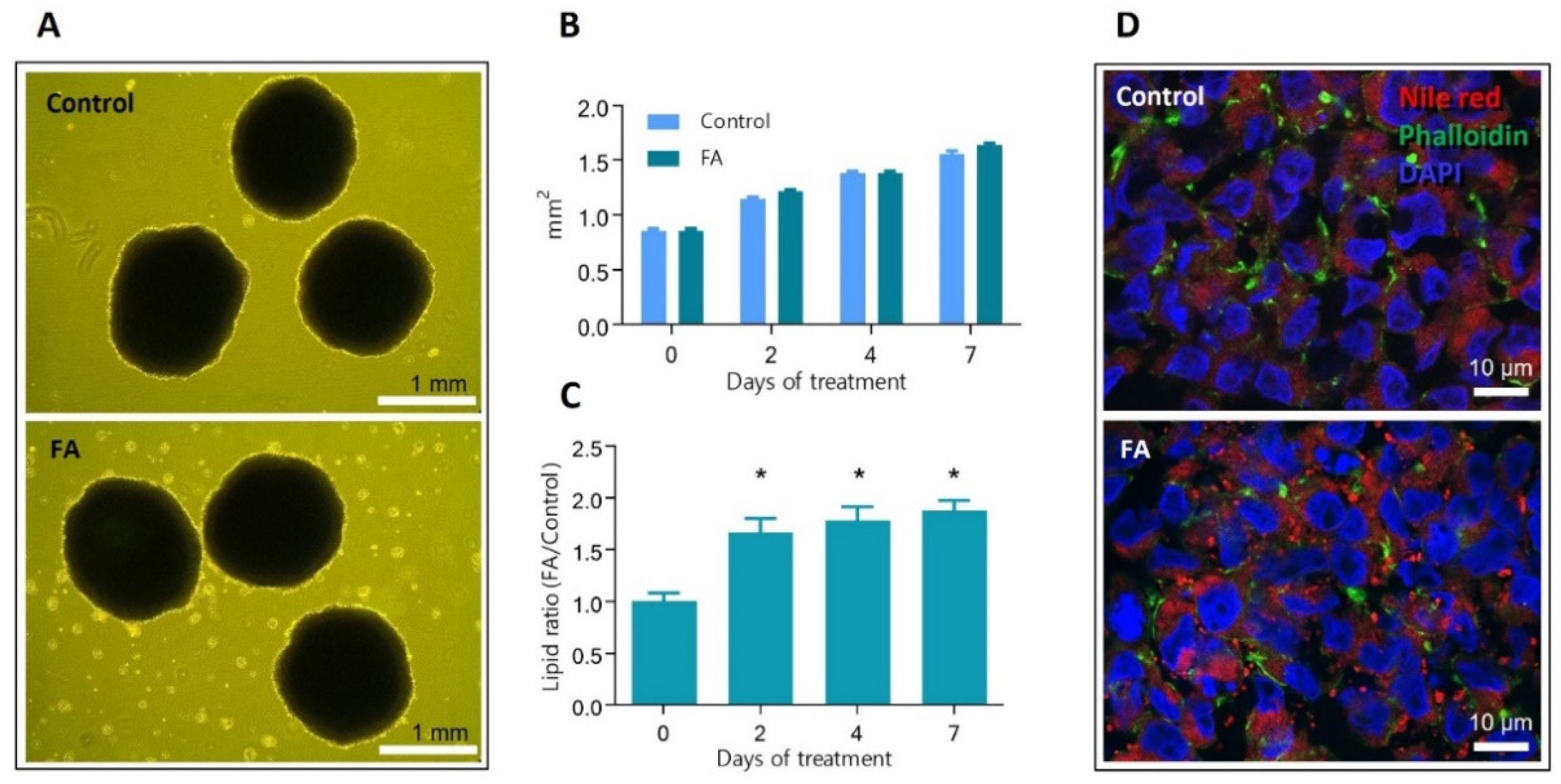
2.6. Quantification of Intracellular Lipid Content
2.7. Glucose Consumption and Glycogen Storage
2.8. Fluorescent Microscopy
2.9. Data Analysis of Functional Assays
2.10. Gene Expression Analysis
2.11. Lipidomics
2.12. Proteomics
3. Results
3.1. Spheroids Exposed to Palmitic and Oleic Acids Show Significantly Increased Levels of Intracellular Lipids
3.2. Hepatocytes in Spheroids Remain Functional despite Slight Changes in ATP Production and Glycogen Storage Management
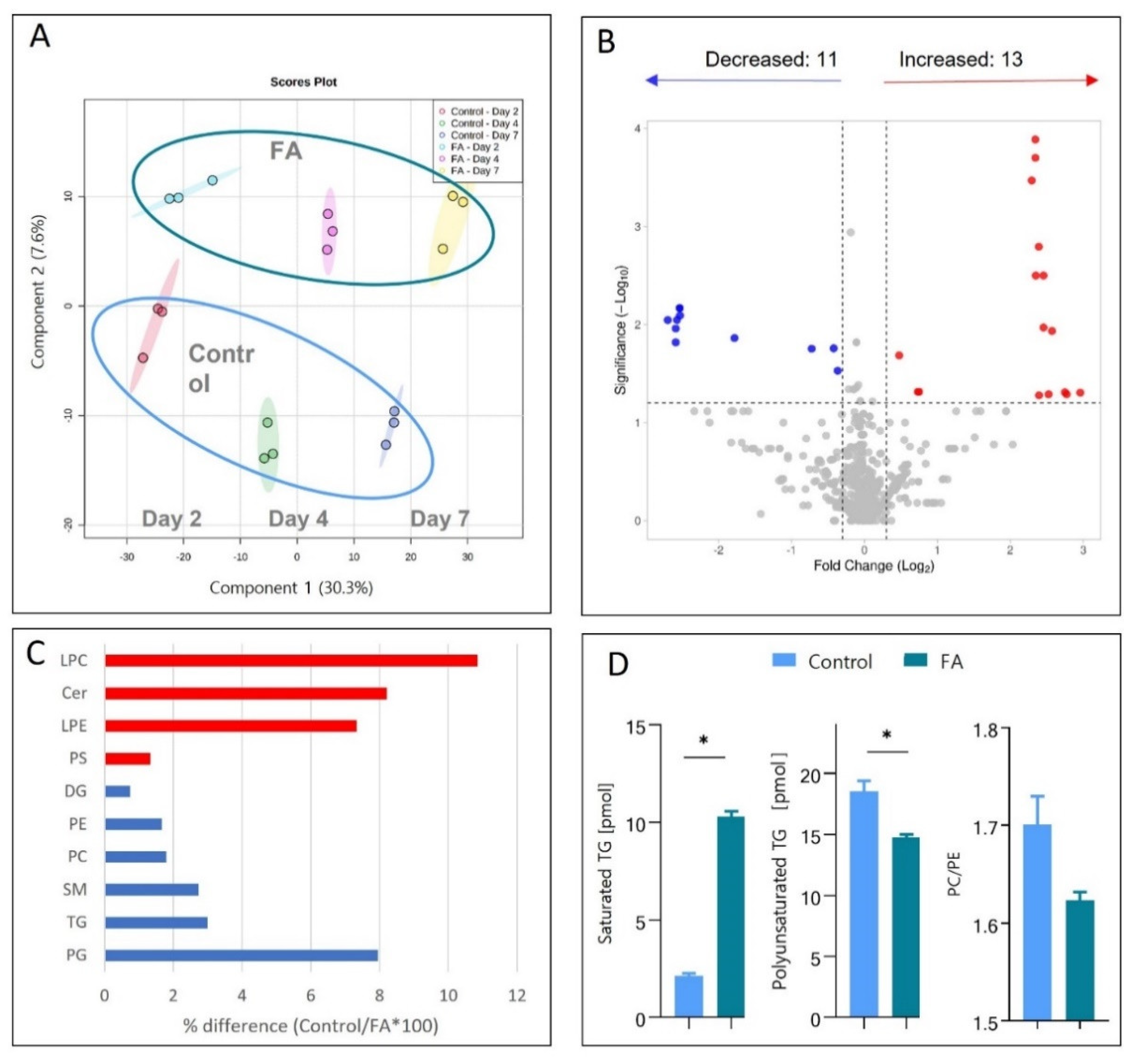
3.3. Palmitic and Oleic Acids Treatment Induce Lipidome Changes Like Those Seen In Vivo
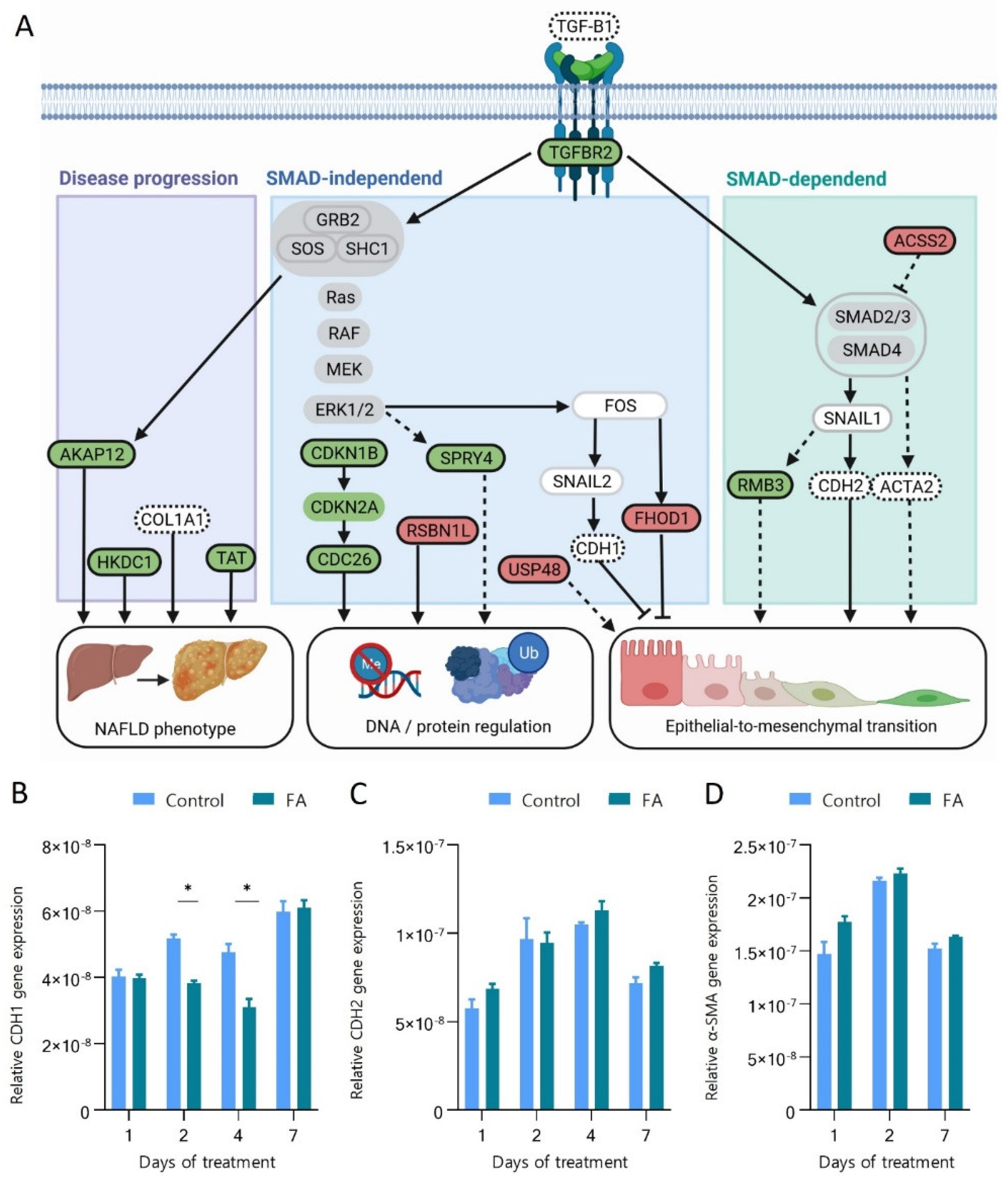
3.4. Protein and Gene Expression Changes Pinpoint the Involvement of the TGF-β Pathway and the Transition of Hepatocytes towards a Mesenchymal Phenotype during Their Early Response to FA Overload
3.5. Fatty Acids Increase Protein Ubiquitination and Degradation and Affect the Regulation of Chromatin Structure
3.6. Hepatic 3D Spheroids Show Similar Protein Regulation as Observed in NAFLD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hallsworth, K.; Adams, L.A. Lifestyle modification in NAFLD/NASH: Facts and figures. JHEP Rep. 2019, 1, 468–479. [Google Scholar] [CrossRef]
- Sanyal, A.J. Past, present and future perspectives in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 377–386. [Google Scholar] [CrossRef]
- Parthasarathy, G.; Revelo, X.; Malhi, H. Pathogenesis of Nonalcoholic Steatohepatitis: An Overview. Hepatol. Commun. 2020, 4, 478–492. [Google Scholar] [CrossRef]
- Pingitore, P.; Sasidharan, K.; Ekstrand, M.; Prill, S.; Lindén, D.; Romeo, S. Human Multilineage 3D Spheroids as a Model of Liver Steatosis and Fibrosis. Int. J. Mol. Sci. 2019, 20, 1629. [Google Scholar] [CrossRef]
- Yki-Järvinen, H. Liver Fat in the Pathogenesis of Insulin Resistance and Type 2 Diabetes. Dig. Dis. 2010, 28, 203–209. [Google Scholar] [CrossRef]
- Dam, T.V.; Toft, N.I.; Grøntved, L. Cell-Type Resolved Insights into the Cis-Regulatory Genome of NAFLD. Cells 2022, 11, 870. [Google Scholar] [CrossRef]
- Sumida, Y.; Nakajima, A.; Itoh, Y. Limitations of liver biopsy and non-invasive diagnostic tests for the diagnosis of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J. Gastroenterol. 2014, 20, 475–485. [Google Scholar] [CrossRef]
- Ahmed, M. Non-alcoholic fatty liver disease in 2015. World J. Hepatol. 2015, 7, 1450–1459. [Google Scholar] [CrossRef]
- Alwahsh, S.M.; Gebhardt, R. Dietary fructose as a risk factor for non-alcoholic fatty liver disease (NAFLD). Arch. Toxicol. 2017, 91, 1545–1563. [Google Scholar] [CrossRef]
- Lian, C.-Y.; Zhai, Z.-Z.; Li, Z.-F.; Wang, L. High fat diet-triggered non-alcoholic fatty liver disease: A review of proposed mechanisms. Chem. Interact. 2020, 330, 109199. [Google Scholar] [CrossRef]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-Dimensional Cell Culture: A Breakthrough in Vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Lau, J.K.C.; Zhang, X.; Yu, J. Animal models of non-alcoholic fatty liver disease: Current perspectives and recent advances. J. Pathol. 2017, 241, 36–44. [Google Scholar] [CrossRef]
- Mazzoleni, G.; Di Lorenzo, D.; Steimberg, N. Modelling tissues in 3D: The next future of pharmaco-toxicology and food research? Genes Nutr. 2009, 4, 13–22. [Google Scholar] [CrossRef]
- Misra, B.B.; Langefeld, C.D.; Olivier, M.; Cox, L.A. Integrated Omics: Tools, Advances, and Future Approaches. J. Mol. Endocrinol. 2018, 62, R21–R45. [Google Scholar] [CrossRef]
- Day, C.; Anstee, Q. The Genetics of Nonalcoholic Fatty Liver Disease: Spotlight on PNPLA3 and TM6SF2. Semin. Liver Dis. 2015, 35, 270–290. [Google Scholar] [CrossRef]
- Perakakis, N.; Stefanakis, K.; Mantzoros, C.S. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metabolism 2020, 111, 154320. [Google Scholar] [CrossRef]
- Fey, S.J.; Wrzesinski, K. Determination of Drug Toxicity Using 3D Spheroids Constructed from an Immortal Human Hepatocyte Cell Line. Toxicol. Sci. 2012, 127, 403–411. [Google Scholar] [CrossRef]
- Wrzesinski, K.; Fey, S.J. Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors. Bioengineering 2018, 5, 22. [Google Scholar] [CrossRef]
- de Almeida, I.T.; Cortez-Pinto, H.; Fidalgo, G.; Rodrigues, D.; Camilo, M. Plasma total and free fatty acids composition in human non-alcoholic steatohepatitis. Clin. Nutr. 2002, 21, 219–223. [Google Scholar] [CrossRef]
- Gori, M.; Simonelli, M.C.; Giannitelli, S.M.; Businaro, L.; Trombetta, M.; Rainer, A. Investigating Nonalcoholic Fatty Liver Disease in a Liver-on-a-Chip Microfluidic Device. PLoS ONE 2016, 11, e0159729. [Google Scholar] [CrossRef]
- Kostrzewski, T.; Cornforth, T.; A Snow, S.; Ouro-Gnao, L.; Rowe, C.; Large, E.M.; Hughes, D.J. Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J. Gastroenterol. 2017, 23, 204–215. [Google Scholar] [CrossRef]
- Kozyra, M.; Johansson, I.; Nordling, Å.; Ullah, S.; Lauschke, V.M.; Ingelman-Sundberg, M. Human hepatic 3D spheroids as a model for steatosis and insulin resistance. Sci. Rep. 2018, 8, 14297. [Google Scholar] [CrossRef]
- Wrzesinski, K.; Frandsen, H.S.; Calitz, C.; Gouws, C.; Korzeniowska, B.; Fey, S.J. Clinostat 3D Cell Culture: Protocols for the Preparation and Functional Analysis of Highly Reproducible, Large, Uniform Spheroids and Organoids. Methods Mol. Biol. 2021, 2273, 17–62. [Google Scholar] [CrossRef]
- Feaver, R.E.; Cole, B.K.; Lawson, M.J.; Hoang, S.A.; Marukian, S.; Blackman, B.R.; Figler, R.A.; Sanyal, A.J.; Wamhoff, B.R.; Dash, A. Development of an in vitro human liver system for interrogating nonalcoholic steatohepatitis. JCI Insight 2016, 1, e90954. [Google Scholar] [CrossRef]
- Dave, T.; Tilles, A.W.; Vemula, M. A Cell-Based Assay to Investigate Hypolipidemic Effects of Nonalcoholic Fatty Liver Disease Therapeutics. SLAS Discov. Adv. Sci. Drug Discov. 2018, 23, 274–282. [Google Scholar] [CrossRef]
- Štampar, M.; Frandsen, H.S.; Rogowska-Wrzesinska, A.; Wrzesinski, K.; Filipič, M.; Žegura, B. Hepatocellular carcinoma (HepG2/C3A) cell-based 3D model for genotoxicity testing of chemicals. Sci. Total Environ. 2021, 755, 143255. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- Hartler, J.; Triebl, A.; Ziegl, A.; Trötzmüller, M.; Rechberger, G.N.; A Zeleznik, O.; A Zierler, K.; Torta, F.; Cazenave-Gassiot, A.; Wenk, M.R.; et al. Deciphering lipid structures based on platform-independent decision rules. Nat. Methods 2017, 14, 1171–1174. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef]
- Wang, W.-Q.; Jensen, O.N.; Møller, I.M.; Hebelstrup, K.H.; Rogowska-Wrzesinska, A. Evaluation of sample preparation methods for mass spectrometry-based proteomic analysis of barley leaves. Plant Methods 2018, 14, 1–13. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Gaugaz, F.Z. Fast and Sensitive Total Protein and Peptide Assays for Proteomic Analysis. Anal. Chem. 2015, 87, 4110–4116. [Google Scholar] [CrossRef]
- Højrup, P. Analysis of Peptides and Conjugates by Amino Acid Analysis. In Peptide Antibodies; Humana Press: New York, NY, USA, 2015; Volume 1348, pp. 65–76. [Google Scholar] [CrossRef]
- Kovalchuk, S.; Jensen, O.; Rogowska-Wrzesinska, A. FlashPack: Fast and simple preparation of ultra-high performance capillary columns for LC-MS. bioRxiv 2018, 426678. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Schwämmle, V.; Hagensen, C.E.; Rogowska-Wrzesinska, A.; Jensen, O.N. PolySTest: Robust Statistical Testing of Proteomics Data with Missing Values Improves Detection of Biologically Relevant Features. Mol. Cell. Proteom. 2020, 19, 1396–1408. [Google Scholar] [CrossRef]
- Michalak, W.; Tsiamis, V.; Schwämmle, V.; Rogowska-Wrzesińska, A. ComplexBrowser: A Tool for Identification and Quantification of Protein Complexes in Large-scale Proteomics Datasets. Mol. Cell. Proteom. 2019, 18, 2324–2334. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner-Martinez, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 1–9. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 Update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Wrzesinski, K.; Fey, S.J. After trypsinisation, 3D spheroids of C3A hepatocytes need 18 days to re-establish similar levels of key physiological functions to those seen in the liver. Toxicol. Res. 2013, 2, 123–135. [Google Scholar] [CrossRef]
- Kartsoli, S.; E Kostara, C.; Tsimihodimos, V.; Bairaktari, E.T.; Christodoulou, D.K. Lipidomics in non-alcoholic fatty liver disease. World J. Hepatol. 2020, 12, 436–450. [Google Scholar] [CrossRef]
- Gorden, D.; Myers, D.S.; Ivanova, P.T.; Fahy, E.; Maurya, M.R.; Gupta, S.; Min, J.; Spann, N.J.; McDonald, J.G.; Kelly, S.L.; et al. Biomarkers of NAFLD progression: A lipidomics approach to an epidemic. J. Lipid Res. 2015, 56, 722–736. [Google Scholar] [CrossRef]
- Marra, F.; Svegliati-Baroni, G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J. Hepatol. 2018, 68, 280–295. [Google Scholar] [CrossRef]
- Ma, D.W.L.; Arendt, B.M.; Hillyer, L.M.; Fung, S.K.; McGilvray, I.; Guindi, M.; Allard, J.P. Plasma phospholipids and fatty acid composition differ between liver biopsy-proven nonalcoholic fatty liver disease and healthy subjects. Nutr. Diabetes 2016, 6, e220. [Google Scholar] [CrossRef]
- Grefhorst, A.; van de Peppel, I.P.; Larsen, L.E.; Jonker, J.W.; Holleboom, A.G. The Role of Lipophagy in the Development and Treatment of Non-Alcoholic Fatty Liver Disease. Front. Endocrinol. 2021, 11, 601627. [Google Scholar] [CrossRef]
- Waite, K.A.; Cabilio, N.R.; Vance, D.E. Choline Deficiency–Induced Liver Damage Is Reversible in Pemt−/− Mice. J. Nutr. 2002, 132, 68–71. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Sun, H.; Liu, X.; Zheng, Y.; Xu, D.; Wang, J.; Jia, D.; Han, X.; Liu, F.; et al. Defective Phosphatidylglycerol Remodeling Causes Hepatopathy, Linking Mitochondrial Dysfunction to Hepatosteatosis. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 763–781. [Google Scholar] [CrossRef]
- Peng, K.-Y.; Watt, M.J.; Rensen, S.S.; Greve, J.W.; Huynh, K.; Jayawardana, K.S.; Meikle, P.J.; Meex, R.C.R. Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J. Lipid Res. 2018, 59, 1977–1986. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Lujan, P.V.; Esmel, E.V.; Meseguer, E.S. Overview of Non-Alcoholic Fatty Liver Disease (NAFLD) and the Role of Sugary Food Consumption and Other Dietary Components in Its Development. Nutrients 2021, 13, 1442. [Google Scholar] [CrossRef] [PubMed]
- Arendt, B.M.; Ma, D.W.; Simons, B.; Noureldin, S.A.; Therapondos, G.; Guindi, M.; Sherman, M.; Allard, J.P. Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Appl. Physiol. Nutr. Metab. 2013, 38, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Tretbar, S.; Krausbeck, P.; Müller, A.; Friedrich, M.; Vaxevanis, C.; Bukur, J.; Jasinski-Bergner, S.; Seliger, B. TGF-β inducible epithelial-to-mesenchymal transition in renal cell carcinoma. Oncotarget 2019, 10, 1507–1524. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Zhou, R.-B.; Lu, X.-L.; Zhang, C.-Y.; Yin, D.-C. RNA binding motif protein 3: A potential biomarker in cancer and thera-peutic target in neuroprotection. Available online: www.impactjournals.com/oncotarget (accessed on 1 February 2022).
- Pilotte, J.; Kiosses, W.; Chan, S.W.; Makarenkova, H.P.; Dupont-Versteegden, E.; Vanderklish, P.W. Morphoregulatory functions of the RNA-binding motif protein 3 in cell spreading, polarity and migration. Sci. Rep. 2018, 8, 1–19. [Google Scholar] [CrossRef]
- Moffett, J.R.; Puthillathu, N.; Vengilote, R.; Jaworski, D.M.; Namboodiri, A.M. Acetate Revisited: A Key Biomolecule at the Nexus of Metabolism, Epigenetics and Oncogenesis—Part 1: Acetyl-CoA, Acetogenesis and Acyl-CoA Short-Chain Synthetases. Front. Physiol. 2020, 11, 580167. [Google Scholar] [CrossRef]
- Xiong, J.; Kawagishi, H.; Yan, Y.; Liu, J.; Wells, Q.S.; Edmunds, L.R.; Fergusson, M.M.; Yu, Z.-X.; Rovira, I.I.; Brittain, E.L.; et al. A Metabolic Basis for Endothelial-to-Mesenchymal Transition. Mol. Cell 2018, 69, 689–698.e7. [Google Scholar] [CrossRef]
- Sureshbabu, A.; Muhsin, S.A.; Choi, M.E. TGF-β signaling in the kidney: Profibrotic and protective effects. Am. J. Physiol. Physiol. 2016, 310, F596–F606. [Google Scholar] [CrossRef]
- Vander Ark, A.; Cao, J.; Li, X. TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 2018, 52, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Iskratsch, T.; Yu, C.-H.; Mathur, A.; Liu, S.; Stévenin, V.; Dwyer, J.; Hone, J.; Ehler, E.; Sheetz, M. FHOD1 Is Needed for Directed Forces and Adhesion Maturation during Cell Spreading and Migration. Dev. Cell 2013, 27, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Kobielak, A.; Pasolli, H.A.; Fuchs, E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 2004, 6, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Gomez, S.J.; Maziveyi, M.; Alahari, S.K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 2016, 15, 1–14. [Google Scholar] [CrossRef]
- Chu, I.M.; Hengst, L.; Slingerland, J.M. The Cdk inhibitor p27 in human cancer: Prognostic potential and relevance to anticancer therapy. Nat. Cancer 2008, 8, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Razavipour, S.F.; Harikumar, K.B.; Slingerland, J.M. p27 as a Transcriptional Regulator: New Roles in Development and Cancer. Cancer Res. 2020, 80, 3451–3458. [Google Scholar] [CrossRef] [PubMed]
- Masoumi-Moghaddam, S.; Amini, A.; Wei, A.-Q.; Robertson, G.; Morris, D.L. Sprouty 2 protein, but not Sprouty 4, is an independent prognostic biomarker for human epithelial ovarian cancer. Int. J. Cancer 2015, 137, 560–570. [Google Scholar] [CrossRef]
- Sun, M.; Huang, F.; Yu, D.; Zhang, Y.; Xu, H.; Zhang, L.; Li, L.; Dong, L.; Guo, L.; Wang, S. Autoregulatory loop between TGF-β1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma modulates proliferation and differentiation. Cell Death Dis. 2015, 6, e1859. [Google Scholar] [CrossRef]
- Lee, S.H.; Schloss, D.J.; Jarvis, L.; Krasnow, M.A.; Swain, J.L. Inhibition of Angiogenesis by a Mouse Sprouty Protein. J. Biol. Chem. 2001, 276, 4128–4133. [Google Scholar] [CrossRef]
- Du, L.; Li, Y.; Kang, M.; Feng, M.; Ren, Y.; Dai, H.; Wang, Y.; Wang, Y.; Tang, B. USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 2021, 81, 3822–3834. [Google Scholar] [CrossRef]
- Silvestrini, V.C.; Thomé, C.H.; Albuquerque, D.; Palma, C.D.S.; Ferreira, G.A.; Lanfredi, G.P.; Masson, A.P.; Delsin, L.E.A.; Ferreira, F.U.; de Souza, F.C.; et al. Proteomics analysis reveals the role of ubiquitin specific protease (USP47) in Epithelial to Mesenchymal Transition (EMT) induced by TGFβ2 in breast cells. J. Proteom. 2020, 219, 103734. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, D.; Zhao, J.; Weathington, N.M.; Shang, D.; Zhao, Y. The deubiquitinating enzyme USP48 stabilizes TRAF2 and reduces E-cadherin-mediated adherens junctions. FASEB J. 2018, 32, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef]
- Loh, C.-Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Nieman, M.T.; Prudoff, R.S.; Johnson, K.R.; Wheelock, M.J. N-Cadherin Promotes Motility in Human Breast Cancer Cells Regardless of Their E-Cadherin Expression. J. Cell Biol. 1999, 147, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Nouchi, T.; Tanaka, Y.; Tsukada, T.; Sato, C.; Marumo, F. Appearance of α-smooth-muscle-actin-positive cells in hepatic fibrosis. Liver Int. 1991, 11, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Zachariae, W.; Nasmyth, K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes Dev. 1999, 13, 2039–2058. Available online: www.genesdev.org (accessed on 1 March 2022). [CrossRef]
- Cetkovská, K.; Šustová, H.; Uldrijan, S. Ubiquitin-specific peptidase 48 regulates Mdm2 protein levels independent of its deubiquitinase activity. Sci. Rep. 2017, 7, srep43180. [Google Scholar] [CrossRef]
- Lei, X.; Li, X.; Chen, H.; Liu, Z. USP48 Sustains Chemoresistance and Metastasis in Ovarian Cancer. Curr. Cancer Drug Targets 2020, 20, 689–699. [Google Scholar] [CrossRef]
- Zhao, D.; Besser, A.H.; A Wander, S.; Sun, J.; Zhou, W.; Wang, B.; Ince, T.; Durante, M.; Guo, W.; Mills, G.; et al. Cytoplasmic p27 promotes epithelial–mesenchymal transition and tumor metastasis via STAT3-mediated Twist1 upregulation. Oncogene 2015, 34, 5447–5459. [Google Scholar] [CrossRef]
- Yoon, H.; Kim, M.; Jang, K.; Shin, M.; Besser, A.; Xiao, X.; Zhao, D.; Wander, S.A.; Briegel, K.; Morey, L.; et al. p27 transcriptionally coregulates cJun to drive programs of tumor progression. Proc. Natl. Acad. Sci. USA 2019, 116, 7005–7014. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Ramani, K.; Tomasi, M.L.; Berlind, J.; Mavila, N.; Sun, Z. Role of A-Kinase Anchoring Protein Phosphorylation in Alcohol-Induced Liver Injury and Hepatic Stellate Cell Activation. Am. J. Pathol. 2018, 188, 640–655. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Banton, S.; Tran, V.T.; Konomi, J.V.; Li, S.; Jones, D.P.; Vos, M.B. Amino Acid Metabolism is Altered in Adolescents with Nonalcoholic Fatty Liver Disease—An Untargeted, High Resolution Metabolomics Study. J. Pediatr. 2016, 172, 14–19.e5. [Google Scholar] [CrossRef]
- Pusec, C.M.; De Jesus, A.; Khan, W.; Terry, A.R.; E Ludvik, A.; Xu, K.; Giancola, N.; Pervaiz, H.; Smith, E.D.; Ding, X.; et al. Hepatic HKDC1 Expression Contributes to Liver Metabolism. Endocrinology 2018, 160, 313–330. [Google Scholar] [CrossRef] [PubMed]
- Kuchay, M.S.; Choudhary, N.S.; Mishra, S.K. Pathophysiological mechanisms underlying MAFLD. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1875–1887. [Google Scholar] [CrossRef]
- Duriez, M.; Jacquet, A.; Hoet, L.; Roche, S.; Bock, M.-D.; Rocher, C.; Haussy, G.; Vigé, X.; Bocskei, Z.; Slavnic, T.; et al. A 3D Human Liver Model of Nonalcoholic Steatohepatitis. J. Clin. Transl. Hepatol. 2020, 8, 359–370. [Google Scholar] [CrossRef]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered Liver-On-A-Chip Platform to Mimic Liver Functions and Its Biomedical Applications: A Review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef]
- Van der Vusse, G.J. Albumin as Fatty Acid Transporter. Drug Metab. Pharmacokinet. 2009, 24, 300–307. [Google Scholar] [CrossRef]
- Thompson, A.L.; Lim-Fraser, M.Y.-C.; Kraegen, E.W.; Cooney, G.J. Effects of individual fatty acids on glucose uptake and glycogen synthesis in soleus muscle in vitro. Am. J. Physiol. Metab. 2000, 279, E577–E584. [Google Scholar] [CrossRef]
- Ishii, M.; Maeda, A.; Tani, S.; Akagawa, M. Palmitate induces insulin resistance in human HepG2 hepatocytes by enhancing ubiquitination and proteasomal degradation of key insulin signaling molecules. Arch. Biochem. Biophys. 2015, 566, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Cassader, M.; Paschetta, E.; Gambino, R. Bioactive Lipid Species and Metabolic Pathways in Progression and Resolution of Nonalcoholic Steatohepatitis. Gastroenterology 2018, 155, 282–302.e8. [Google Scholar] [CrossRef] [PubMed]
- Paradies, G.; Paradies, V.; Ruggiero, F.M.; Petrosillo, G. Role of Cardiolipin in Mitochondrial Function and Dynamics in Health and Disease: Molecular and Pharmacological Aspects. Cells 2019, 8, 728. [Google Scholar] [CrossRef] [PubMed]
- Nassir, F.; Ibdah, J.A. Role of Mitochondria in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2014, 15, 8713–8742. [Google Scholar] [CrossRef] [PubMed]
- Simões, I.C.M.; Fontes, A.; Pinton, P.; Zischka, H.; Wieckowski, M.R. Mitochondria in non-alcoholic fatty liver disease. Int. J. Biochem. Cell Biol. 2018, 95, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Ye, H. Overview of Lipidomic Analysis Triglyceride Molecular Species in Biological Lipid Extracts. J. Agric. Food Chem. 2021, 69, 8895–8909. [Google Scholar] [CrossRef] [PubMed]
- Ludvik, A.E.; Pusec, C.M.; Priyadarshini, M.; Angueira, A.R.; Guo, C.; Lo, A.; Hershenhouse, K.S.; Yang, G.-Y.; Ding, X.; Reddy, T.E.; et al. HKDC1 Is a Novel Hexokinase Involved in Whole-Body Glucose Use. Endocrinology 2016, 157, 3452–3461. [Google Scholar] [CrossRef]
- Niu, L.; E Geyer, P.; Albrechtsen, N.J.W.; Gluud, L.L.; Santos, A.; Doll, S.; Treit, P.V.; Holst, J.J.; Knop, F.K.; Vilsbøll, T.; et al. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease. Mol. Syst. Biol. 2019, 15, e8793. [Google Scholar] [CrossRef]
- Nair, B.; Nath, L.R. Inevitable role of TGF-β1 in progression of nonalcoholic fatty liver disease. J. Recept. Signal Transduct. 2020, 40, 195–200. [Google Scholar] [CrossRef]
- Yang, L.; Roh, Y.S.; Song, J.; Zhang, B.; Liu, C.; Loomba, R.; Seki, E. Transforming growth factor beta signaling in hepatocytes participates in steatohepatitis through regulation of cell death and lipid metabolism in mice. Hepatology 2013, 59, 483–495. [Google Scholar] [CrossRef]
- Cuvellier, M.; Ezan, F.; Oliveira, H.; Rose, S.; Fricain, J.-C.; Langouët, S.; Legagneux, V.; Baffet, G. 3D culture of HepaRG cells in GelMa and its application to bioprinting of a multicellular hepatic model. Biomaterials 2020, 269, 120611. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2018, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, I.; Caballero-Díaz, D. Transforming Growth Factor-β-Induced Cell Plasticity in Liver Fibrosis and Hepatocarcinogenesis. Front. Oncol. 2018, 8, 357. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Zhelnin, L.; Sanfiz, A.; Pan, J.; Li, Z.; Yarde, M.; McCarty, J.; Jarai, G. Development and validation of an in vitro 3D model of NASH with severe fibrotic phenotype. Am. J. Transl. Res. 2019, 11, 1531–1540. [Google Scholar]
- Hurrell, T.; Kastrinou-Lampou, V.; Fardellas, A.; Hendriks, D.F.G.; Nordling, A.; Johansson, I.; Baze, A.; Parmentier, C.; Richert, L.; Ingelman-Sundberg, M. Human Liver Spheroids as a Model to Study Aetiology and Treatment of Hepatic Fibrosis. Cells 2020, 9, 964. [Google Scholar] [CrossRef]
- Song, Y.; Kim, S.; Heo, J.; Shum, D.; Lee, S.-Y.; Lee, M.; Kim, A.-R.; Seo, H.R. Identification of hepatic fibrosis inhibitors through morphometry analysis of a hepatic multicellular spheroids model. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Di Gregorio, J.; Robuffo, I.; Spalletta, S.; Giambuzzi, G.; De Iuliis, V.; Toniato, E.; Martinotti, S.; Conti, P.; Flati, V. The Epithelial-to-Mesenchymal Transition as a Possible Therapeutic Target in Fibrotic Disorders. Front. Cell Dev. Biol. 2020, 8, 607483. [Google Scholar] [CrossRef]
- Dooley, S.; Hamzavi, J.; Ciuclan, L.; Godoy, P.; Ilkavets, I.; Ehnert, S.; Ueberham, E.; Gebhardt, R.; Kanzler, S.; Geier, A.; et al. Hepatocyte-Specific Smad7 Expression Attenuates TGF-β–Mediated Fibrogenesis and Protects Against Liver Damage. Gastroenterology 2008, 135, 642–659.e46. [Google Scholar] [CrossRef]
- Xie, G.; Diehl, A.M. Evidence for and against epithelial-to-mesenchymal transition in the liver. Am. J. Physiol. Liver Physiol. 2013, 305, G881–G890. [Google Scholar] [CrossRef]
- Nitta, T.; Kim, J.-S.; Mohuczy, D.; Behrns, K.E. Murine cirrhosis induces hepatocyte epithelial mesenchymal transition and alterations in survival signaling pathways. Hepatology 2008, 48, 909–919. [Google Scholar] [CrossRef]
- Zeisberg, M.; Yang, C.; Martino, M.; Duncan, M.B.; Rieder, F.; Tanjore, H.; Kalluri, R. Fibroblasts Derive from Hepatocytes in Liver Fibrosis via Epithelial to Mesenchymal Transition. J. Biol. Chem. 2007, 282, 23337–23347. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.S.; Diehl, A.M. Epithelial-to-mesenchymal transitions in the liver. Hepatology 2009, 50, 2007–2013. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yu, K.; Li, Q.; Shi, G. Involvement of epithelial-mesenchymal transition in liver fibrosis. Saudi J. Gastroenterol. 2018, 24, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Taura, K.; Miura, K.; Iwaisako, K.; Österreicher, C.H.; Kodama, Y.; Penz-Österreicher, M.; Brenner, D.A. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology 2010, 51, 1027–1036. [Google Scholar] [CrossRef]
- Taylor, R.S.; Taylor, R.J.; Bayliss, S.; Hagström, H.; Nasr, P.; Schattenberg, J.M.; Ishigami, M.; Toyoda, H.; Wong, V.W.-S.; Peleg, N.; et al. Association Between Fibrosis Stage and Outcomes of Patients with Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology 2020, 158, 1611–1625.e12. [Google Scholar] [CrossRef] [PubMed]
- Knowles, B.B.; Howe, C.C.; Aden, D.P. Human Hepatocellular Carcinoma Cell Lines Secrete the Major Plasma Proteins and Hepatitis B Surface Antigen. Science 1980, 209, 497–499. [Google Scholar] [CrossRef]
- Wrzesinski, K. From 2D to 3D—A New Dimension for Modelling the Effect of Natural Products on Human Tissue. Curr. Pharm. Des. 2015, 21, 5605–5616. [Google Scholar] [CrossRef]
- Wolffe, A.P.; Tata, J.R. Primary culture, cellular stress and differentiated function Tissue dtiaggregation Cellular phe-notype Hormonal responsiveness Heat shock protein Culture shock. FEBS Lett. 1886, 176, 8–15. [Google Scholar] [CrossRef]
- Zeilinger, K.; Freyer, N.; Damm, G.; Seehofer, D.; Knöspel, F. Cell sources for in vitro human liver cell culture models. Exp. Biol. Med. 2016, 241, 1684–1698. [Google Scholar] [CrossRef]
- Zhou, Z.; Xu, M.-J.; Gao, B. Hepatocytes: A key cell type for innate immunity. Cell. Mol. Immunol. 2015, 13, 301–315. [Google Scholar] [CrossRef]
- Fey, S.J.; Korzeniowska, B.; Wrzesinski, K. Response to and recovery from treatment in human liver-mimetic clinostat spheroids: A model for assessing repeated-dose drug toxicity. Toxicol. Res. 2020, 9, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Wrzesinski, K.; Magnone, M.C.; Hansen, L.V.; Kruse, M.E.; Bergauer, T.; Bobadilla, M.; Gubler, M.; Mizrahi, J.; Zhang, K.; Andreasen, C.M.; et al. HepG2/C3A 3D spheroids exhibit stable physiological functionality for at least 24 days after recovering from trypsinisation. Toxicol. Res. 2013, 2, 163–172. [Google Scholar] [CrossRef]


| Class | Name | Fold Change | p-Value |
|---|---|---|---|
| Ceramides (Cer) | CE (22:5) | 1.57 | 0.27 |
| Cer (d28:2) | 6.83 | 0.05 | |
| Cer (30:0) | 1.46 | 0.34 | |
| Diacylglycerol (DG) | DG (42:9) | 5.48 | 0.01 |
| DG (36:6) | −1.11 | 0.05 | |
| Lysophosphatidylcholine (LPC) | LPC (24:2) | 2.6 | 0.00 |
| Lysophosphatidylethanolamine (LPE) | LPE (13:0) | 1.68 | 0.05 |
| LPE (26:2) | 1.09 | 0.02 | |
| Phosphatidylcholine (PC) | PC (46:8) | 5.94 | 0.01 |
| PC (42:12) | 5.48 | 0.00 | |
| PC (31:2) | −1.08 | 0.02 | |
| PC (44:11) | −1.08 | 0.04 | |
| PC (33:5) | −1.14 | 0.00 | |
| PC (16:0/18:2) | −5.79 | 0.01 | |
| Glycerophosphoethanolamines (PE) | PE (43:6) | 5.09 | 0.00 |
| PE (16:0/18:2) | −1.08 | 0.02 | |
| PE (37:2) | −5.79 | 0.01 | |
| Phosphoglycerols (PG) | PG (28:2) | 5.24 | 0.00 |
| PG (44:2) | −3.09 | 0.01 | |
| PG (47:4) | −5.94 | 0.01 | |
| Phosphatidylserine (PS) | PS (37:4) | 7.77 | 0.05 |
| PS (35:1) | −6.48 | 0.03 | |
| Triacylglycerol (TG) | TG (18:0/18:2/20:0) | 5.25 | 0.00 |
| TG (45:0) | 5.07 | 0.00 | |
| TG (47:2) | −1.05 | 0.04 | |
| TG (43:1) | −1.16 | 0.05 | |
| TG (43:2) | −1.29 | 0.03 | |
| TG (47:4) | −1.34 | 0.02 | |
| TG (55:0) | −1.65 | 0.02 | |
| TG (42:4) | −6.01 | 0.01 | |
| TG (50:7) | −6.01 | 0.02 |
| Name | Gene Name | Fold Change | Q-Value |
|---|---|---|---|
| DNA and protein regulation | |||
| Anaphase-promoting complex subunit CDC26 | CDC26 | 1.74 | 0.0169 |
| Ubiquitin carboxyl-terminal hydrolase 48 | USP48 | −1.53 | 0.0369 |
| Lysine-specific demethylase RSBN1L | RSBN1L | −1.99 | 0.0277 |
| NAFLD phenotype | |||
| Hexokinase HKDC1 | HKDC1 | 3.72 | 0.0002 |
| A-kinase anchor protein 12 | AKAP12 | 2.58 | 0.0002 |
| Tyrosine aminotransferase | TAT | 1.93 | 0.0035 |
| EMT transition | |||
| Cyclin-dependent kinase inhibitor 1B | CDKN1B | 3.17 | 0.0279 |
| RNA-binding protein 3 | RBM3 | 2.10 | 0.0011 |
| TGF-beta receptor type-2 | TGFBR2 | 1.95 | 0.0035 |
| Protein sprouty homolog 4 | SPRY4 | 1.75 | 0.0084 |
| Acetyl-coenzyme A synthetase | ACSS2 | −1.53 | 0.0080 |
| FH1/FH2 domain-containing protein 1 | FHOD1 | −1.69 | 0.0370 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frandsen, H.S.; Vej-Nielsen, J.M.; Smith, L.E.; Sun, L.; Mikkelsen, K.L.; Thulesen, A.P.; Hagensen, C.E.; Yang, F.; Rogowska-Wrzesinska, A. Mapping Proteome and Lipidome Changes in Early-Onset Non-Alcoholic Fatty Liver Disease Using Hepatic 3D Spheroids. Cells 2022, 11, 3216. https://doi.org/10.3390/cells11203216
Frandsen HS, Vej-Nielsen JM, Smith LE, Sun L, Mikkelsen KL, Thulesen AP, Hagensen CE, Yang F, Rogowska-Wrzesinska A. Mapping Proteome and Lipidome Changes in Early-Onset Non-Alcoholic Fatty Liver Disease Using Hepatic 3D Spheroids. Cells. 2022; 11(20):3216. https://doi.org/10.3390/cells11203216
Chicago/Turabian StyleFrandsen, Helle Sedighi, Joel Mario Vej-Nielsen, Lauren Elizabeth Smith, Lang Sun, Karoline Lindgaard Mikkelsen, Annemette Præstegaard Thulesen, Christina Erika Hagensen, Fuquan Yang, and Adelina Rogowska-Wrzesinska. 2022. "Mapping Proteome and Lipidome Changes in Early-Onset Non-Alcoholic Fatty Liver Disease Using Hepatic 3D Spheroids" Cells 11, no. 20: 3216. https://doi.org/10.3390/cells11203216
APA StyleFrandsen, H. S., Vej-Nielsen, J. M., Smith, L. E., Sun, L., Mikkelsen, K. L., Thulesen, A. P., Hagensen, C. E., Yang, F., & Rogowska-Wrzesinska, A. (2022). Mapping Proteome and Lipidome Changes in Early-Onset Non-Alcoholic Fatty Liver Disease Using Hepatic 3D Spheroids. Cells, 11(20), 3216. https://doi.org/10.3390/cells11203216






