Glutathione S-Transferases S1, Z1 and A1 Serve as Prognostic Factors in Glioblastoma and Promote Drug Resistance through Antioxidant Pathways
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Data and Bioinformatics Analyses
2.2. Cell Culture and Western Blotting
2.3. Cell Counting Kit 8 (CCK8) Assays
2.4. Quantification of GSH and 4-Hydroxynonanol (4-HNE)
2.5. Cell Proliferation Assay
2.6. Apoptosis Assay
2.7. Data Analysis
3. Results
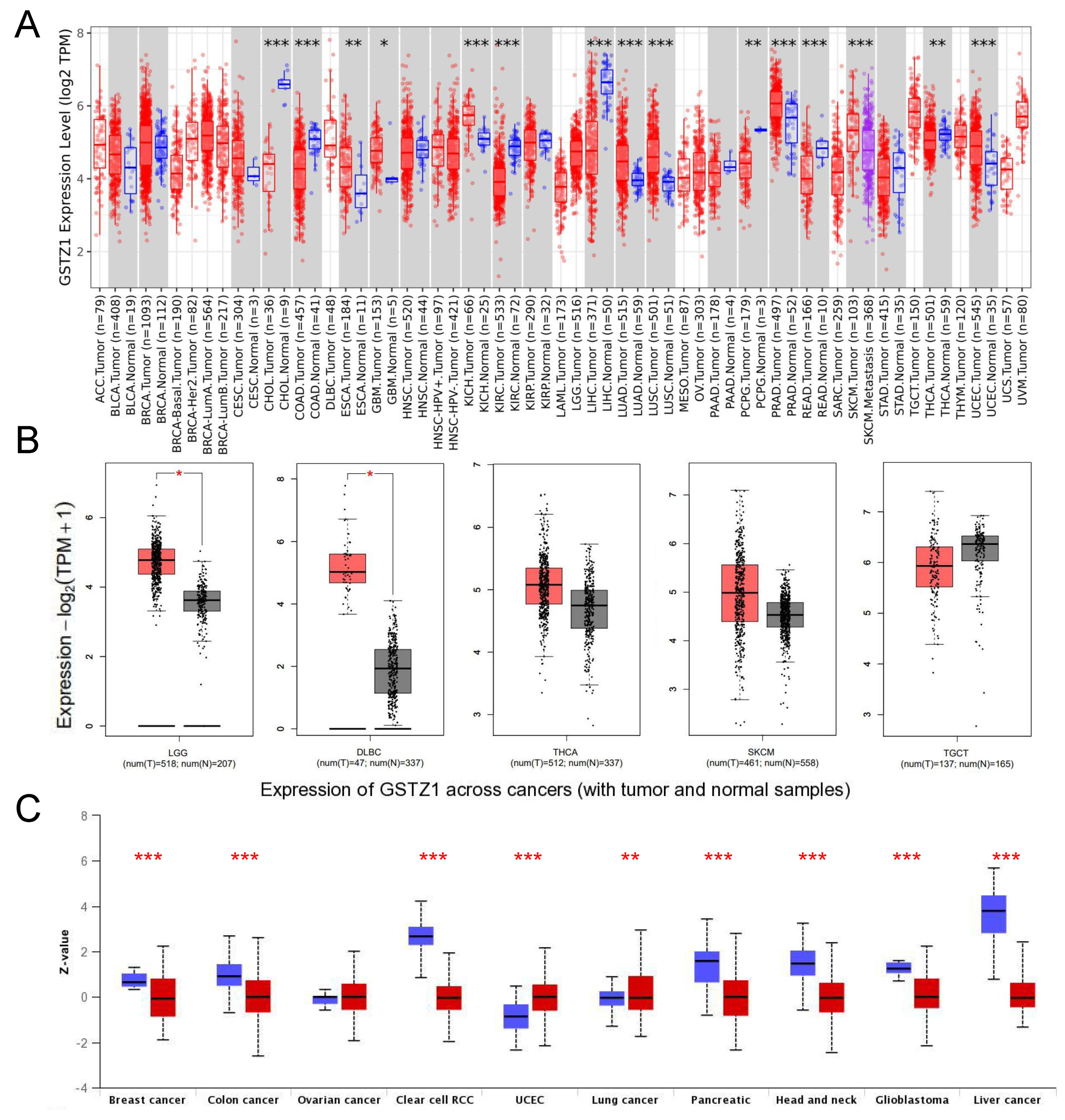
3.1. HPGDS, GSTZ1, and GSTA1 Are Abnormally Expressed in Various Cancers
3.2. Abnormally Expressed GSTs Are Significantly Correlated with the Prognosis of Patients with Various Cancers
3.3. Single Nucleotide Polymorphisms Associated with Cancer Affect the Structure and Function of GSTs
3.4. GSTs Are Related to Tumor Immunity and Participate in Multiple Biological Processes
3.5. High HPGDS Expression Predicts Poor Prognosis in Glioma Patients
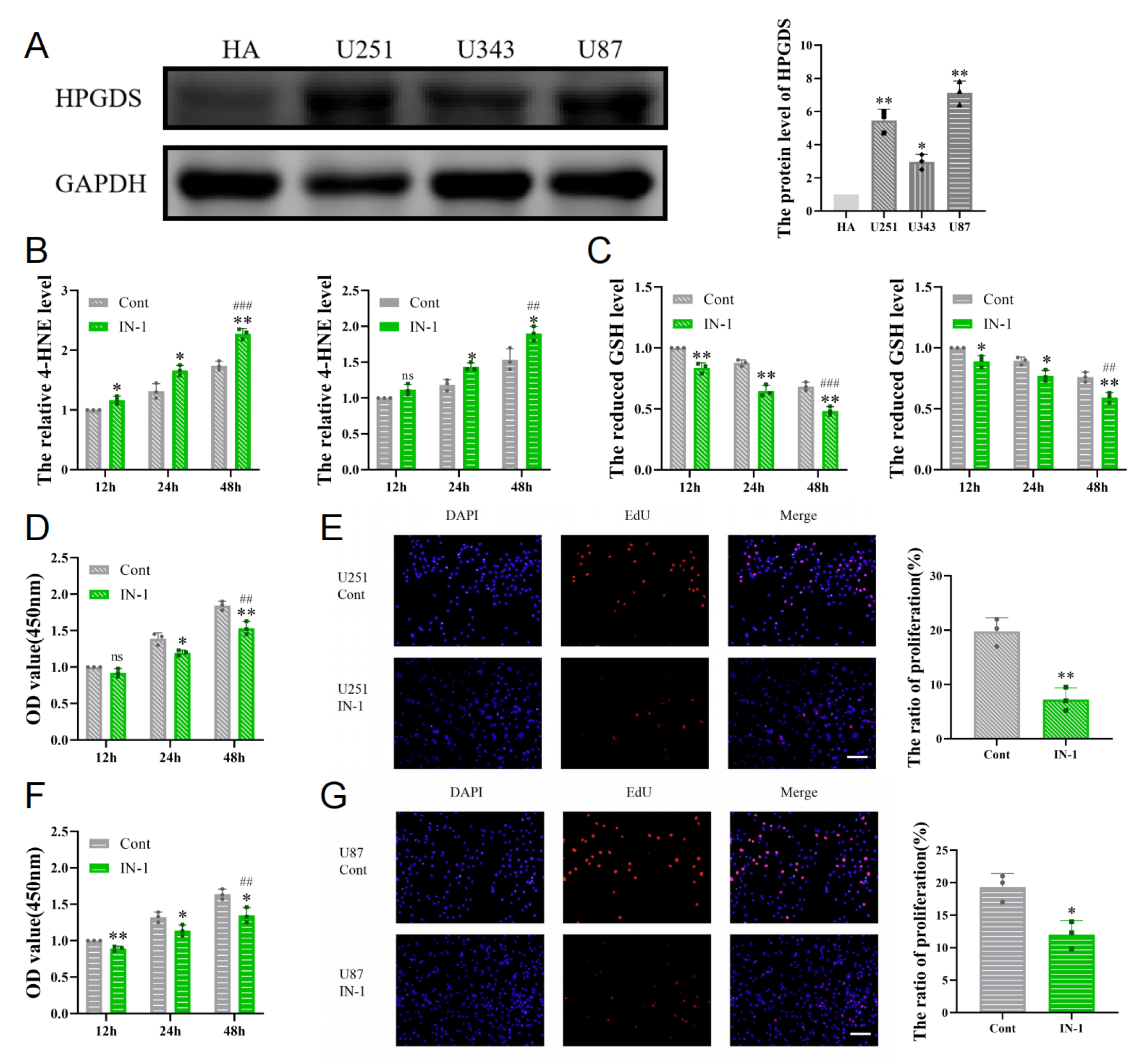
3.6. Inhibition of HPGDS Lowers GSH Levels and Decreases Proliferation of GBM Cells
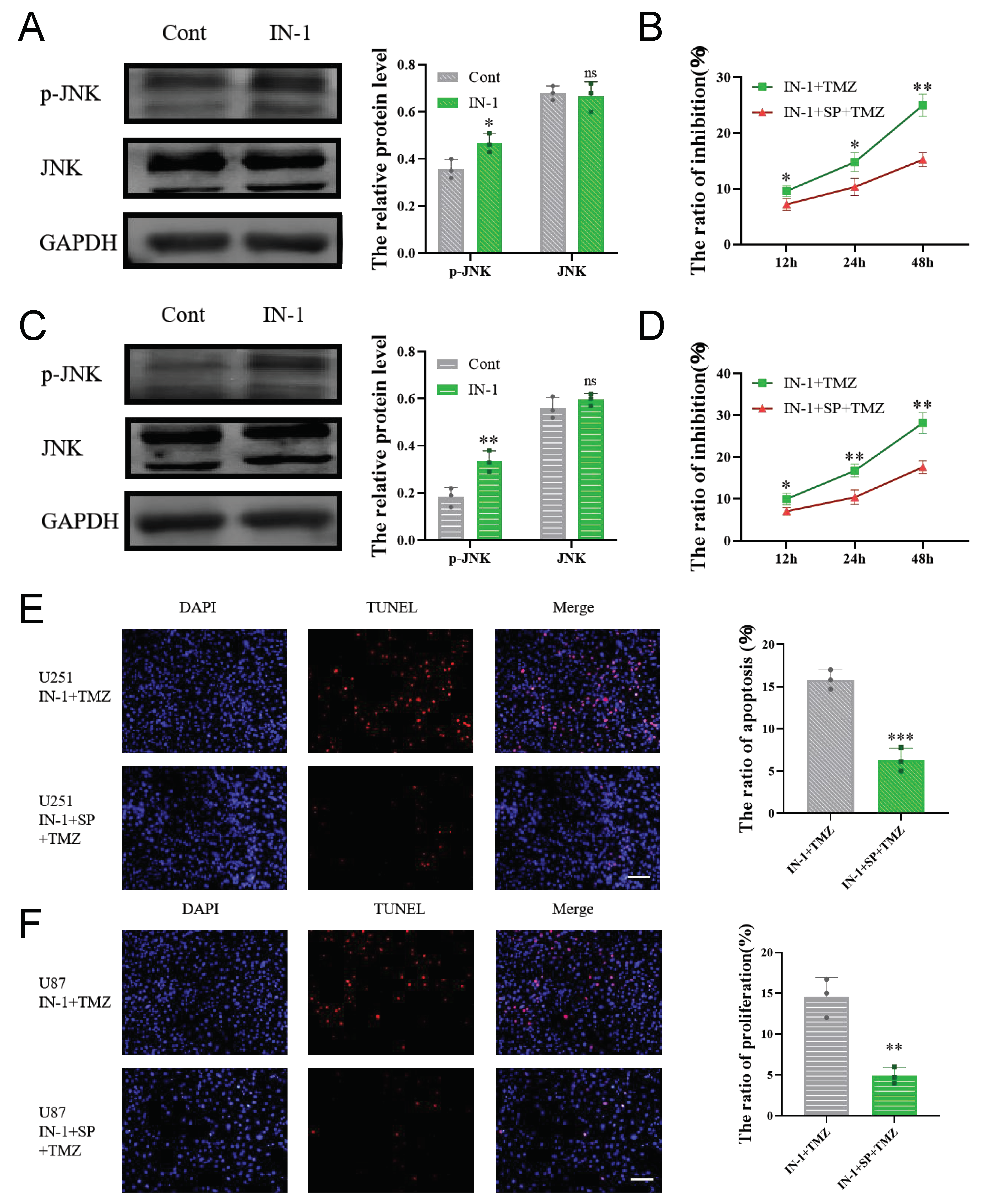
3.7. Inhibition of HPGDS Reduces Drug Resistance in GBM
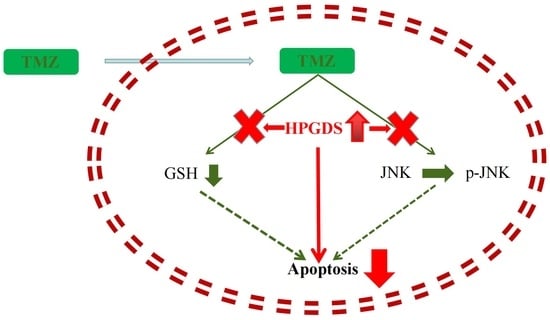
3.8. HPGDS Mediates Drug Resistance of GBM via the JNK Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adhikaree, J.; Moreno-Vicente, J.; Kaur, A.P.; Jackson, A.M.; Patel, P.M. Resistance Mechanisms and Barriers to Successful Immunotherapy for Treating Glioblastoma. Cells 2020, 9, 263. [Google Scholar] [CrossRef]
- Ma, Q.; Long, W.; Xing, C.; Chu, J.; Luo, M.; Wang, H.Y.; Liu, Q.; Wang, R.F. Cancer Stem Cells and Immunosuppressive Microenvironment in Glioma. Front. Immunol. 2018, 9, 2924. [Google Scholar] [CrossRef]
- Broekman, M.L.; Maas, S.L.N.; Abels, E.R.; Mempel, T.R.; Krichevsky, A.M.; Breakefield, X.O. Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 2018, 14, 482–495. [Google Scholar] [CrossRef] [PubMed]
- De Cordova, S.; Shastri, A.; Tsolaki, A.G.; Yasmin, H.; Klein, L.; Singh, S.K.; Kishore, U. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front. Immunol. 2020, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Muir, M.; Gopakumar, S.; Traylor, J.; Lee, S.; Rao, G. Glioblastoma multiforme: Novel therapeutic targets. Expert. Opin. Ther. Targets 2020, 24, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Lauko, A.; Lo, A.; Ahluwalia, M.S.; Lathia, J.D. Cancer cell heterogeneity & plasticity in glioblastoma and brain tumors. Semin. Cancer Biol. 2022, 82, 162–175. [Google Scholar] [PubMed]
- Haar, C.P.; Hebbar, P.; Wallace, G.C.; Das, A.; Vandergrift, W.A., III; Smith, J.A.; Giglio, P.; Patel, S.J.; Ray, S.K.; Banik, N.L. Drug resistance in glioblastoma: A mini review. Neurochem. Res. 2012, 37, 1192–1200. [Google Scholar] [CrossRef]
- Johannessen, T.C.; Bjerkvig, R.; Tysnes, B.B. DNA repair and cancer stem-like cells--potential partners in glioma drug resistance? Cancer Treat. Rev. 2008, 34, 558–567. [Google Scholar] [CrossRef]
- Niu, B.; Liao, K.; Zhou, Y.; Wen, T.; Quan, G.; Pan, X.; Wu, C. Application of glutathione depletion in cancer therapy: Enhanced ROS-based therapy, ferroptosis, and chemotherapy. Biomaterials 2021, 277, 121110. [Google Scholar] [CrossRef]
- Tew, K.D. Glutathione-Associated Enzymes in Anticancer Drug Resistance. Cancer Res. 2016, 76, 7–9. [Google Scholar] [CrossRef]
- Chatterjee, A.; Gupta, S. The multifaceted role of glutathione S-transferases in cancer. Cancer Lett. 2018, 433, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Fruehauf, J.P.; Brem, H.; Brem, S.; Sloan, A.; Barger, G.; Huang, W.; Parker, R. In vitro drug response and molecular markers associated with drug resistance in malignant gliomas. Clin. Cancer Res. 2006, 12, 4523–4532. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Chen, N.F.; Wen, Z.H.; Yao, Z.K.; Tsui, K.H.; Kuo, H.M.; Chen, W.F. Glutathione S-Transferase M3 Is Associated with Glycolysis in Intrinsic Temozolomide-Resistant Glioblastoma Multiforme Cells. Int. J. Mol. Sci. 2021, 22, 7080. [Google Scholar] [CrossRef] [PubMed]
- Moltgen, S.; Piumatti, E.; Massafra, G.M.; Metzger, S.; Jaehde, U.; Kalayda, G.V. Cisplatin Protein Binding Partners and Their Relevance for Platinum Drug Sensitivity. Cells 2020, 9, 1322. [Google Scholar] [CrossRef]
- Dong, X.; Yang, Y.; Zhou, Y.; Bi, X.; Zhao, N.; Zhang, Z.; Li, L.; Hang, Q.; Zhang, R.; Chen, D.; et al. Glutathione S-transferases P1 protects breast cancer cell from adriamycin-induced cell death through promoting autophagy. Cell Death Differ. 2019, 26, 2086–2099. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Xie, L.; Lu, Y.; Hu, Z.; Chang, J. miR-133b reverses cisplatin resistance by targeting GSTP1 in cisplatin-resistant lung cancer cells. Int. J. Mol. Med. 2018, 41, 2050–2058. [Google Scholar] [CrossRef]
- Feng, Q.; Li, X.; Sun, W.; Sun, M.; Li, Z.; Sheng, H.; Xie, F.; Zhang, S.; Shan, C. Targeting G6PD reverses paclitaxel resistance in ovarian cancer by suppressing GSTP1. Biochem. Pharmacol. 2020, 178, 114092. [Google Scholar] [CrossRef]
- Rittchen, S.; Heinemann, A. Therapeutic Potential of Hematopoietic Prostaglandin D2 Synthase in Allergic Inflammation. Cells 2019, 8, 619. [Google Scholar] [CrossRef]
- Tippin, B.L.; Kwong, A.M.; Inadomi, M.J.; Lee, O.J.; Park, J.M.; Materi, A.M.; Buslon, V.S.; Lin, A.M.; Kudo, L.C.; Karsten, S.L.; et al. Intestinal tumor suppression in ApcMin/+ mice by prostaglandin D2 receptor PTGDR. Cancer Med. 2014, 3, 1041–1051. [Google Scholar] [CrossRef]
- Tippin, B.L.; Kwong, A.M.; Inadomi, M.J.; Lee, O.J.; Park, J.M.; Materi, A.M.; Buslon, V.S.; Lin, A.M.; Kudo, L.C.; Karsten, S.L.; et al. Anti-inflammatory Effects of Enhanced Recovery Programs on Early-Stage Colorectal Cancer Surgery. World J. Surg. 2018, 42, 953–964. [Google Scholar]
- DelGiorno, K.E.; Chung, C.Y.; Vavinskaya, V.; Maurer, H.C.; Novak, S.W.; Lytle, N.K.; Ma, Z.; Giraddi, R.R.; Wang, D.; Fang, L.; et al. Tuft Cells Inhibit Pancreatic Tumorigenesis in Mice by Producing Prostaglandin D2. Gastroenterology 2020, 159, 1866–1881.e8. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.; Ding, L. Comprehensive Analysis to Identify Enhancer-Regulated Inflammation-Associated Genes in Lung Adenocarcinoma. Cancer Manag. Res. 2021, 13, 7115–7129. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Wang, Q.; Tang, N.; Wang, K. GSTZ1-1 downregulates Wnt/beta-catenin signalling in hepatocellular carcinoma cells. FEBS Open Bio 2020, 10, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Q.; Yang, Y.; Lei, C.; Yang, F.; Liang, L.; Chen, C.; Xia, J.; Wang, K.; Tang, N. GSTZ1 deficiency promotes hepatocellular carcinoma proliferation via activation of the KEAP1/NRF2 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 438. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bin, C.; Xue, Q.; Gao, Q.; Huang, A.; Wang, K.; Tang, N. GSTZ1 sensitizes hepatocellular carcinoma cells to sorafenib-induced ferroptosis via inhibition of NRF2/GPX4 axis. Cell Death Dis. 2021, 12, 426. [Google Scholar] [CrossRef]
- Yogev, O.; Barker, K.; Sikka, A.; Almeida, G.S.; Hallsworth, A.; Smith, L.M.; Jamin, Y.; Ruddle, R.; Koers, A.; Webber, H.T.; et al. p53 Loss in MYC-Driven Neuroblastoma Leads to Metabolic Adaptations Supporting Radioresistance. Cancer Res. 2016, 76, 3025–3035. [Google Scholar] [CrossRef]
- Liu, X.; Sui, X.; Zhang, C.; Wei, K.; Bao, Y.; Xiong, J.; Zhou, Z.; Chen, Z.; Wang, C.; Zhu, H.; et al. Glutathione S-transferase A1 suppresses tumor progression and indicates better prognosis of human primary hepatocellular carcinoma. J. Cancer 2020, 11, 83–91. [Google Scholar] [CrossRef]
- Zou, M.; Hu, X.; Xu, B.; Tong, T.; Jing, Y.; Xi, L.; Zhou, W.; Lu, J.; Wang, X.; Yang, X.; et al. Glutathione Stransferase isozyme alpha 1 is predominantly involved in the cisplatin resistance of common types of solid cancer. Oncol. Rep. Feb. 2019, 41, 989–998. [Google Scholar]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic. Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]
- Asare-Werehene, M.; Communal, L.; Carmona, E.; Han, Y.; Song, Y.S.; Burger, D.; Mes-Masson, A.M.; Tsang, B.K. Plasma Gelsolin Inhibits CD8(+) T-cell Function and Regulates Glutathione Production to Confer Chemoresistance in Ovarian Cancer. Cancer Res. 2020, 80, 3959–3971. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Ye, G.; Shao, Y. High expression of the ferroptosis-associated MGST1 gene in relation to poor outcome and maladjusted immune cell infiltration in uterine corpus endometrial carcinoma. J. Clin. Lab. Anal. 2022, 36, e24317. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Zhong, Y.; Yin, Y.; Qin, K.; Yang, L.; Li, D.; Yu, W.; Yang, C.; Deng, Z.; Hong, K. A marine-derived small molecule induces immunogenic cell death against triple-negative breast cancer through ER stress-CHOP pathway. Int. J. Biol. Sci. 2022, 18, 2898–2913. [Google Scholar] [CrossRef]
- Chen, S.; Su, X.; Mi, H.; Dai, X.; Li, S.; Chen, S.; Zhang, S. Comprehensive analysis of glutathione peroxidase-1 (GPX1) expression and prognostic value in three different types of renal cell carcinoma. Transl. Androl. Urol. 2020, 9, 2737–2750. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Zhuo, M.; Li, M.; Chen, Q.; Mo, P.; Yu, C. Histone demethylase JMJD2D activates HIF1 signaling pathway via multiple mechanisms to promote colorectal cancer glycolysis and progression. Oncogene 2020, 39, 7076–7091. [Google Scholar] [CrossRef]
- Weller, M.; Weber, R.; Willscher, E.; Riehmer, V.; Hentschel, B.; Kreuz, M.; Felsberg, J.; Beyer, U.; Löffler-Wirth, H.; Kaulich, K.; et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015, 129, 679–693. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Hu, J.; Fang, T.; Tang, W.; Lv, B.; Yang, B.; Xia, J. Protein convertase subtilisin/Kexin type 9 inhibits hepatocellular carcinoma growth by interacting with GSTP1 and suppressing the JNK signaling pathway. Cancer Biol. Med. 2021, 18, 1–14. [Google Scholar] [CrossRef]
- Yadav, P.; Banerjee, A.; Boruah, N.; Singh, C.S.; Chatterjee, P.; Mukherjee, S.; Dakhar, H.; Nongrum, H.B.; Bhattacharjee, A.; Chatterjee, A. Glutathione S-transferasesP1 AA (105Ile) allele increases oral cancer risk, interacts strongly with c-Jun Kinase and weakly detoxifies areca-nut metabolites. Sci. Rep. 2020, 10, 6032. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, K.; Zhang, J.; Hai, Y.; Wang, P.; Wang, H.; Liu, Q.; Wong, C.C.L.; Yao, J.; Gao, Y.; et al. c-Jun NH2-Terminal Protein Kinase Phosphorylates the Nrf2-ECH Homology 6 Domain of Nuclear Factor Erythroid 2-Related Factor 2 and Downregulates Cytoprotective Genes in Acetaminophen-Induced Liver Injury in Mice. Hepatology 2020, 71, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, B.; Cheng, S.; Fan, H.; Liu, S.; Zhou, B.; Liu, W.; Liang, R.; Tang, Y.; Zhang, Y. KDELR2 knockdown synergizes with temozolomide to induce glioma cell apoptosis through the CHOP and JNK/p38 pathways. Transl. Cancer Res. 2021, 10, 3491–3506. [Google Scholar] [CrossRef]
- Chang, C.Y.; Pan, P.H.; Wu, C.C.; Liao, S.L.; Chen, W.Y.; Kuan, Y.H.; Wang, W.Y.; Chen, C.J. Endoplasmic Reticulum Stress Contributes to Gefitinib-Induced Apoptosis in Glioma. Int. J. Mol. Sci. 2021, 22, 3934. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liu, Z.; Xu, W.; Wang, Q.; Zhang, C.; Ding, Y.; Nie, W.; Lai, J.; Chen, Y.; Huang, H. Pterostilbene promotes mitochondrial apoptosis and inhibits proliferation in glioma cells. Sci. Rep. 2021, 11, 6381. [Google Scholar] [CrossRef]
- Murakami, Y.; Akahoshi, T.; Hayashi, I.; Endo, H.; Hashimoto, A.; Kono, S.; Kondo, H.; Kawai, S.; Inoue, M.; Kitasato, H. Inhibition of monosodium urate monohydrate crystal-induced acute inflammation by retrovirally transfected prostaglandin D synthase. Arthritis Rheum. 2003, 48, 2931–2941. [Google Scholar] [CrossRef]
- Korbecki, J.; Baranowska-Bosiacka, I.; Gutowska, I.; Chlubek, D. The effect of reactive oxygen species on the synthesis of prostanoids from arachidonic acid. J. Physiol. Pharmacol. 2013, 64, 409–421. [Google Scholar] [PubMed]
- Ishii, T. Close teamwork between Nrf2 and peroxiredoxins 1 and 6 for the regulation of prostaglandin D2 and E2 production in macrophages in acute inflammation. Free Radic. Biol. Med. 2015, 88, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Q.; Su, Z.; Sun, Q.; Zhao, Y.; Feng, T.; Jiang, J.; Zhang, F.; Ma, H. Lipid metabolism gene-wide profile and survival signature of lung adenocarcinoma. Lipids Health Dis. 2020, 19, 222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-F.; Li, Y.-K.; Chen, C.-Y.; Zhang, X.-D.; Cao, L.; Quan, F.-F.; Zeng, X.; Wang, J.; Liu, J. Identification and validation of a prognostic index based on a metabolic-genomic landscape analysis of ovarian cancer. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Schutz, B.; Ruppert, A.L.; Strobel, O.; Lazarus, M.; Urade, Y.; Buchler, M.W.; Weihe, E. Distribution pattern and molecular signature of cholinergic tuft cells in human gastro-intestinal and pancreatic-biliary tract. Sci. Rep. 2019, 9, 17466. [Google Scholar] [CrossRef]
- Valente, A.L.; Kane, J.L.; Ellsworth, D.L.; Shriver, C.D.; Ellsworth, R.E. Molecular response of the axillary lymph node microenvironment to metastatic colonization. Clin. Exp. Metastasis 2014, 31, 565–572. [Google Scholar] [CrossRef]
- Liu, X.; Song, B.; Li, S.; Wang, N.; Yang, H. Identification and functional analysis of the risk microRNAs associated with cerebral low-grade glioma prognosis. Mol. Med. Rep. 2017, 16, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Hou, X.; Mei, L. Dihydrotanshinone I inhibits human glioma cell proliferation via the activation of ferroptosis. Oncol. Lett. 2020, 20, 122. [Google Scholar] [CrossRef]
- Krylova, N.G.; Drobysh, M.S.; Semenkova, G.N.; Kulahava, T.A.; Pinchuk, S.V.; Shadyro, O.I. Cytotoxic and antiproliferative effects of thymoquinone on rat C6 glioma cells depend on oxidative stress. Mol. Cell Biochem. 2019, 462, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, C.; Lu, S.; Wang, X.; Wang, L.; Liang, S.; Wang, X.; Piao, M.; Cui, J.; Chi, G.; et al. Autophagy activated by silibinin contributes to glioma cell death via induction of oxidative stress-mediated BNIP3-dependent nuclear translocation of AIF. Cell Death Dis. 2020, 11, 630. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, D.C.; Rafique, J.; Saba, S.; Grinevicius, V.M.A.S.; Filho, D.W.; Zamoner, A.; Braga, A.L.; Pedrosa, R.C.; Ourique, F. IP-Se-06, a Selenylated Imidazo[1,2-a]pyridine, Modulates Intracellular Redox State and Causes Akt/mTOR/HIF-1alpha and MAPK Signaling Inhibition, Promoting Antiproliferative Effect and Apoptosis in Glioblastoma Cells. Oxid. Med. Cell Longev. 2022, 3710449. [Google Scholar]
- Su, Z.; Han, S.; Jin, Q.; Zhou, N.; Lu, J.; Shangguan, F.; Yu, S.; Liu, Y.; Wang, L.; Lu, J.; et al. Ciclopirox and bortezomib synergistically inhibits glioblastoma multiforme growth via simultaneously enhancing JNK/p38 MAPK and NF-kappaB signaling. Cell Death Dis. 2021, 12, 251. [Google Scholar] [CrossRef]
- Shi, L.; Sun, G.; Zhu, H. Demethoxycurcumin analogue DMC-BH inhibits orthotopic growth of glioma stem cells by targeting JNK/ERK signaling. Aging (Albany NY) 2020, 12, 14718–14735. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, B.; Wang, Y.; Ayanlaja, A.A.; Zhu, J.; Kambey, P.A.; Qiu, Z.; Zhang, C.; Hu, W. Glutathione S-Transferases S1, Z1 and A1 Serve as Prognostic Factors in Glioblastoma and Promote Drug Resistance through Antioxidant Pathways. Cells 2022, 11, 3232. https://doi.org/10.3390/cells11203232
Cheng B, Wang Y, Ayanlaja AA, Zhu J, Kambey PA, Qiu Z, Zhang C, Hu W. Glutathione S-Transferases S1, Z1 and A1 Serve as Prognostic Factors in Glioblastoma and Promote Drug Resistance through Antioxidant Pathways. Cells. 2022; 11(20):3232. https://doi.org/10.3390/cells11203232
Chicago/Turabian StyleCheng, Bo, Yu Wang, Abiola Abdulrahman Ayanlaja, Jing Zhu, Piniel Alphayo Kambey, Ziqiang Qiu, Caiyi Zhang, and Wei Hu. 2022. "Glutathione S-Transferases S1, Z1 and A1 Serve as Prognostic Factors in Glioblastoma and Promote Drug Resistance through Antioxidant Pathways" Cells 11, no. 20: 3232. https://doi.org/10.3390/cells11203232
APA StyleCheng, B., Wang, Y., Ayanlaja, A. A., Zhu, J., Kambey, P. A., Qiu, Z., Zhang, C., & Hu, W. (2022). Glutathione S-Transferases S1, Z1 and A1 Serve as Prognostic Factors in Glioblastoma and Promote Drug Resistance through Antioxidant Pathways. Cells, 11(20), 3232. https://doi.org/10.3390/cells11203232