The Highly Efficient Expression System of Recombinant Human Prolidase and the Effect of N-Terminal His-Tag on the Enzyme Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Expressionof rhPEPD (Optimized)
2.2. Purification, Activation, and His-Tag Removal of rhPEPD
2.3. Protein Electrophoresis (SDS-PAGE)
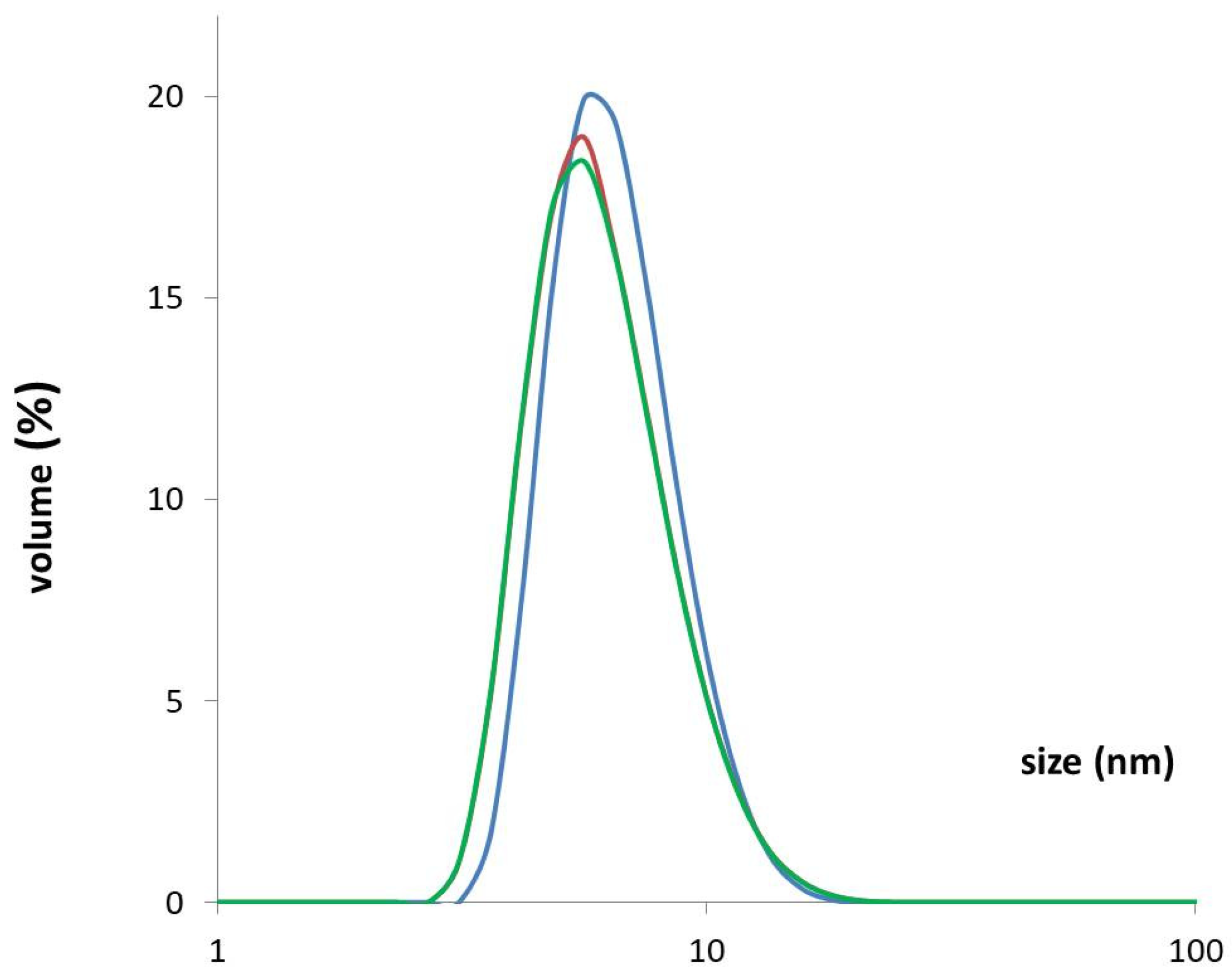
2.4. Dynamic Light Scattering (DLS)
2.5. Mass Spectrometry
2.6. HaCaT Cell Cultures
2.7. HaCaT Treatment
2.8. Preparation of Lysates
2.9. Western Immunoblotting
2.10. Determination of ProlidaseActivity
2.11. Statistical Analysis
3. Results
3.1. Enzyme Expressionand Purification
3.2. His-Tag Removal
3.3. Mass Spectrometry and SDS-PAGE
3.4. DLS—Confirmation of the Stability and Quality of the Purified rhPEPD Preparation
3.5. Effect on the Enzymatic Activity of the N-Terminal-Fused His-Tag
3.6. Effect on the Biological Activity of the N-Terminal-Fused His-Tag
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Namiduru, E.S. Prolidase. Bratisl. Lek. Listy 2016, 117, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Myara, I.; Charpentier, C.; Lemonnier, A. Prolidase and prolidase deficiency. Life Sci. 1984, 34, 1985–1998. [Google Scholar] [CrossRef]
- Misiura, M.; Guszczyn, T.; Oscilowska, I.; Baszanowska, W.; Palka, J.; Miltyk, W. Platelet-Rich Plasma Promotes the Proliferation of Human Keratinocytes via a Progression of the Cell Cycle. A Role of Prolidase. Int. J. Mol. Sci. 2021, 22, 936. [Google Scholar] [CrossRef] [PubMed]
- Guszczyn, T.; Surażyński, A.; Zaręba, I.; Rysiak, E.; Popko, J.; Pałka, J. Differential Effect of Platelet-Rich Plasma Fractions on β1-integrin Signaling, Collagen Biosynthesis, and Prolidase Activity in Human Skin Fibroblasts. Drug Des. Devel. Ther. 2017, 11, 1849–1857. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y.; Zhang, Y. Identification of Prolidase as a High Affinity Ligand of the ErbB2 Receptor and its Regulation of ErbB2 Signaling and Cell Growth. Cell Death Dis. 2014, 5, e1211. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y.; Ding, Y.; Choi, K.S.; Kazim, A.L.; Zhang, Y. Prolidase Directly Binds and Activates Epidermal Growth Factor Receptor and Stimulates Downstream Signaling. J. Biol. Chem. 2013, 288, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, Y.; Bhattacharya, A.; Zhang, Y. PEPD is a Pivotal Regulator of p53 Tumor Suppressor. Nat. Commun. 2017, 8, 2052. [Google Scholar] [CrossRef]
- Lubick, K.J.; Robertson, S.J.; McNally, K.L.; Freedman, B.A.; Rasmussen, A.L.; Taylor, R.T.; Walts, A.D.; Tsuruda, S.; Sakai, M.; Ishizuka, M.; et al. Flavivirus Antagonism of Type I Interferon Signaling Reveals Prolidase as a Regulator of IFNAR1 Surface Expression. Cell Host Microbe 2015, 18, 61–74. [Google Scholar] [CrossRef]
- Wilk, P.; Uehlein, M.; Piwowarczyk, R.; Dobbek, H.; Mueller, U.; Weiss, M.S. Structural Basis for Prolidase Deficiency Disease Mechanisms. FEBS J. 2018, 285, 3422–3441. [Google Scholar] [CrossRef]
- Besio, R.; Gioia, R.; Cossu, F.; Monzani, E.; Nicolis, S.; Cucca, L.; Profumo, A.; Casella, L.; Tenni, R.; Bolognesi, M.; et al. Kinetic and Structural Evidences on Human Prolidase Pathological Mutants Suggest Strategies for Enzyme Functional Rescue. PLoS One 2013, 8, e58792. [Google Scholar] [CrossRef]
- Alberto, M.E.; Leopoldini, M.; Russo, N. Can human Prolidase Enzyme Use Different Metals for Full Catalytic Activity? Inorg. Chem. 2011, 50, 3394–3403. [Google Scholar] [CrossRef]
- Demir, Y.; Beydemir, S. Purification, Refolding, and Characterization of Recombinant Human Paraoxonase-1. Turk. J. Chem. 2015, 39, 764–776. [Google Scholar] [CrossRef]
- Lupi, A.; Della Torre, S.; Campari, E.; Tenni, R.; Cetta, G.; Rossi, A.; Forlino, A. Human Recombinant Prolidase from Eukaryotic and Prokaryotic Sources. FEBS J. 2006, 273, 5466–5478. [Google Scholar] [CrossRef]
- Wang, S.H.; Zhi, Q.W.; Sun, M.J. Dual Activities of Human Prolidase. Toxicol. Vitr. 2006, 20, 71–77. [Google Scholar] [CrossRef]
- Wang, S.H.; Zhi, Q.W.; Sun, M.J. Purification and Characterization of Recombinant Human Liver Prolidase Expressed in Saccharomyces Cerevisiae. Arch. Toxicol. 2005, 79, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Uramatsu, S.; Liu, G.; Yang, Q.; Uramatsu, M.; Chi, H.; Lu, J.; Yamashita, K.; Kodama, H. Characterization of Prolidase I and Ii Purified from Normal Human Erythrocytes: Comparison with Prolidase in Erythrocytes from a Patient with Prolidase Deficiency. Amino Acids 2009, 37, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Endo, F.; Matsuda, I.; Ogata, A.; Tanaka, S. Human Erythrocyte Prolidase and Prolidase Deficiency. Pediatr. Res. 1982, 16, 227–231. [Google Scholar] [CrossRef]
- Boright, A.P.; Scriver, C.R.; Lancaster, G.A.; Choy, F. Prolidase deficiency: Biochemical Classification of Alleles. Am. J. Hum. Genet. 1989, 44, 731–740. [Google Scholar] [PubMed]
- Sjöström, H.; Norén, O.; Josefsson, L. Purification and Specificity of Pig Intestinal Prolidase. Biochim. Biophys. Acta (BBA)—Enzym. 1973, 327, 457–470. [Google Scholar] [CrossRef]
- Endo, F.; Tanoue, A.; Nakai, H.; Hata, A.; Indo, Y.; Titani, K.; Matsuda, I. Primary Structure and Gene Localization of Human Prolidase. J. Biol. Chem. 1989, 264, 4476–4481. [Google Scholar] [CrossRef]
- Fernández-Esplá, M.D.; Martín-Hernández, M.C.; Fox, P.F. Purification and Characterization of a Prolidase from Lactobacillus casei subsp. casei IFPL 731. Appl. Environ. Microbiol. 1997, 63, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Besio, R.; Monzani, E.; Gioia, R.; Nicolis, S.; Rossi, A.; Casella, L.; Forlino, A. Improved prolidase activity assay allowed enzyme kinetic characterization and faster prolidase deficiency diagnosis. Clin. Chim. Acta 2011, 412, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Timasheff, S.N. Protein Hydration, Thermodynamic Binding, and Preferential Hydration. Biochemistry 2002, 41, 13473–13482. [Google Scholar] [CrossRef] [PubMed]
- Kohyama, K.; Matsumoto, T.; Imoto, T. Refolding of an Unstable Lysozyme by Gradient Removal of a Solubilizer and Gradient Addition of a Stabilizer. J. Biochem. 2010, 147, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Nizioł, M.; Ościłowska, I.; Baszanowska, W.; Pałka, J.; Besio, R.; Forlino, A.; Miltyk, W. Recombinant Prolidase Activates EGFR-Dependent Cell Growth in an Experimental Model of Inflammation in HaCaT Keratinocytes. Implication for Wound Healing. Front. Mol. Biosci. 2022, 9, 876348. [Google Scholar] [CrossRef]






| Primary Antibodies | Dilution | Vendor |
|---|---|---|
| p44/42 MAPK (ERK1/2) Rabbit mAb | 1:1000 | Cell Signaling Technology |
| EGF Receptor Rabbit mAb | 1:1000 | Cell Signaling Technology |
| Phospho-EGF Receptor (Tyr1068) Rabbit mAb | 1:1000 | Cell Signaling Technology |
| Phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) Rabbit mAb | 1:1000 | Cell Signaling Technology |
| Akt Rabbit mAb | 1:2000 | Cell Signaling Technology |
| Phospho-Akt (Ser473) Rabbit mAb | 1:1000 | Cell Signaling Technology |
| STAT3Rabbit mAb | 1:1000 | Cell Signaling Technology |
| Phospho-STAT3 (Tyr705) Rabbit mAb | 1:1000 | Cell Signaling Technology |
| GAPDH Rabbit mAb | 1:1000 | Cell Signaling Technology |
| Attempt | IPTG Final Concentration (mM) | Temperature (°C) | Time (h) | Protein Production | Lysis Buffers Additional Components | The Yield of Protein Production Insoluble Fraction |
|---|---|---|---|---|---|---|
| 1 | 1 | 37 | 2 | protein in inclusion bodies (in the pellet after centrifugation) | 300 mM NaCl 20 mM Tris-HCl pH 8.0 20 mMimidazole 1 mM EDTA 10% glycerol 100 μg·mL−1 lysozyme 1–1.5% N-laurylsarcosine 2% Triton X-100 | low |
| 2 | 30 | 5 | low | |||
| 3 | 25 | 10 | low | |||
| 4 | 18 | 15 | low | |||
| 5 | 16 | 15 | low | |||
| 6 | 0.2 | 30 | 5 | protein in the soluble fraction | 300 mM NaCl 20 mM Tris-HCl pH 8.0 20 mMimidazole 1 mM EDTA 10% glycerol 100 μg·mL−1 lysozyme 1 mM TCEP | low |
| 7 | 25 | 10 | low | |||
| 8 | 18 | 10 | low | |||
| 9 | 18 | 18 | high | |||
| 10 | 18 | 15 | high |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czyrko-Horczak, J.; Nizioł, M.; Forlino, A.; Besio, R.; Miltyk, W. The Highly Efficient Expression System of Recombinant Human Prolidase and the Effect of N-Terminal His-Tag on the Enzyme Activity. Cells 2022, 11, 3284. https://doi.org/10.3390/cells11203284
Czyrko-Horczak J, Nizioł M, Forlino A, Besio R, Miltyk W. The Highly Efficient Expression System of Recombinant Human Prolidase and the Effect of N-Terminal His-Tag on the Enzyme Activity. Cells. 2022; 11(20):3284. https://doi.org/10.3390/cells11203284
Chicago/Turabian StyleCzyrko-Horczak, Justyna, Magdalena Nizioł, Antonella Forlino, Roberta Besio, and Wojciech Miltyk. 2022. "The Highly Efficient Expression System of Recombinant Human Prolidase and the Effect of N-Terminal His-Tag on the Enzyme Activity" Cells 11, no. 20: 3284. https://doi.org/10.3390/cells11203284
APA StyleCzyrko-Horczak, J., Nizioł, M., Forlino, A., Besio, R., & Miltyk, W. (2022). The Highly Efficient Expression System of Recombinant Human Prolidase and the Effect of N-Terminal His-Tag on the Enzyme Activity. Cells, 11(20), 3284. https://doi.org/10.3390/cells11203284







