RNA Editing Alterations Define Disease Manifestations in the Progression of Experimental Autoimmune Encephalomyelitis (EAE)
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Editing Analysis
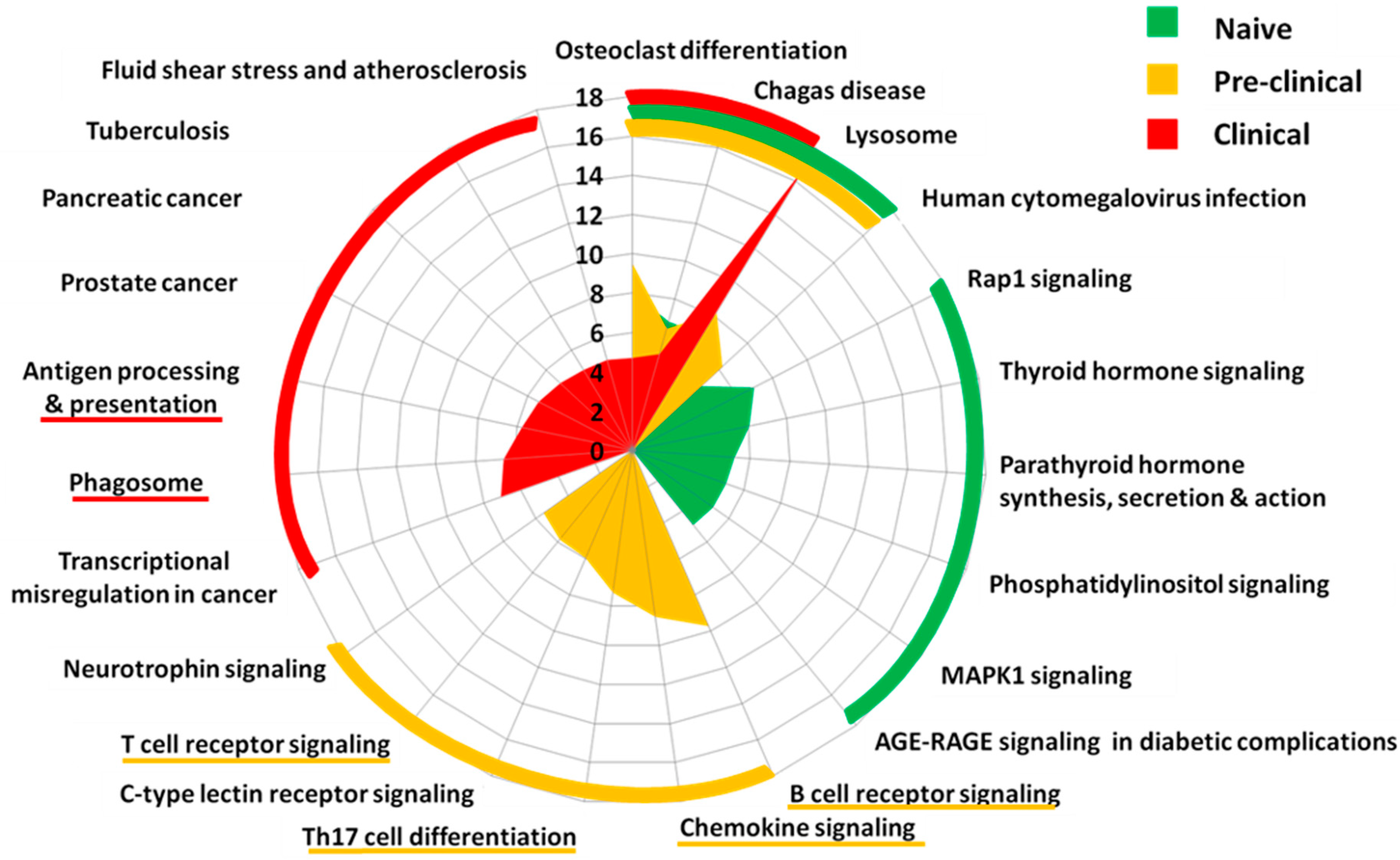
2.2. Pathway Analysis
2.3. RNA Editing Experimental Validations
2.3.1. Microglia Isolation
2.3.2. Microglia RNA Extraction and Reverse Transcription
2.3.3. Brain Tissue RNA Extraction and Reverse Transcription
2.3.4. Genomic DNA Isolation
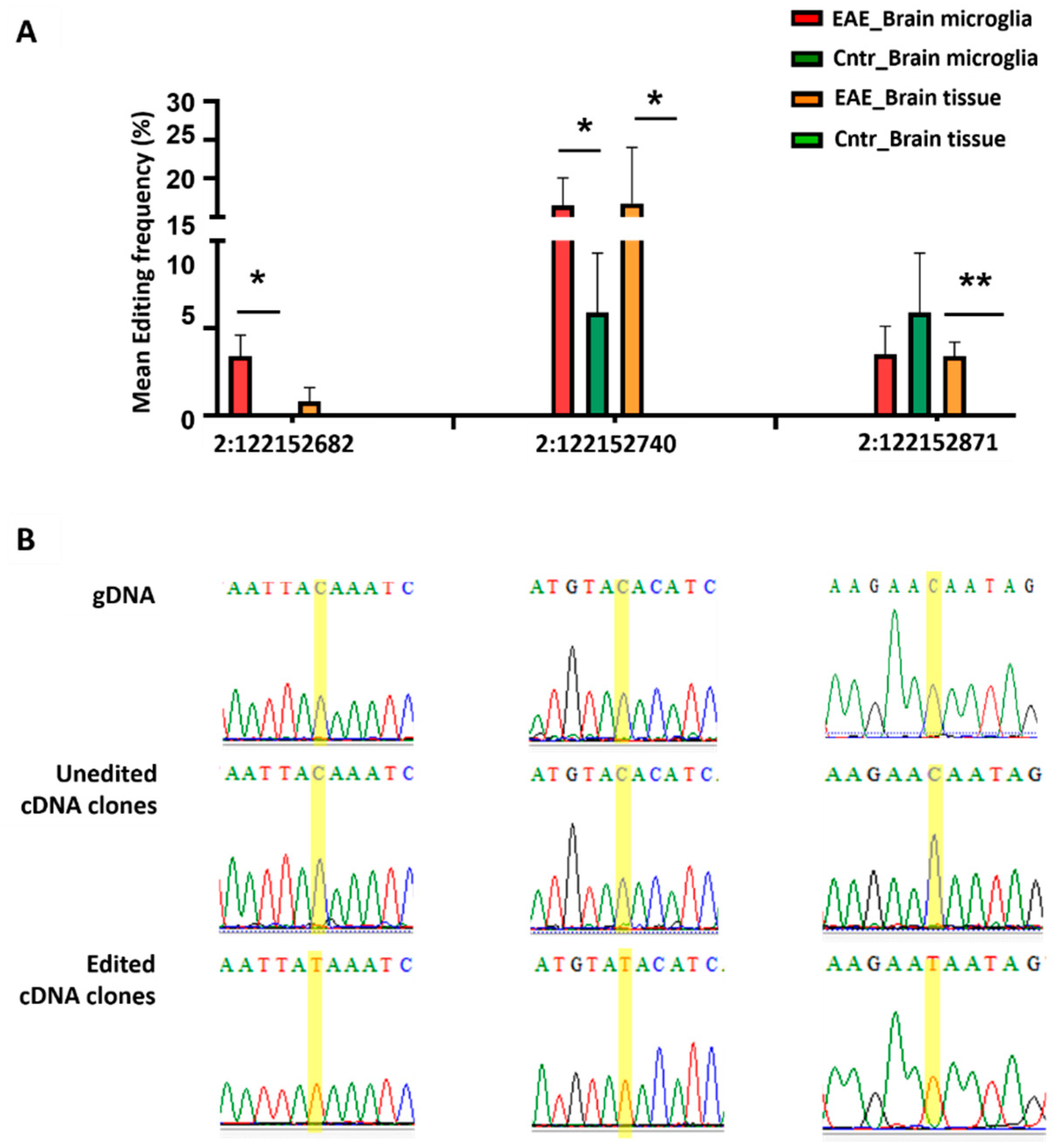
2.3.5. RNA Editing Validations
2.4. Gene Expression Validations
2.5. In Vivo Study
2.5.1. Animals Used for The In Vivo Study
2.5.2. EAE Induction
2.5.3. Tissue Staining
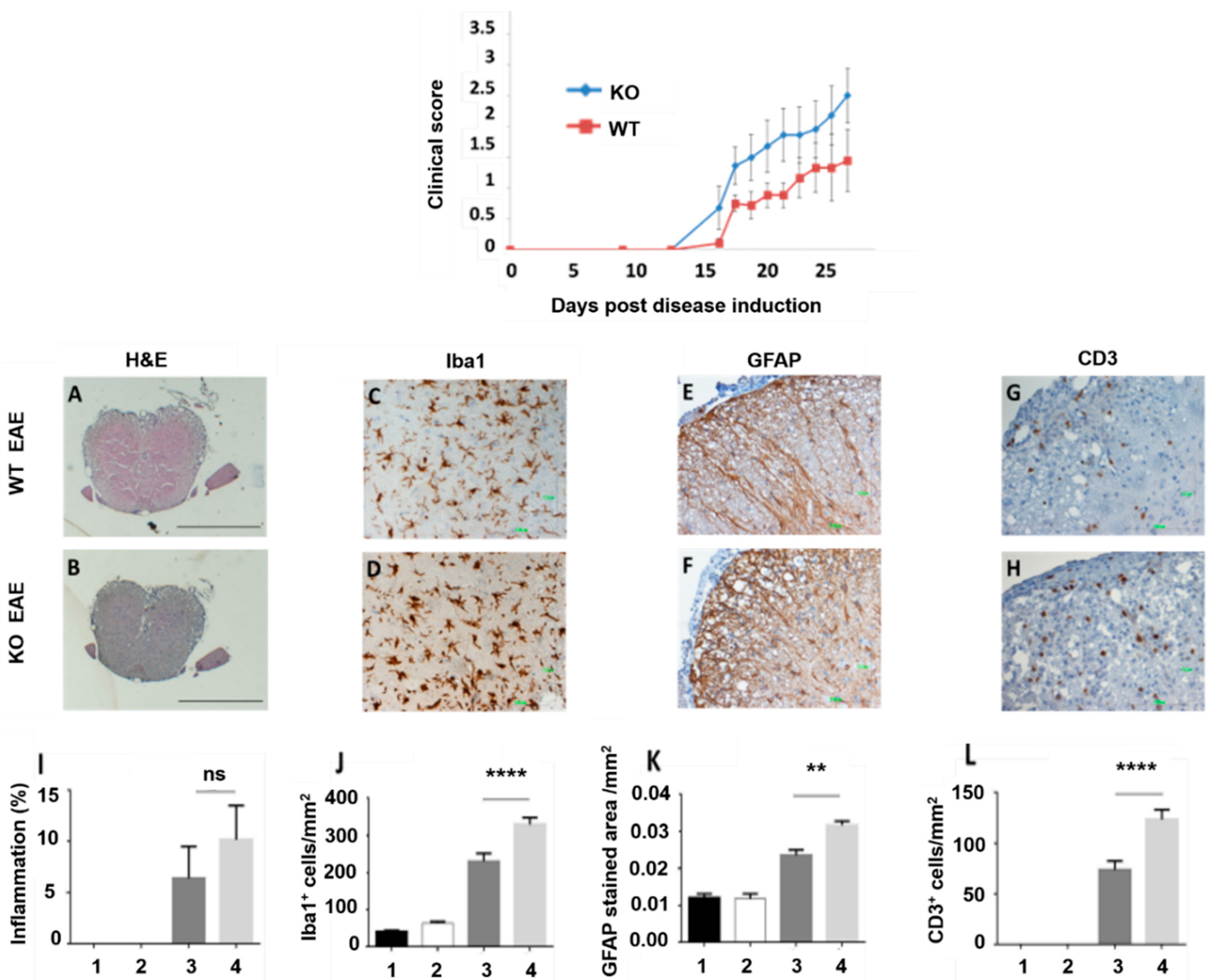
2.5.4. Immunohistochemistry
2.6. Statistical Analysis
3. Results
3.1. Microglia Editomes during EAE Progression
3.2. Experimental Verification of RNA Editing Alterations
3.3. In Vivo Study
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miedema, A.; Gerrits, E.; Brouwer, N.; Jiang, Q.; Kracht, L.; Meijer, M.; Nutma, E.; Peferoen-Baert, R.; Pijnacker, A.T.E.; Wesseling, E.M.; et al. Brain Macrophages Acquire Distinct Transcriptomes in Multiple Sclerosis Lesions and Normal Appearing White Matter. Acta Neuropathol. Commun. 2022, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, H. Experimental Models of Multiple Sclerosis. Rev. Neurol. 2007, 163, 651–655. [Google Scholar] [CrossRef]
- Sawcer, S.; Franklin, R.J.M.; Ban, M. Multiple Sclerosis Genetics. Lancet Neurol. 2014, 13, 700–709. [Google Scholar] [CrossRef]
- Santiago, J.A.; Bottero, V.; Potashkin, J.A. Biological and Clinical Implications of Comorbidities in Parkinson’s Disease. Front. Aging Neurosci. 2017, 9, 394. [Google Scholar] [CrossRef]
- Karagianni, K.; Pettas, S.; Christoforidou, G.; Kanata, E.; Bekas, N.; Xanthopoulos, K.; Dafou, D.; Sklaviadis, T. A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations. Biomolecules 2022, 12, 465. [Google Scholar] [CrossRef] [PubMed]
- Farajollahi, S.; Maas, S. Molecular Diversity through RNA Editing: A Balancing Act. Trends Genet. 2010, 26, 221–230. [Google Scholar] [CrossRef] [Green Version]
- Nishikura, K. A-to-I Editing of Coding and Non-Coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2015, 17, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Licht, K.; Jantsch, M.F. Rapid and Dynamic Transcriptome Regulation by RNA Editing and RNA Modifications. J. Cell Biol. 2016, 213, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic Landscape and Regulation of RNA Editing in Mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef] [Green Version]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016, 41, 578. [Google Scholar] [CrossRef]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA Editing in Human Tissues: Towards the Inosinome Atlas. Sci. Rep. 2015, 5, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tossberg, J.T.; Heinrich, R.M.; Farley, V.M.; Crooke, P.S.; Aune, T.M. Adenosine-to-Inosine RNA Editing of Alu Double-Stranded (Ds)RNAs Is Markedly Decreased in Multiple Sclerosis and Unedited Alu DsRNAs Are Potent Activators of Proinflammatory Transcriptional Responses. J. Immunol. 2020, 205, 2606–2617. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.K.; Bagnati, M.; Delahaye-Duriez, A.; Ko, J.H.; Rotival, M.; Langley, S.R.; Shkura, K.; Mazzuferi, M.; Danis, B.; Van Eyll, J.; et al. Genome-Wide Analysis of Differential RNA Editing in Epilepsy. Genome Res. 2017, 27, 440–450. [Google Scholar] [CrossRef] [Green Version]
- Tran, S.S.; Jun, H.I.; Bahn, J.H.; Azghadi, A.; Ramaswami, G.; Van Nostrand, E.L.; Nguyen, T.B.; Hsiao, Y.H.E.; Lee, C.; Pratt, G.A.; et al. Widespread RNA Editing Dysregulation in Brains from Autistic Individuals. Nat. Neurosci. 2018, 22, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Flomen, R.; Makoff, A. Increased RNA Editing in EAAT2 Pre-MRNA from Amyotrophic Lateral Sclerosis Patients: Involvement of a Cryptic Polyadenylation Site. Neurosci. Lett. 2011, 497, 139–143. [Google Scholar] [CrossRef]
- Akbarian, S.; Smith, M.A.; Jones, E.G. Editing for an AMPA Receptor Subunit RNA in Prefrontal Cortex and Striatum in Alzheimer’s Disease, Huntington’s Disease and Schizophrenia. Brain Res. 1995, 699, 297–304. [Google Scholar] [CrossRef]
- Gaisler-Salomon, I.; Kravitz, E.; Feiler, Y.; Safran, M.; Biegon, A.; Amariglio, N.; Rechavi, G. Hippocampus-Specific Deficiency in RNA Editing of GluA2 in Alzheimer’s Disease. Neurobiol. Aging 2014, 35, 1785–1791. [Google Scholar] [CrossRef]
- Khermesh, K.; D’Erchia, A.M.; Barak, M.; Annese, A.; Wachtel, C.; Levanon, E.Y.; Picardi, E.; Eisenberg, E. Reduced Levels of Protein Recoding by A-to-I RNA Editing in Alzheimer’s Disease. RNA 2016, 22, 290–302. [Google Scholar] [CrossRef] [Green Version]
- Kanata, E.; Llorens, F.; Dafou, D.; Dimitriadis, A.; Thüne, K.; Xanthopoulos, K.; Bekas, N.; Espinosa, J.C.; Schmitz, M.; Marín-Moreno, A.; et al. RNA Editing Alterations Define Manifestation of Prion Diseases. Proc. Natl. Acad. Sci. USA 2019, 116, 19727–19735. [Google Scholar] [CrossRef] [Green Version]
- Cole, D.C.; Chung, Y.; Gagnidze, K.; Hajdarovic, K.H.; Rayon-Estrada, V.; Harjanto, D.; Bigio, B.; Gal-Toth, J.; Milner, T.A.; McEwen, B.S.; et al. Loss of APOBEC1 RNA-Editing Function in Microglia Exacerbates Age-Related CNS Pathophysiology. Proc. Natl. Acad. Sci. USA 2017, 114, 13272–13277. [Google Scholar] [CrossRef]
- Guo, X.; Wiley, C.A.; Steinman, R.A.; Sheng, Y.; Ji, B.; Wang, J.; Zhang, L.; Wang, T.; Zenatai, M.; Billiar, T.R.; et al. Aicardi-Goutières Syndrome-Associated Mutation at ADAR1 Gene Locus Activates Innate Immune Response in Mouse Brain. J. Neuroinflammation 2021, 18, 1–16. [Google Scholar] [CrossRef]
- Rice, G.I.; Kasher, P.R.; Forte, G.M.A.; Mannion, N.M.; Greenwood, S.M.; Szynkiewicz, M.; Dickerson, J.E.; Bhaskar, S.S.; Zampini, M.; Briggs, T.A.; et al. Mutations in ADAR1 Cause Aicardi-Goutières Syndrome Associated with a Type I Interferon Signature. Nat. Genet. 2012, 44, 1243–1248. [Google Scholar] [CrossRef] [Green Version]
- Mannion, N.M.; Greenwood, S.M.; Young, R.; Cox, S.; Brindle, J.; Read, D.; Nellåker, C.; Vesely, C.; Ponting, C.P.; McLaughlin, P.J.; et al. The RNA-Editing Enzyme ADAR1 Controls Innate Immune Responses to RNA. Cell Rep. 2014, 9, 1482–1494. [Google Scholar] [CrossRef] [Green Version]
- Liddicoat, B.J.; Piskol, R.; Chalk, A.M.; Ramaswami, G.; Higuchi, M.; Hartner, J.C.; Li, J.B.; Seeburg, P.H.; Walkley, C.R. RNA Editing by ADAR1 Prevents MDA5 Sensing of Endogenous DsRNA as Nonself. Science 2015, 349, 1115–1120. [Google Scholar] [CrossRef] [Green Version]
- Pestal, K.; Funk, C.C.; Snyder, J.M.; Price, N.D.; Treuting, P.M.; Stetson, D.B. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-Organ Development. Immunity 2015, 43, 933–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA Editing—Immune Protector and Transcriptome Diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Adenosine Deaminase Acting on RNA (ADAR1), a Suppressor of Double-Stranded RNA–Triggered Innate Immune Responses. J. Biol. Chem. 2019, 294, 1710–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Gloudemans, M.J.; Geisinger, J.M.; Fan, B.; Aguet, F.; Sun, T.; Ramaswami, G.; Li, Y.I.; Ma, J.-B.; Pritchard, J.K.; et al. RNA Editing Underlies Genetic Risk of Common Inflammatory Diseases. Nature 2022, 608, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Alqassim, E.Y.; Sharma, S.; Khan, A.N.M.N.H.; Emmons, T.R.; Cortes Gomez, E.; Alahmari, A.; Singel, K.L.; Mark, J.; Davidson, B.A.; Robert McGray, A.J.; et al. RNA Editing Enzyme APOBEC3A Promotes Pro-Inflammatory M1 Macrophage Polarization. Commun. Biol. 2021, 4, 102. [Google Scholar] [CrossRef]
- Xiang, R.; Liu, Y.; Fan, L.; Jiang, B.; Wang, F. RNA Adenosine Deaminase (ADAR1) Alleviates High-Fat Diet-Induced Nonalcoholic Fatty Liver Disease by Inhibiting NLRP3 Inflammasome. Lab. Investig. 2022, 102, 1088–1100. [Google Scholar] [CrossRef]
- Pettas, S.; Karagianni, K.; Kanata, E.; Chatziefstathiou, A.; Christoudia, N.; Xanthopoulos, K.; Sklaviadis, T.; Dafou, D. Profiling Microglia through Single-Cell RNA Sequencing over the Course of Development, Aging, and Disease. Cells 2022, 11, 2383. [Google Scholar] [CrossRef] [PubMed]
- Jordão, M.J.C.; Sankowski, R.; Brendecke, S.M.; Sagar; Locatelli, G.; Tai, Y.H.; Tay, T.L.; Schramm, E.; Armbruster, S.; Hagemeyer, N.; et al. Single-Cell Profiling Identifies Myeloid Cell Subsets with Distinct Fates during Neuroinflammation. Science 2019, 363, eaat7554. [Google Scholar] [CrossRef] [PubMed]
- Voet, S.; Prinz, M.; van Loo, G. Microglia in Central Nervous System Inflammation and Multiple Sclerosis Pathology. Trends Mol. Med. 2019, 25, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Barres, B.A. Microglia and Macrophages in Brain Homeostasis and Disease. Nat. Rev. Immunol. 2017, 18, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Jian, C.; Liao, Y.; Huang, Q.; Wu, Y.; Liu, X.; Zou, D.; Wu, Y. The Role of Microglia in Multiple Sclerosis. Neuropsychiatr. Dis. Treat. 2017, 13, 1661. [Google Scholar] [CrossRef] [Green Version]
- Lewis, N.D.; Hill, J.D.; Juchem, K.W.; Stefanopoulos, D.E.; Modis, L.K. RNA Sequencing of Microglia and Monocyte-Derived Macrophages from Mice with Experimental Autoimmune Encephalomyelitis Illustrates a Changing Phenotype with Disease Course. J. Neuroimmunol. 2014, 277, 26–38. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Hirano, K.I.; Young, S.G.; Farese, R.V.; Ng, J.; Sande, E.; Warburton, C.; Powell-Braxton, L.M.; Davidson, N.O. Targeted Disruption of the Mouse Apobec-1 Gene Abolishes Apolipoprotein B MRNA Editing and Eliminates Apolipoprotein B48. J. Biol. Chem. 1996, 271, 9887–9890. [Google Scholar] [CrossRef] [Green Version]
- Grigoriadis, N.; Lourbopoulos, A.; Lagoudaki, R.; Frischer, J.M.; Polyzoidou, E.; Touloumi, O.; Simeonidou, C.; Deretzi, G.; Kountouras, J.; Spandou, E.; et al. Variable Behavior and Complications of Autologous Bone Marrow Mesenchymal Stem Cells Transplanted in Experimental Autoimmune Encephalomyelitis. Exp. Neurol. 2011, 230, 78–89. [Google Scholar] [CrossRef]
- Theotokis, P.; Kleopa, K.A.; Touloumi, O.; Lagoudaki, R.; Lourbopoulos, A.; Nousiopoulou, E.; Kesidou, E.; Poulatsidou, K.N.; Dardiotis, E.; Hadjigeorgiou, G.; et al. Connexin43 and Connexin47 Alterations after Neural Precursor Cells Transplantation in Experimental Autoimmune Encephalomyelitis. Glia 2015, 63, 1772–1783. [Google Scholar] [CrossRef] [PubMed]
- Theotokis, P.; Touloumi, O.; Lagoudaki, R.; Nousiopoulou, E.; Kesidou, E.; Siafis, S.; Tselios, T.; Lourbopoulos, A.; Karacostas, D.; Grigoriadis, N.; et al. Nogo Receptor Complex Expression Dynamics in the Inflammatory Foci of Central Nervous System Experimental Autoimmune Demyelination. J. Neuroinflammation 2016, 13, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardner, O.K.; Wang, L.; van Booven, D.; Whitehead, P.L.; Hamilton-Nelson, K.L.; Adams, L.D.; Starks, T.D.; Hofmann, N.K.; Vance, J.M.; Cuccaro, M.L.; et al. RNA Editing Alterations in a Multi-Ethnic Alzheimer Disease Cohort Converge on Immune and Endocytic Molecular Pathways. Hum. Mol. Genet. 2019, 28, 3053–3061. [Google Scholar] [CrossRef]
- Dick, A.L.W.; Khermesh, K.; Paul, E.; Stamp, F.; Levanon, E.Y.; Chen, A. Adenosine-to-Inosine RNA Editing within Corticolimbic Brain Regions Is Regulated in Response to Chronic Social Defeat Stress in Mice. Front. Psychiatry 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Roth, S.H.; Danan-Gotthold, M.; Ben-Izhak, M.; Rechavi, G.; Cohen, C.J.; Louzoun, Y.; Levanon, E.Y. Increased RNA Editing May Provide a Source for Autoantigens in Systemic Lupus Erythematosus. Cell Rep. 2018, 23, 50–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, S.; Alsop, E.; Lorenzini, I.; Starr, A.; Rabichow, B.E.; Mendez, E.; Levy, J.L.; Burciu, C.; Reiman, R.; Chew, J.; et al. ADAR2 Mislocalization and Widespread RNA Editing Aberrations in C9orf72-Mediated ALS/FTD. Acta Neuropathol. 2019, 138, 49–65. [Google Scholar] [CrossRef]
- Vlachogiannis, N.I.; Gatsiou, A.; Silvestris, D.A.; Stamatelopoulos, K.; Tektonidou, M.G.; Gallo, A.; Sfikakis, P.P.; Stellos, K. Increased Adenosine-to-Inosine RNA Editing in Rheumatoid Arthritis. J. Autoimmun. 2020, 106, 102329. [Google Scholar] [CrossRef]
- Linker, R.A.; Reinhardt, M.; Bendszus, M.; Ladewig, G.; Briel, A.; Schirner, M.; Mäurer, M.; Hauff, P. In Vivo Molecular Imaging of Adhesion Molecules in Experimental Autoimmune Encephalomyelitis (EAE). J. Autoimmun. 2005, 25, 199–205. [Google Scholar] [CrossRef]
- Beeston, T.; Smith, T.R.F.; Maricic, I.; Tang, X.; Kumar, V. Involvement of IFN-γ and Perforin, but Not Fas/FasL Interactions in Regulatory T Cell-Mediated Suppression of Experimental Autoimmune Encephalomyelitis. J. Neuroimmunol. 2010, 229, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Chalk, A.M.; Taylor, S.; Heraud-Farlow, J.E.; Walkley, C.R. The Majority of A-to-I RNA Editing Is Not Required for Mammalian Homeostasis. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Danecek, P.; Nellåker, C.; McIntyre, R.E.; Buendia-Buendia, J.E.; Bumpstead, S.; Ponting, C.P.; Flint, J.; Durbin, R.; Keane, T.M.; Adams, D.J. High Levels of RNA-Editing Site Conservation amongst 15 Laboratory Mouse Strains. Genome Biol. 2012, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Harjanto, D.; Papamarkou, T.; Oates, C.J.; Rayon-Estrada, V.; Papavasiliou, F.N.; Papavasiliou, A. RNA Editing Generates Cellular Subsets with Diverse Sequence within Populations. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picardi, E.; Horner, D.S.; Pesole, G. Single-Cell Transcriptomics Reveals Specific RNA Editing Signatures in the Human Brain. RNA 2017, 23, 860–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, T.; Park, C.K.; Leung, A.K.L.; Gao, Y.; Hyde, T.M.; Kleinman, J.E.; Rajpurohit, A.; Tao, R.; Shin, J.H.; Weinberger, D.R. Dynamic Regulation of RNA Editing in Human Brain Development and Disease. Nat. Neurosci. 2016, 19, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Quinones-Valdez, G.; Tran, S.S.; Jun, H.I.; Bahn, J.H.; Yang, E.W.; Zhan, L.; Brümmer, A.; Wei, X.; Van Nostrand, E.L.; Pratt, G.A.; et al. Regulation of RNA Editing by RNA-Binding Proteins in Human Cells. Commun. Biol. 2019, 2, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayon-Estrada, V.; Harjanto, D.; Hamilton, C.E.; Berchiche, Y.A.; Gantman, E.C.; Sakmar, T.P.; Bulloch, K.; Gagnidze, K.; Harroch, S.; McEwen, B.S.; et al. Epitranscriptomic Profiling across Cell Types Reveals Associations between APOBEC1-Mediated RNA Editing, Gene Expression Outcomes, and Cellular Function. Proc. Natl. Acad. Sci. USA 2017, 114, 13296–13301. [Google Scholar] [CrossRef] [Green Version]
- Pujantell, M.; Riveira-Muñoz, E.; Badia, R.; Castellví, M.; Garcia-Vidal, E.; Sirera, G.; Puig, T.; Ramirez, C.; Clotet, B.; Esté, J.A.; et al. RNA Editing by ADAR1 Regulates Innate and Antiviral Immune Functions in Primary Macrophages. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Chen, T.; Yu, Z.; Zhu, X.; Yang, M.; Xie, B.; Li, N.; Cao, X.; Wang, J. RasGRP3 Limits Toll-like Receptor-Triggered Inflammatory Response in Macrophages by Activating Rap1 Small GTPase. Nat. Commun. 2014, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Ma, Y.; Zeng, Z.; Yin, Q.; Hong, Y.; Hou, X.; Liu, X. RAGE-Specific Inhibitor FPS-ZM1 Attenuates AGEs-Induced Neuroinflammation and Oxidative Stress in Rat Primary Microglia. Neurochem. Res. 2017, 42, 2902–2911. [Google Scholar] [CrossRef]
- Bachstetter, A.D.; Xing, B.; de Almeida, L.; Dimayuga, E.R.; Watterson, D.M.; Van Eldik, L.J. Microglial P38α MAPK Is a Key Regulator of Proinflammatory Cytokine Up-Regulation Induced by Toll-like Receptor (TLR) Ligands or Beta-Amyloid (Aβ). J. Neuroinflamm. 2011, 8, 1–12. [Google Scholar] [CrossRef]
- Mori, Y.; Tomonaga, D.; Kalashnikova, A.; Furuya, F.; Akimoto, N.; Ifuku, M.; Okuno, Y.; Beppu, K.; Fujita, K.; Katafuchi, T.; et al. Effects of 3,3′,5-Triiodothyronine on Microglial Functions. Glia 2015, 63, 906–920. [Google Scholar] [CrossRef] [PubMed]
- Prowse, N.; Hayley, S. Microglia and BDNF at the Crossroads of Stressor Related Disorders: Towards a Unique Trophic Phenotype. Neurosci. Biobehav. Rev. 2021, 131, 135–163. [Google Scholar] [CrossRef] [PubMed]
- Pöyhönen, S.; Er, S.; Domanskyi, A.; Airavaara, M. Effects of Neurotrophic Factors in Glial Cells in the Central Nervous System: Expression and Properties in Neurodegeneration and Injury. Front. Physiol. 2019, 10, 486. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, C.; Yao, H.; Li, X.; Yang, N.; Qiao, J.; Xu, K.; Zeng, L. Identification of Suitable Reference Genes for MRNA Studies in Bone Marrow in a Mouse Model of Hematopoietic Stem Cell Transplantation. Transplant. Proc. 2016, 48, 2826–2832. [Google Scholar] [CrossRef]
- Martin, N.A.; Nawrocki, A.; Molnar, V.; Elkjaer, M.L.; Thygesen, E.K.; Palkovits, M.; Acs, P.; Sejbaek, T.; Nielsen, H.H.; Hegedus, Z.; et al. Orthologous Proteins of Experimental De- and Remyelination Are Differentially Regulated in the CSF Proteome of Multiple Sclerosis Subtypes. PLoS ONE 2018, 13, e0202530. [Google Scholar] [CrossRef] [Green Version]
- Starkey, H.D.V.; Van Kirk, C.A.; Bixler, G.V.; Imperio, C.G.; Kale, V.P.; Serfass, J.M.; Farley, J.A.; Yan, H.; Warrington, J.P.; Han, S.; et al. Neuroglial Expression of the Mhci Pathway and Pirb Receptor Is Upregulated in the Hippocampus with Advanced Aging. J. Mol. Neurosci. 2012, 48, 111–126. [Google Scholar] [CrossRef] [Green Version]
- Yousef, H.; Conboy, M.J.; Morgenthaler, A.; Schlesinger, C.; Bugaj, L.; Paliwal, P.; Greer, C.; Conboy, I.M.; Schaffer, D. Systemic Attenuation of the TGF-β Pathway by a Single Drug Simultaneously Rejuvenates Hippocampal Neurogenesis and Myogenesis in the Same Old Mammal. Oncotarget 2015, 6, 11959. [Google Scholar] [CrossRef] [Green Version]
- Hammond, T.R.R.; McEllin, B.; Morton, P.D.D.; Raymond, M.; Dupree, J.; Gallo, V. Endothelin-B Receptor Activation in Astrocytes Regulates the Rate of Oligodendrocyte Regeneration during Remyelination. Cell Rep. 2015, 13, 2090–2097. [Google Scholar] [CrossRef] [Green Version]
- Kopacek, J.; Sakaguchi, S.; Shigematsu, K.; Nishida, N.; Atarashi, R.; Nakaoke, R.; Moriuchi, R.; Niwa, M.; Katamine, S. Upregulation of the Genes Encoding Lysosomal Hydrolases, a Perforin-Like Protein, and Peroxidases in the Brains of Mice Affected with an Experimental Prion Disease. J. Virol. 2000, 74, 411–417. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dafou, D.; Kanata, E.; Pettas, S.; Bekas, N.; Dimitriadis, A.; Kempapidou, G.; Lagoudaki, R.; Theotokis, P.; Touloumi, O.; Delivanoglou, N.; et al. RNA Editing Alterations Define Disease Manifestations in the Progression of Experimental Autoimmune Encephalomyelitis (EAE). Cells 2022, 11, 3582. https://doi.org/10.3390/cells11223582
Dafou D, Kanata E, Pettas S, Bekas N, Dimitriadis A, Kempapidou G, Lagoudaki R, Theotokis P, Touloumi O, Delivanoglou N, et al. RNA Editing Alterations Define Disease Manifestations in the Progression of Experimental Autoimmune Encephalomyelitis (EAE). Cells. 2022; 11(22):3582. https://doi.org/10.3390/cells11223582
Chicago/Turabian StyleDafou, Dimitra, Eirini Kanata, Spyros Pettas, Nikolaos Bekas, Athanasios Dimitriadis, Garyfalia Kempapidou, Roza Lagoudaki, Paschalis Theotokis, Olga Touloumi, Nikoleta Delivanoglou, and et al. 2022. "RNA Editing Alterations Define Disease Manifestations in the Progression of Experimental Autoimmune Encephalomyelitis (EAE)" Cells 11, no. 22: 3582. https://doi.org/10.3390/cells11223582
APA StyleDafou, D., Kanata, E., Pettas, S., Bekas, N., Dimitriadis, A., Kempapidou, G., Lagoudaki, R., Theotokis, P., Touloumi, O., Delivanoglou, N., Kesidou, E., Xanthopoulos, K., Grigoriadis, N., Papavasiliou, F. N., & Sklaviadis, T. (2022). RNA Editing Alterations Define Disease Manifestations in the Progression of Experimental Autoimmune Encephalomyelitis (EAE). Cells, 11(22), 3582. https://doi.org/10.3390/cells11223582






