o-Vanillin Modulates Cell Phenotype and Extracellular Vesicles of Human Mesenchymal Stem Cells and Intervertebral Disc Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Tissue Collection
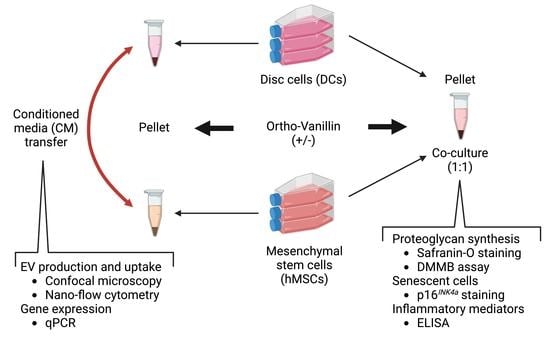
2.2. Cell Culture
2.3. Histology Assessments
2.4. Dimethylmethylene Blue (DMMB) Assay
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Plasma Membrane Labeling and Confocal Microscope
2.7. Nano-Flow Cytometry
2.8. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
2.9. Statistical Analysis and Illustrations
3. Results
3.1. GAG Deposition and Release in DCs:hMSCs Co-Cultures
3.2. Presence of Senescent Cells and Inflammatory Cytokine Release in DCs:hMSCs Co-Cultures
3.3. Extracellular Vesicle Generation and Release
3.4. Extracellular Vesicle Transfer and Uptake
3.5. hMSC Differentiation and DC Phenotype in Response to CM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vos, T.; Allen, C.; Arora, M.; Barber, R.M.; Bhutta, Z.A.; Brown, A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1545–1602. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.F. The prevalence of low back pain: A systematic review of the literature from 1966 to 1998. J. Spinal Disord. 2000, 13, 205–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussières, A.E.; Stewart, G.; Al-Zoubi, F.; Decina, P.; Descarreaux, M.; Haskett, D.; Hincapié, C.; Pageé, I.; Passmore, S.; Srbely, J.; et al. Spinal Manipulative Therapy and Other Conservative Treatments for Low Back Pain: A Guideline From the Canadian Chiropractic Guideline Initiative. J. Manip. Physiol. Ther. 2018, 41, 265–293. [Google Scholar] [CrossRef]
- Chang, D.; Zygourakis, C.C.; Wadhwa, H.; Kahn, J.G. Systematic Review of Cost-Effectiveness Analyses in U.S. Spine Surgery. World Neurosurg. 2020, 142, e32–e57. [Google Scholar] [CrossRef] [PubMed]
- Vadalà, G.; Ambrosio, L.; Russo, F.; Papalia, R.; Denaro, V. Interaction between Mesenchymal Stem Cells and Intervertebral Disc Microenvironment: From Cell Therapy to Tissue Engineering. Stem Cells Int. 2019, 2019, 2376172. [Google Scholar] [CrossRef]
- Vadalà, G.; Ambrosio, L.; Russo, F.; Papalia, R.; Denaro, V. Stem Cells and Intervertebral Disc Regeneration Overview-What They Can and Can’t Do. Int. J. Spine Surg. 2021, 15, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sun, T.; Li, Y.; Yang, M.; Zhao, Y.; Liu, J.; Li, Z. Application of stem cells in the repair of intervertebral disc degeneration. Stem Cell Res. Ther. 2022, 13, 70. [Google Scholar] [CrossRef]
- Williams, R.J.; Tryfonidou, M.A.; Snuggs, J.W.; Le Maitre, C.L. Cell sources proposed for nucleus pulposus regeneration. JOR Spine 2021, 4, e1175. [Google Scholar] [CrossRef]
- Croft, A.S.; Illien-Jünger, S.; Grad, S.; Guerrero, J.; Wangler, S.; Gantenbein, B. The Application of Mesenchymal Stromal Cells and Their Homing Capabilities to Regenerate the Intervertebral Disc. Int. J. Mol. Sci. 2021, 22, 3519. [Google Scholar] [CrossRef]
- Hingert, D.; Nawilaijaroen, P.; Aldridge, J.; Baranto, A.; Brisby, H. Investigation of the Effect of Secreted Factors from Mesenchymal Stem Cells on Disc Cells from Degenerated Discs. Cells Tissues Organs 2019, 208, 76–88. [Google Scholar] [CrossRef]
- Strassburg, S.; Richardson, S.M.; Freemont, A.J.; Hoyland, J.A. Co-culture induces mesenchymal stem cell differentiation and modulation of the degenerate human nucleus pulposus cell phenotype. Regen. Med. 2010, 5, 701–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, J.; Lee, Y.D.; Wagers, A.J. Stem cell aging: Mechanisms, regulators and therapeutic opportunities. Nat. Med. 2014, 20, 870–880. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Tang, L. Stem Cell Senescence: The Obstacle of the Treatment of Degenerative Disk Disease. Curr. Stem Cell Res. Ther. 2019, 14, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Jurk, D.; Khosla, S.; Kirkland, J.L.; LeBrasseur, N.K.; Miller, J.D.; Passos, J.F.; Pignolo, R.J.; Tchkonia, T.; Niedernhofer, L.J. Senolytic Drugs: Reducing Senescent Cell Viability to Extend Health Span. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 779–803. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, L.; Wang, Y.; Yao, J.; Jin, F.; Tao, T.; Yuan, H.; Shi, L.; Lu, S. Treatment with toll-like receptor 2 inhibitor ortho-vanillin alleviates lipopolysaccharide-induced acute kidney injury in mice. Exp. Ther. Med. 2019, 18, 4829–4837. [Google Scholar] [CrossRef] [Green Version]
- Nasr, S.; Varshosaz, J.; Hajhashemi, V. Ortho-vanillin nanoparticle-doped glucan microspheres exacerbate the anti-arthritic effects of methotrexate in adjuvant-induced arthritis in rats. Pharm. Rep 2020, 72, 680–691. [Google Scholar] [CrossRef]
- Cherif, H.; Bisson, D.G.; Jarzem, P.; Weber, M.; Ouellet, J.A.; Haglund, L. Curcumin and o-Vanillin Exhibit Evidence of Senolytic Activity in Human IVD Cells In Vitro. J. Clin. Med. 2019, 8, 433. [Google Scholar] [CrossRef] [Green Version]
- Cherif, H.; Bisson, D.G.; Mannarino, M.; Rabau, O.; Ouellet, J.A.; Haglund, L. Senotherapeutic drugs for human intervertebral disc degeneration and low back pain. eLife 2020, 9, e54693. [Google Scholar] [CrossRef]
- Shah, B.S.; Burt, K.G.; Jacobsen, T.; Fernandes, T.D.; Alipui, D.O.; Weber, K.T.; Levine, M.; Chavan, S.S.; Yang, H.; Tracey, K.J.; et al. High mobility group box-1 induces pro-inflammatory signaling in human nucleus pulposus cells via toll-like receptor 4-dependent pathway. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2019, 37, 220–231. [Google Scholar] [CrossRef] [Green Version]
- Triller, P.; Bachorz, J.; Synowitz, M.; Kettenmann, H.; Markovic, D. O-Vanillin Attenuates the TLR2 Mediated Tumor-Promoting Phenotype of Microglia. Int. J. Mol. Sci. 2020, 21, 2959. [Google Scholar] [CrossRef]
- Mannarino, M.; Cherif, H.; Li, L.; Sheng, K.; Rabau, O.; Jarzem, P.; Weber, M.H.; Ouellet, J.A.; Haglund, L. Toll-like receptor 2 induced senescence in intervertebral disc cells of patients with back pain can be attenuated by o-vanillin. Arthritis Res. Ther. 2021, 23, 117. [Google Scholar] [CrossRef] [PubMed]
- Marton, A.; Kusz, E.; Kolozsi, C.; Tubak, V.; Zagotto, G.; Buzás, K.; Quintieri, L.; Vizler, C. Vanillin analogues o-vanillin and 2, 4, 6-trihydroxybenzaldehyde inhibit NFĸB activation and suppress growth of A375 human melanoma. Anticancer Res. 2016, 36, 5743–5750. [Google Scholar] [CrossRef] [PubMed]
- Gawri, R.; Rosenzweig, D.H.; Krock, E.; Ouellet, J.A.; Stone, L.S.; Quinn, T.M.; Haglund, L. High mechanical strain of primary intervertebral disc cells promotes secretion of inflammatory factors associated with disc degeneration and pain. Arthritis Res. Ther. 2014, 16, R21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solchaga, L.A.; Penick, K.; Porter, J.D.; Goldberg, V.M.; Caplan, A.I.; Welter, J.F. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J. Cell. Physiol. 2005, 203, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Gardner, O.F.; Alini, M.; Stoddart, M.J. Mesenchymal Stem Cells Derived from Human Bone Marrow. Methods Mol Biol 2015, 1340, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Shihan, M.H.; Novo, S.G.; Le Marchand, S.J.; Wang, Y.; Duncan, M.K. A simple method for quantitating confocal fluorescent images. Biochem. Biophys. Rep. 2021, 25, 100916. [Google Scholar] [CrossRef] [PubMed]
- Krock, E.; Rosenzweig, D.H.; Chabot-Doré, A.J.; Jarzem, P.; Weber, M.H.; Ouellet, J.A.; Stone, L.S.; Haglund, L. Painful, degenerating intervertebral discs up-regulate neurite sprouting and CGRP through nociceptive factors. J. Cell. Mol. Med. 2014, 18, 1213–1225. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bisson, D.G.; Sheng, K.; Kocabas, S.; Krock, E.; Teles, A.; Saran, N.; Ouellet, J.A.; Haglund, L. Toll-like receptor involvement in adolescent scoliotic facet joint degeneration. J. Cell. Mol. Med. 2020, 24, 11355–11365. [Google Scholar] [CrossRef]
- Zhang, X.; Guerrero, J.; Croft, A.S.; Albers, C.E.; Häckel, S.; Gantenbein, B. Spheroid-Like Cultures for Expanding Angiopoietin Receptor-1 (aka. Tie2) Positive Cells from the Human Intervertebral Disc. Int. J. Mol. Sci. 2020, 21, 9423. [Google Scholar] [CrossRef]
- Capossela, S.; Bertolo, A.; Gunasekera, K.; Pötzel, T.; Baur, M.; Stoyanov, J.V. VEGF vascularization pathway in human intervertebral disc does not change during the disc degeneration process. BMC Res. Notes 2018, 11, 333. [Google Scholar] [CrossRef]
- Martin, I.; Jakob, M.; Schäfer, D.; Dick, W.; Spagnoli, G.; Heberer, M. Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthr. Cartil. 2001, 9, 112–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Sakai, D.; Yamamoto, Y.; Iwashina, T.; Serigano, K.; Tamura, F.; Mochida, J. Human nucleus pulposus cells significantly enhanced biological properties in a coculture system with direct cell-to-cell contact with autologous mesenchymal stem cells. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2010, 28, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Le Visage, C.; Kim, S.W.; Tateno, K.; Sieber, A.N.; Kostuik, J.P.; Leong, K.W. Interaction of human mesenchymal stem cells with disc cells: Changes in extracellular matrix biosynthesis. Spine (Phila Pa 1976) 2006, 31, 2036–2042. [Google Scholar] [CrossRef]
- Svanvik, T.; Henriksson, H.B.; Karlsson, C.; Hagman, M.; Lindahl, A.; Brisby, H. Human disk cells from degenerated disks and mesenchymal stem cells in co-culture result in increased matrix production. Cells Tissues Organs 2010, 191, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Liebscher, T.; Haefeli, M.; Wuertz, K.; Nerlich, A.G.; Boos, N. Age-related variation in cell density of human lumbar intervertebral disc. Spine 2011, 36, 153–159. [Google Scholar] [CrossRef]
- Orozco, L.; Soler, R.; Morera, C.; Alberca, M.; Sánchez, A.; García-Sancho, J. Intervertebral disc repair by autologous mesenchymal bone marrow cells: A pilot study. Transplantation 2011, 92, 822–828. [Google Scholar] [CrossRef] [Green Version]
- Elabd, C.; Centeno, C.J.; Schultz, J.R.; Lutz, G.; Ichim, T.; Silva, F.J. Intra-discal injection of autologous, hypoxic cultured bone marrow-derived mesenchymal stem cells in five patients with chronic lower back pain: A long-term safety and feasibility study. J. Transl. Med. 2016, 14, 253. [Google Scholar] [CrossRef]
- Noriega, D.C.; Ardura, F.; Hernández-Ramajo, R.; Martín-Ferrero, M.; Sánchez-Lite, I.; Toribio, B.; Alberca, M.; García, V.; Moraleda, J.M.; Sánchez, A.; et al. Intervertebral Disc Repair by Allogeneic Mesenchymal Bone Marrow Cells: A Randomized Controlled Trial. Transplantation 2017, 101, 1945–1951. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.E.; McConnell, J.C.; Sherratt, M.J.; Derby, B.; Richardson, S.M.; Hoyland, J.A. Growth differentiation factor 6 and transforming growth factor-beta differentially mediate mesenchymal stem cell differentiation, composition, and micromechanical properties of nucleus pulposus constructs. Arthritis Res. Ther. 2014, 16, R67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, S.M.; Walker, R.V.; Parker, S.n.; Rhodes, N.P.; Hunt, J.A.; Freemont, A.J.; Hoyland, J.A. Intervertebral Disc Cell-Mediated Mesenchymal Stem Cell Differentiation. Stem Cells 2006, 24, 707–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frith, J.E.; Thomson, B.; Genever, P.G. Dynamic Three-Dimensional Culture Methods Enhance Mesenchymal Stem Cell Properties and Increase Therapeutic Potential. Tissue Eng. Part C Methods 2009, 16, 735–749. [Google Scholar] [CrossRef]
- Stich, S.; Stolk, M.; Girod, P.P.; Thomé, C.; Sittinger, M.; Ringe, J.; Seifert, M.; Hegewald, A.A. Regenerative and Immunogenic Characteristics of Cultured Nucleus Pulposus Cells from Human Cervical Intervertebral Discs. PLoS ONE 2015, 10, e0126954. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.P.; Pearce, R.H.; Schechter, M.T.; Adams, M.E.; Tsang, I.K.; Bishop, P.B. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine (Phila Pa 1976) 1990, 15, 411–415. [Google Scholar] [CrossRef]
- Rajasekaran, S.; Tangavel, C.; K.S, S.V.A.; Soundararajan, D.C.R.; Nayagam, S.M.; Matchado, M.S.; Raveendran, M.; Shetty, A.P.; Kanna, R.M.; Dharmalingam, K. Inflammaging determines health and disease in lumbar discs—Evidence from differing proteomic signatures of healthy, aging, and degenerating discs. Spine J. 2020, 20, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Sarath Babu, N.; Krishnan, S.; Brahmendra Swamy, C.V.; Venkata Subbaiah, G.P.; Gurava Reddy, A.V.; Idris, M.M. Quantitative proteomic analysis of normal and degenerated human intervertebral disc. Spine J. 2016, 16, 989–1000. [Google Scholar] [CrossRef]
- Huang, Y.C.; Leung, V.Y.; Lu, W.W.; Luk, K.D. The effects of microenvironment in mesenchymal stem cell-based regeneration of intervertebral disc. Spine J. Off. J. North Am. Spine Soc. 2013, 13, 352–362. [Google Scholar] [CrossRef]
- Waterman, R.S.; Tomchuck, S.L.; Henkle, S.L.; Betancourt, A.M. A New Mesenchymal Stem Cell (MSC) Paradigm: Polarization into a Pro-Inflammatory MSC1 or an Immunosuppressive MSC2 Phenotype. PLoS ONE 2010, 5, e10088. [Google Scholar] [CrossRef] [Green Version]
- Reis, M.; Ogonek, J.; Qesari, M.; Borges, N.M.; Nicholson, L.; Preußner, L.; Dickinson, A.M.; Wang, X.N.; Weissinger, E.M.; Richter, A. Recent Developments in Cellular Immunotherapy for HSCT-Associated Complications. Front Immunol. 2016, 7, 500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.; Shojaee, M.; Mitchell Crow, J.; Khanabdali, R. From Mesenchymal Stromal Cells to Engineered Extracellular Vesicles: A New Therapeutic Paradigm. Front Cell Dev. Biol. 2021, 9, 705676. [Google Scholar] [CrossRef] [PubMed]
- Jeyaram, A.; Jay, S.M. Preservation and Storage Stability of Extracellular Vesicles for Therapeutic Applications. Aaps. J. 2017, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Wiley, C.D.; Campisi, J. From Ancient Pathways to Aging Cells-Connecting Metabolism and Cellular Senescence. Cell Metab. 2016, 23, 1013–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairag, R.; Rosenzweig, D.H.; Ramirez-Garcialuna, J.L.; Weber, M.H.; Haglund, L. Three-Dimensional Printed Polylactic Acid Scaffolds Promote Bone-like Matrix Deposition in Vitro. ACS Appl. Mater. Interfaces 2019, 11, 15306–15315. [Google Scholar] [CrossRef] [PubMed]






| Experiments | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor | Age | Gender | Disc pathology | Levels | Safranin-O staining | DMMB assay 1 | p16INK4a staining | ELISA 2 | Fluorescent staining | qPCR 5 | Hoechst assay | |||
| Membrane signal transfer (Co-culture) | EV 3 identification (Confocal microscope) | Membrane signal transfer (CM 4) | EV quantification (Nano-Flow cytometry) | |||||||||||
| 1 | 32 | M | Degenerative disc disease with an annular tear | L5-S1 | ✔ | ✔ | ✔ | |||||||
| 2 | 59 | F | Simple herniation | L5-S1 | ✔ | ✔ | ||||||||
| 3 | 38 | F | Simple herniation | L5-S1 | ✔ | ✔ | ✔ | ✔ | ✔ | |||||
| 4 | 45 | M | Degenerative disc disease | L5-S1 | ✔ | ✔ | ✔ | ✔ | ✔ | |||||
| 5 | 35 | F | Degenerative disc disease | L4-5 | ✔ | ✔ | ✔ | ✔ | ✔ | |||||
| 6 | 42 | F | Recurrent hernia | L5-S1 | ✔ | ✔ | ||||||||
| 7 | 51 | F | Degenerative disc disease | L5-S1 | ✔ | ✔ | ✔ | ✔ | ✔ | |||||
| 8 | 38 | F | Degenerative disc disease | L4-S1 | ✔ | |||||||||
| 9 | 49 | F | Degenerative disc disease | L5-S1 | ✔ | |||||||||
| 10 | 40 | F | Degenerative disc disease | L5-S1 | ✔ | |||||||||
| 11 | 26 | M | Degenerative disc disease | L4-5 | ✔ | ✔ | ✔ | ✔ | ||||||
| 12 | 39 | F | Back pain | L4-S1 | ✔ | ✔ | ||||||||
| 13 | 54 | M | Back pain | L4-5 | ✔ | ✔ | ||||||||
| 14 | 44 | F | Back pain | L4-5 | ✔ | |||||||||
| 15 | 23 | F | Disc herniation | L5-S1 | ✔ | |||||||||
| 16 | 35 | F | Degenerative disc disease | L4-S1 | ||||||||||
| 17 | 54 | M | Degenerative disc disease | L4-S1 | ✔ | |||||||||
| 18 | 50 | F | Spinal stenosis/Spondylo | L5-S1 | ✔ | |||||||||
| 19 | 71 | F | Simple herniation | L3-S1 | ✔ | |||||||||
| 20 | 45 | F | Simple herniation | L4-S1 | ✔ | ✔ | ||||||||
| 21 | 24 | F | Simple herniation | L4-5 | ✔ | |||||||||
| Target Gene | Name/Gene ID | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) | Primer Bank ID | Reference |
|---|---|---|---|---|---|
| beta-ACTIN (Reference gene) | ACTB/ID: 60 | GTC TTC CCC TCC ATC GTG G | AAT CCT TCT GAC CCA TGC C | [29] | |
| CD73 | NT5E/ID: 4907 | CCA GTA CCA GGG CAC TAT CTG | TGG CTC GAT CAG TCC TTC CA | 325651882c2 | |
| CD105 | ENG/ID: 2022 | TGC ACT TGG CCT ACA ATT CCA | AGC TGC CCA CTC AAG GAT CT | 168693646c1 | |
| FOXF1 | FOXF1/ID: 2294 | GCG GCT TCC GAA GGA AAT G | CAA GTG GCC GTT CAT CAT GC | 110735444c1 | |
| PAX1 | PAX1/ID: 5075 | TCG CTA TGG AGC AGA CGT ATG | GCT GCC GAC TGA TGT CAC A | 380036025c1 | |
| TEK | TEK/ID: 7010 | TTA GCC AGC TTA GTT CTC TGT GG | AGC ATC AGA TAC AAG AGG TAG GG | [30] | |
| SOX9 | SOX9/ID: 6662 | AGC GAA CGC ACA TCA AGA C | CTG TAG GCG ATC TGT TGG GG | 182765453c1 | |
| HIF-1⍺ | HIF1A/ID: 3091 | AAG GAA CCT GAT GCT TTA ACT TTG | TGG TCA TCA GTT TCT GTG TCG | [31] | |
| ACAN | ACAN/ID: 176 | TCG AGG ACA GCG AGG CC | TCG AGG GTG TAG CGT GTA GAG A | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Sheng, K.; Mannarino, M.; Jarzem, P.; Cherif, H.; Haglund, L. o-Vanillin Modulates Cell Phenotype and Extracellular Vesicles of Human Mesenchymal Stem Cells and Intervertebral Disc Cells. Cells 2022, 11, 3589. https://doi.org/10.3390/cells11223589
Li L, Sheng K, Mannarino M, Jarzem P, Cherif H, Haglund L. o-Vanillin Modulates Cell Phenotype and Extracellular Vesicles of Human Mesenchymal Stem Cells and Intervertebral Disc Cells. Cells. 2022; 11(22):3589. https://doi.org/10.3390/cells11223589
Chicago/Turabian StyleLi, Li, Kai Sheng, Matthew Mannarino, Peter Jarzem, Hosni Cherif, and Lisbet Haglund. 2022. "o-Vanillin Modulates Cell Phenotype and Extracellular Vesicles of Human Mesenchymal Stem Cells and Intervertebral Disc Cells" Cells 11, no. 22: 3589. https://doi.org/10.3390/cells11223589
APA StyleLi, L., Sheng, K., Mannarino, M., Jarzem, P., Cherif, H., & Haglund, L. (2022). o-Vanillin Modulates Cell Phenotype and Extracellular Vesicles of Human Mesenchymal Stem Cells and Intervertebral Disc Cells. Cells, 11(22), 3589. https://doi.org/10.3390/cells11223589