CRISPR-Cas9 Technology for the Creation of Biological Avatars Capable of Modeling and Treating Pathologies: From Discovery to the Latest Improvements
Abstract
1. Introduction
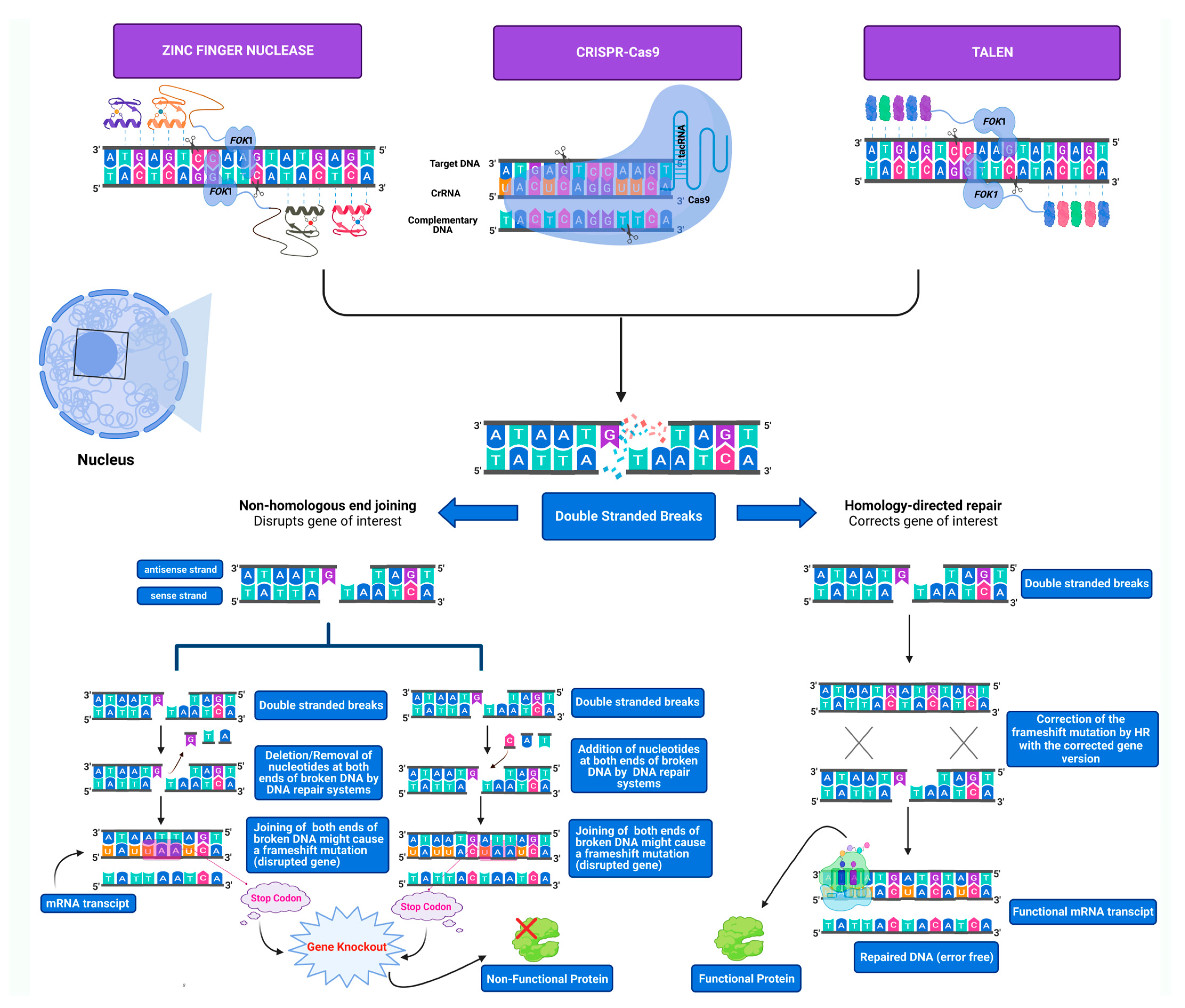
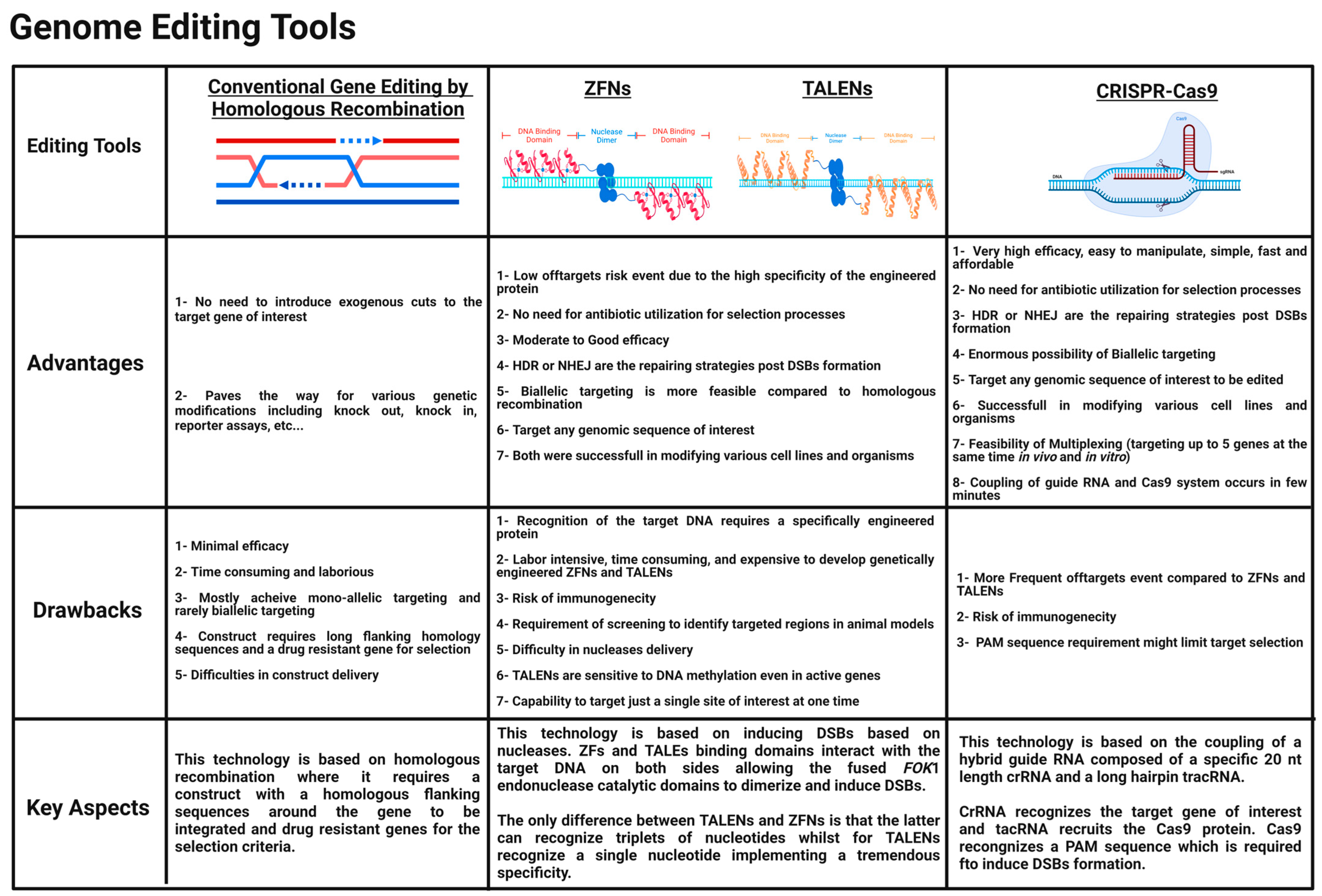
2. Genome Editing Tools
2.1. ZFNs
2.2. TALENs
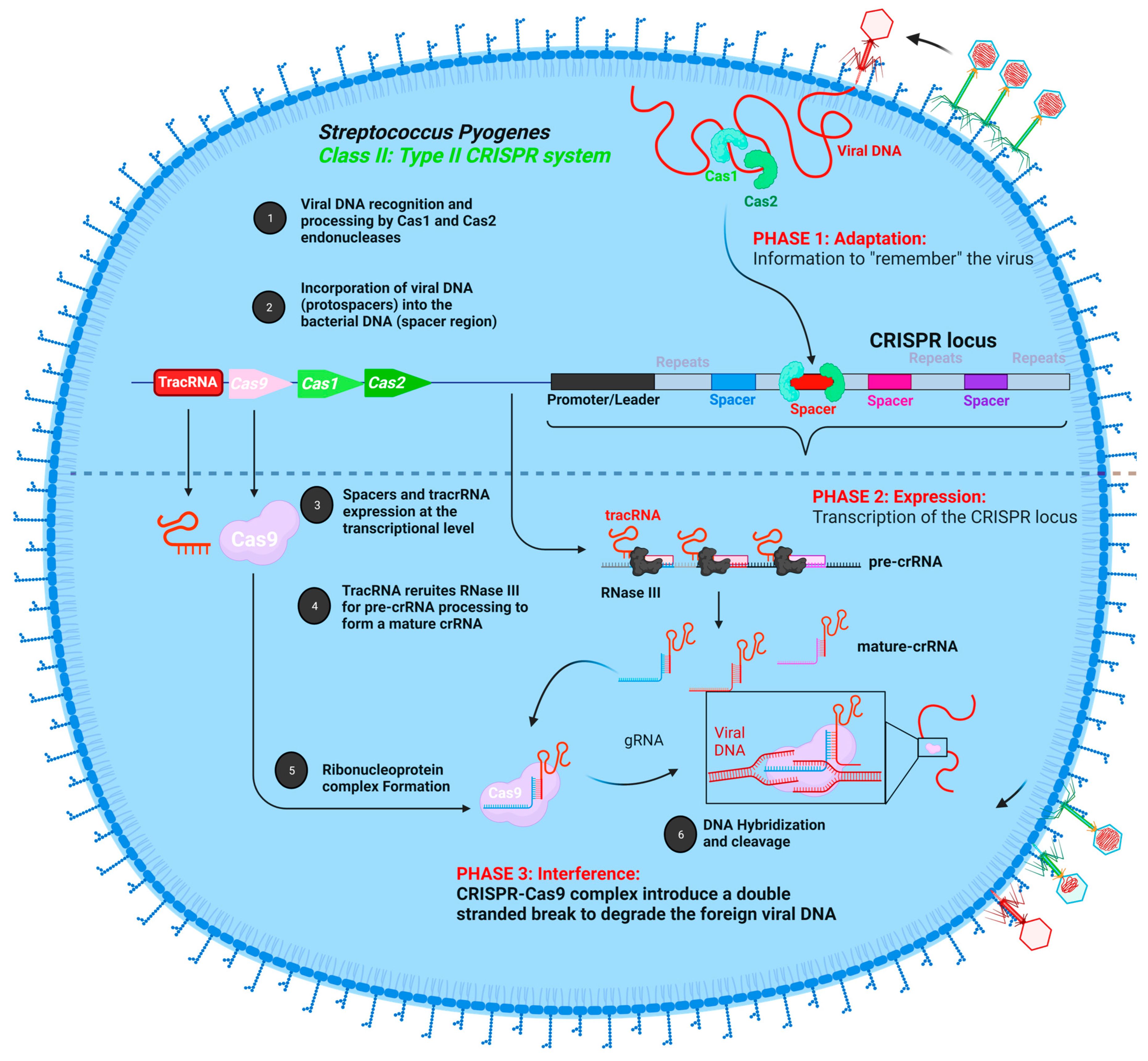
2.3. CRISPR-Cas
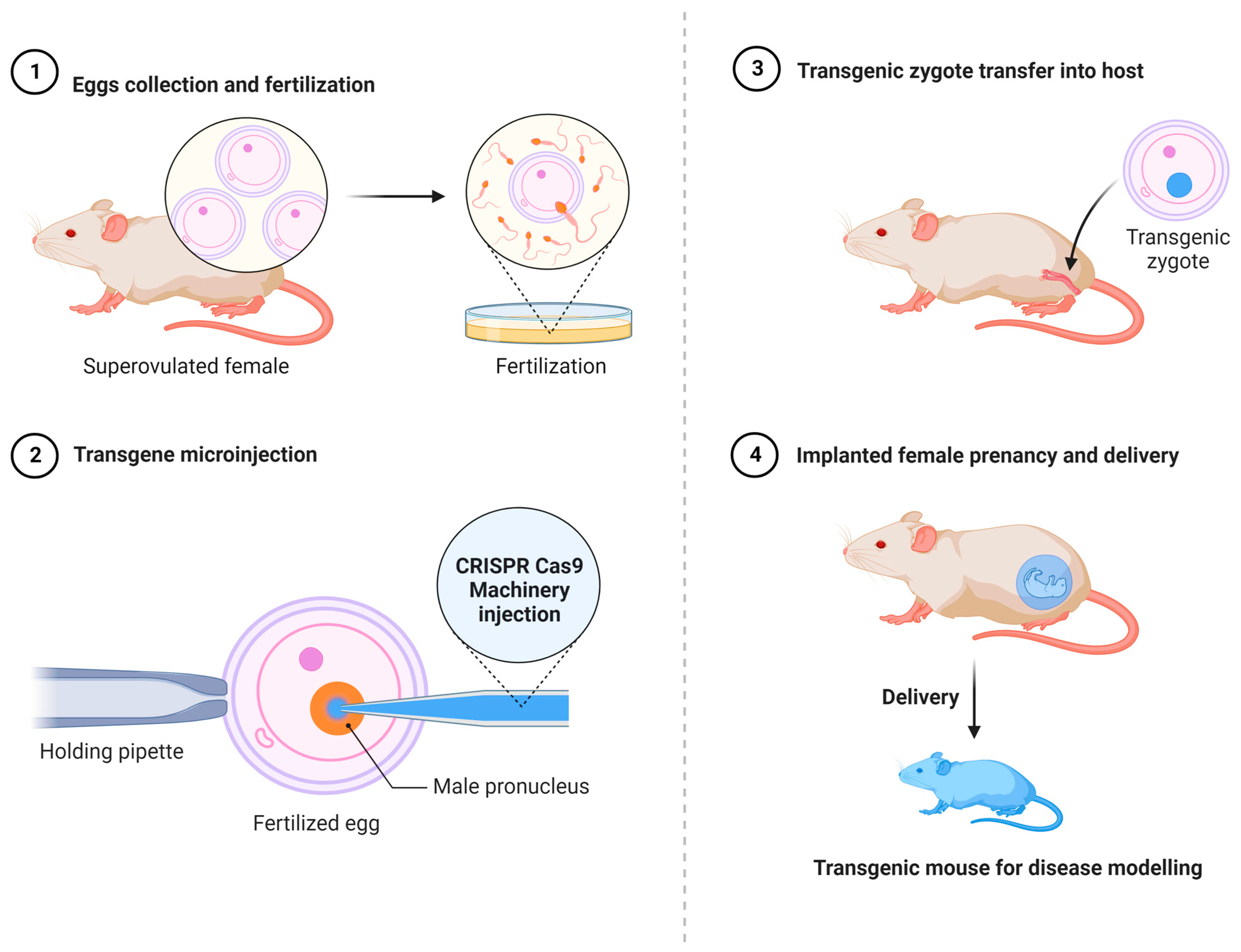
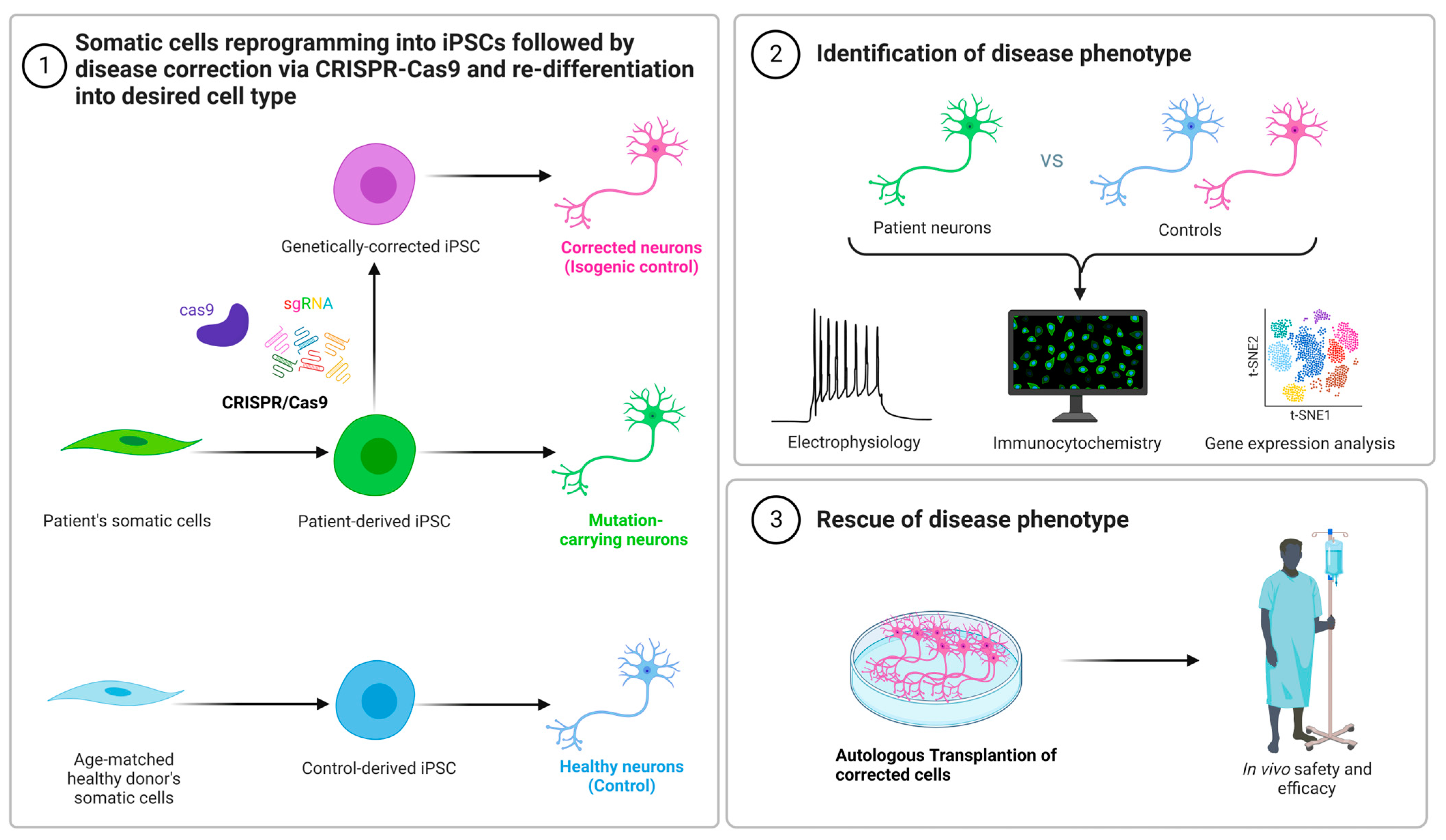
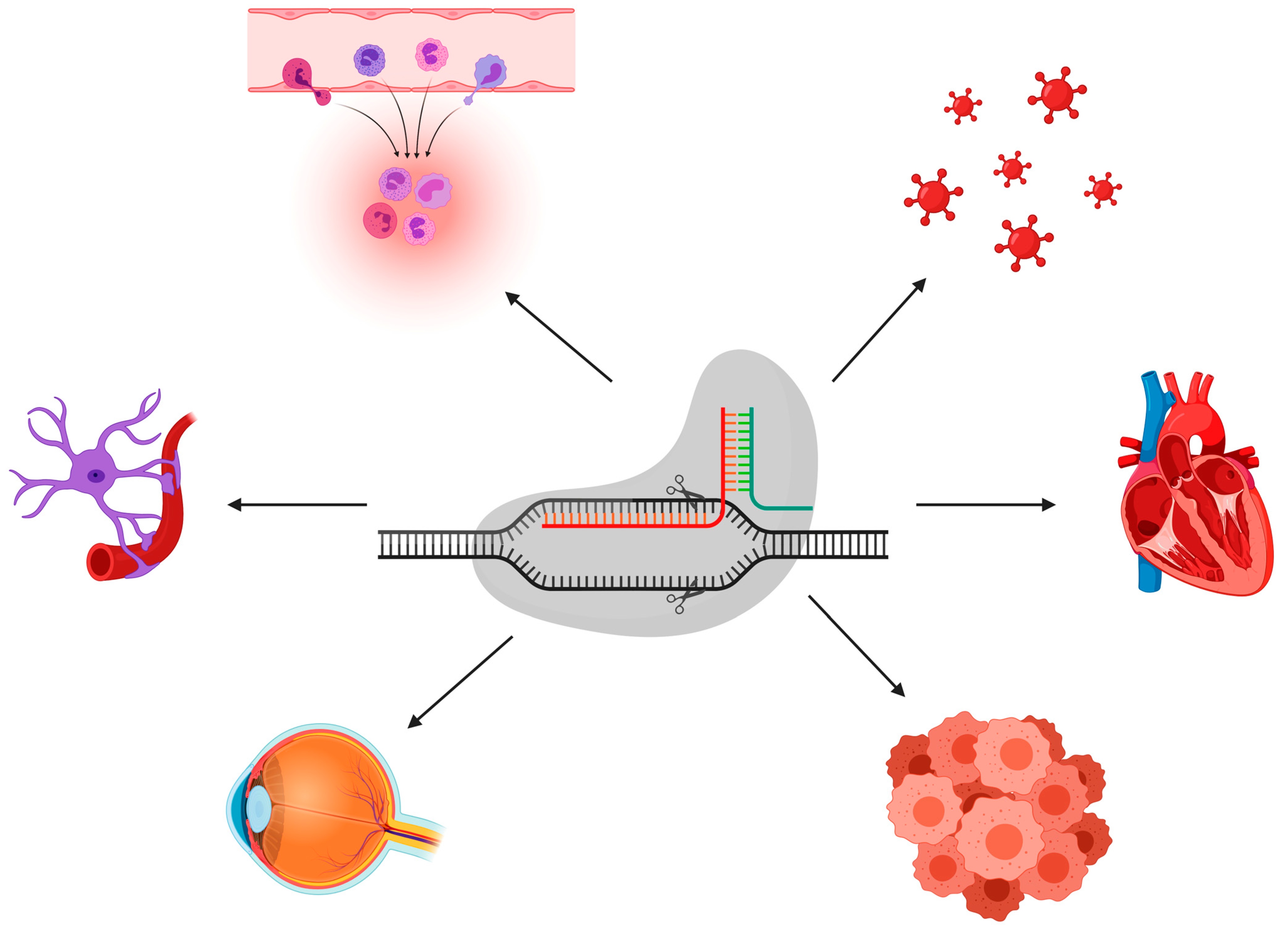
3. Disease Modeling and Gene Therapy Using CRISPR-Cas9
3.1. CRISPR-Cas9 and Cancer Disease Modeling
3.2. CRISPR-Cas9 and Cardiovascular Disease Modeling
3.3. CRISPR-Cas9 and Neurological Disease Modeling
3.4. CRISPR-Cas9 and Hereditary Eye Disease Modeling
3.5. CRISPR-Cas9 and Infectious Disease Modeling
3.6. CRISPR-Cas9 and Immunodeficiency Disease Modeling
4. CRISPR-Cas9 and Gene Therapy
4.1. Monogenic Diseases Correction by CRISPR-Cas9
4.2. Non-Monogenic Disease Correction by CRISPR-Cas9
5. Challenges of CRISPR-Cas9 and Enhanced Specificity
5.1. CRISPR-Cas9 Off-Targets
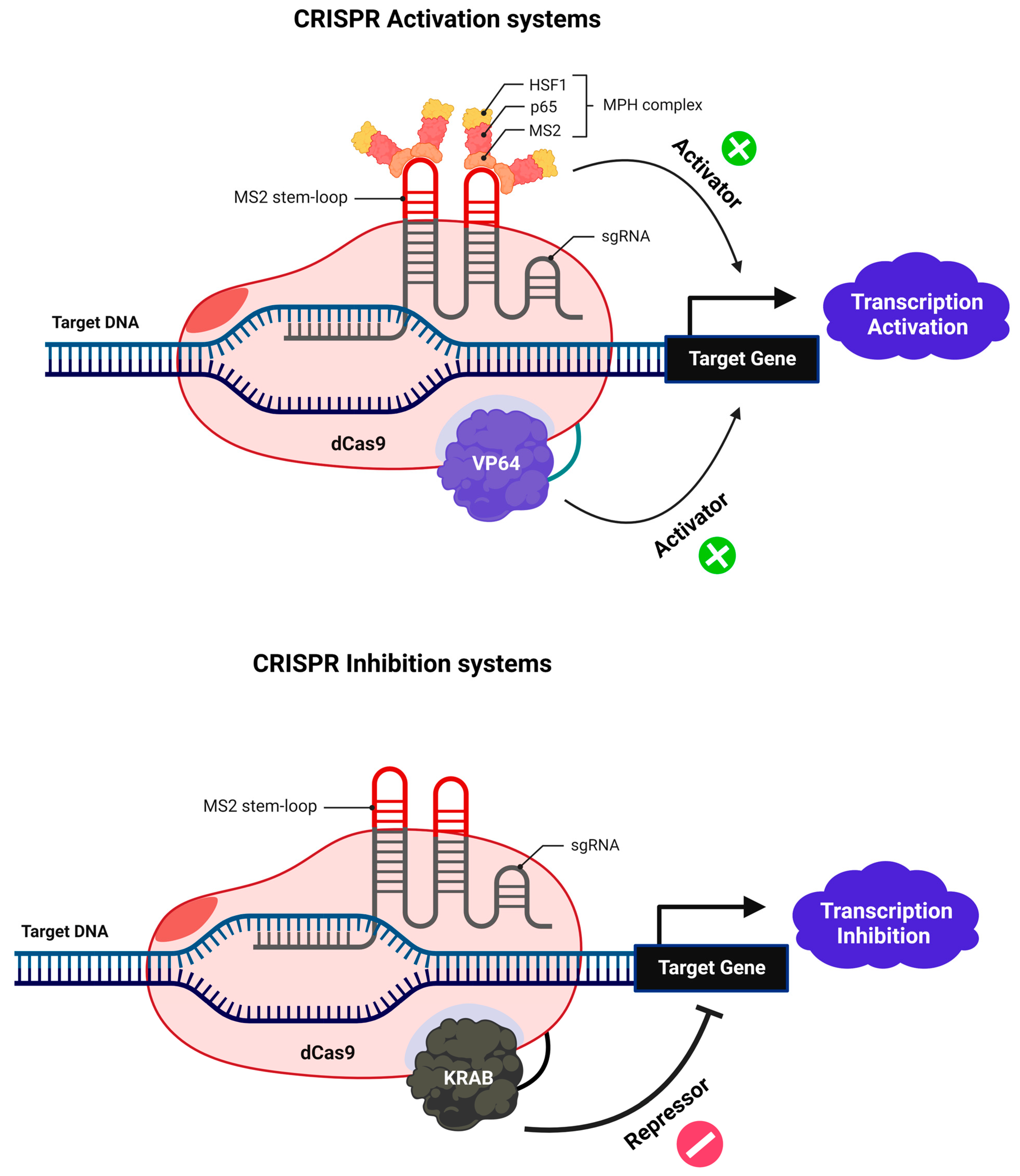
5.2. nCas9 and dCas9
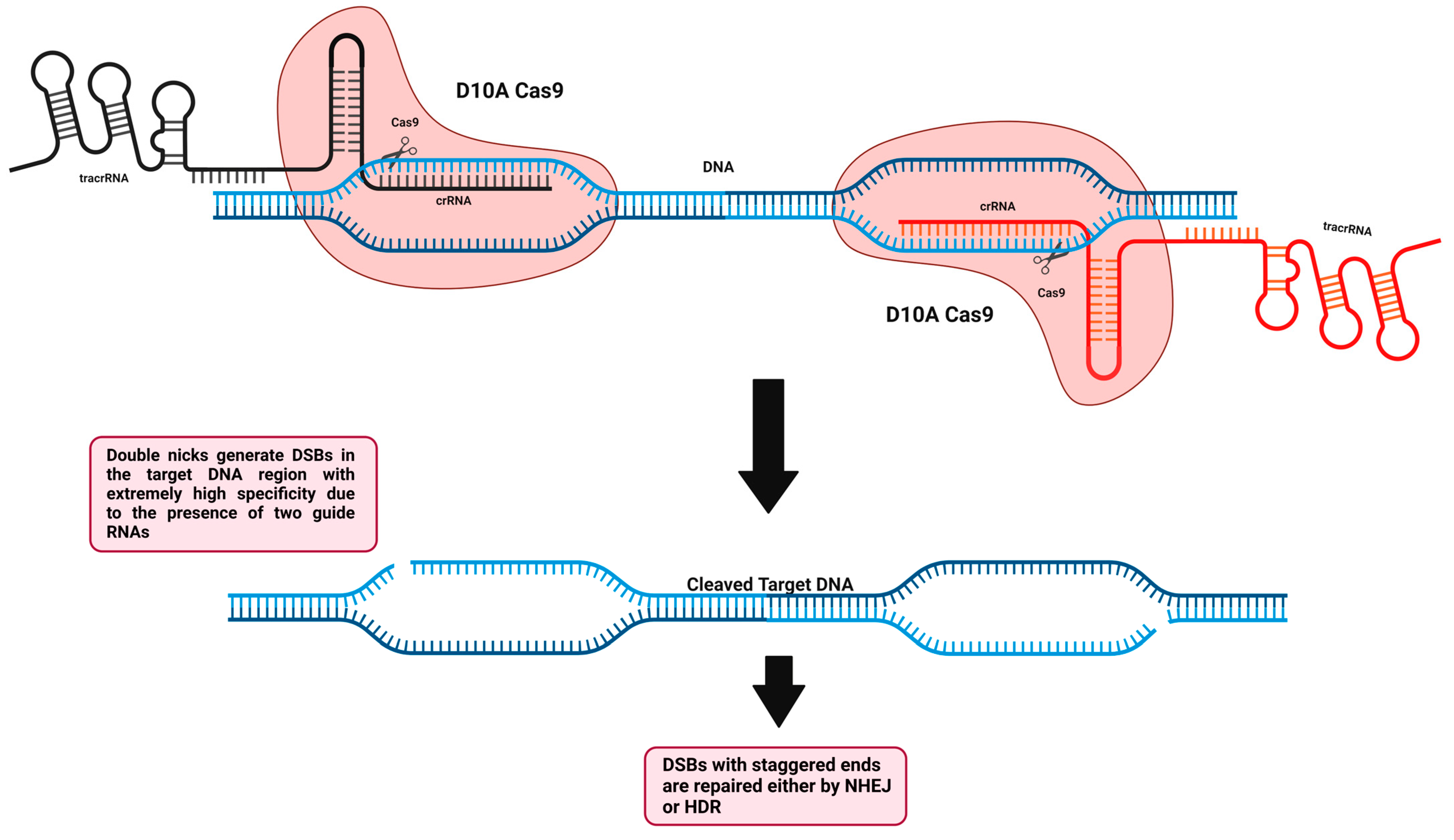
5.2.1. Cas9 Nickase
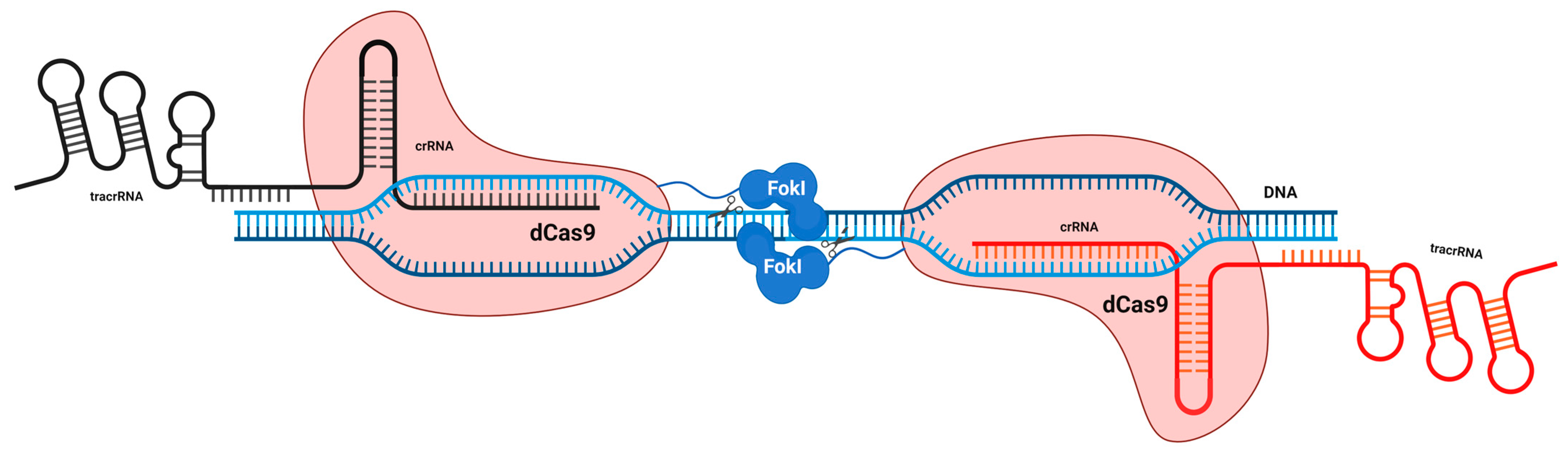
5.2.2. Inactive dCas and Dimeric RNA-Guided FokI Nucleases (RFNs)
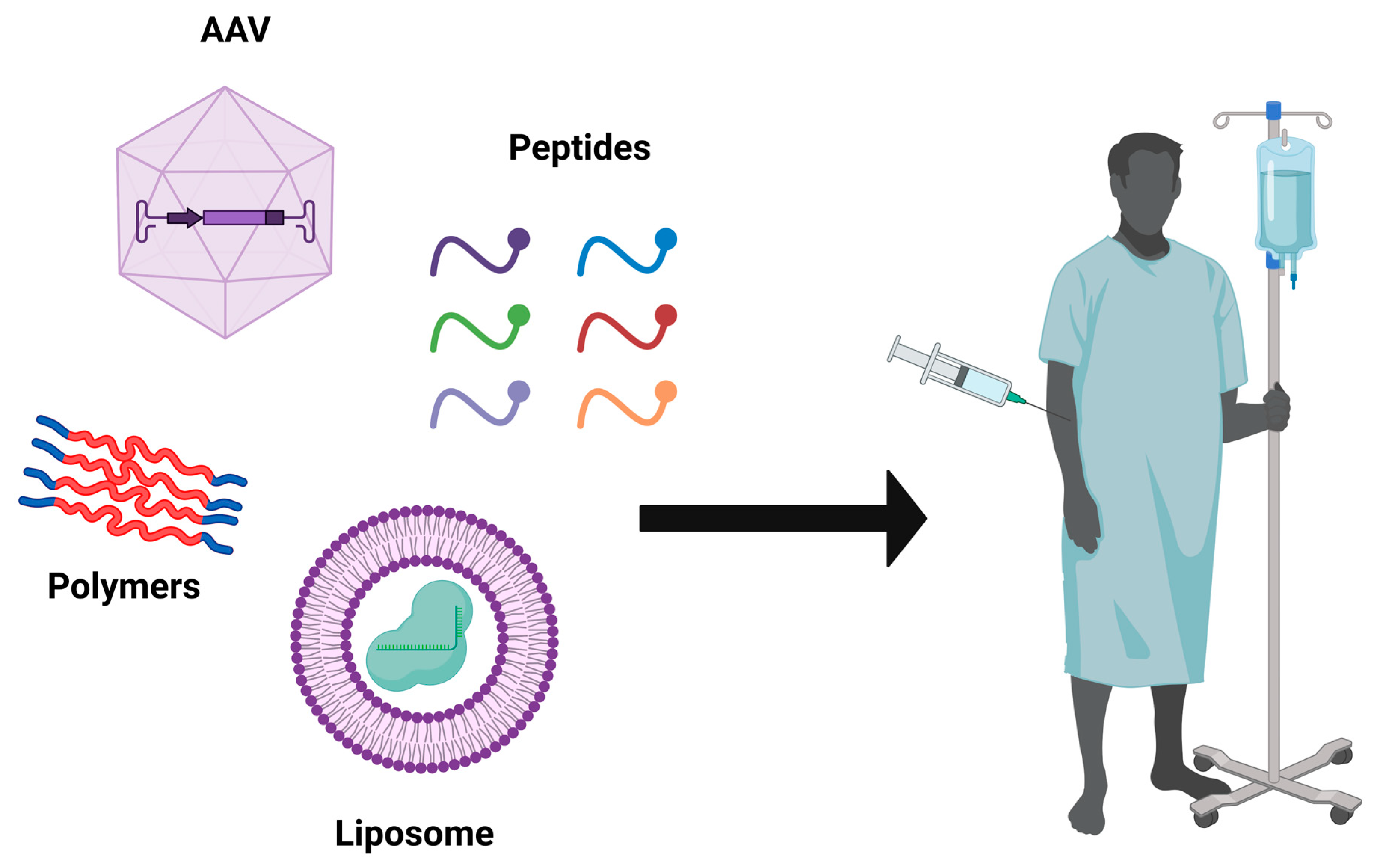
5.3. CRISPR-Cas9 Delivery Strategies
5.4. CRISPR-Cas9 PAM Dependency
5.5. gRNA Synthesis and Modifications
5.6. Immune Response against Cas9 Endonuclease
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.S.; Golic, K.G. Gene Targeting by Homologous Recombination in Drosophila. Science 2000, 288, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.J.; Golic, K.G. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 2003, 100, 2556–2561. [Google Scholar] [CrossRef]
- Smith, H.O.; Welcox, K. A Restriction enzyme from Hemophilus influenzae: I. Purification and general properties. J. Mol. Biol. 1970, 51, 379–391. [Google Scholar] [CrossRef]
- Li, J.; Sun, H.; Huang, Y.; Wang, Y.; Liu, Y.; Chen, X. Pathways and assays for DNA double-strand break repair by homologous recombination. Acta Biochim. Biophys. Sin. 2019, 51, 879–889. [Google Scholar] [CrossRef]
- Zhao, X.; Wei, C.; Li, J.; Xing, P.; Li, J.; Zheng, S.; Chen, X. Cell cycle-dependent control of homologous recombination. Acta Biochim. Biophys. Sin. 2017, 49, 655–668. [Google Scholar] [CrossRef]
- Aylon, Y.; Liefshitz, B.; Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004, 23, 4868–4875. [Google Scholar] [CrossRef]
- Ira, G.; Pellicioli, A.; Balijja, A.; Wang, X.; Fiorani, S.; Carotenuto, W.; Liberi, G.; Bressan, D.; Wan, L.; Hollingsworth, N.M.; et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 2004, 431, 1011–1017. [Google Scholar] [CrossRef]
- Haber, J.E. A Life Investigating Pathways That Repair Broken Chromosomes. Annu. Rev. Genet. 2016, 50, 1–28. [Google Scholar] [CrossRef]
- Kowalczykowski, S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016410. [Google Scholar] [CrossRef]
- Chakraborty, A.; Tapryal, N.; Venkova, T.; Horikoshi, N.; Pandita, R.K.; Sarker, A.H.; Sarkar, P.S.; Pandita, T.K.; Hazra, T.K. Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 2016, 7, 13049. [Google Scholar] [CrossRef]
- Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. Methodologies for Improving HDR Efficiency. Front. Genet. 2019, 9, 691. [Google Scholar] [CrossRef]
- Carroll, D. Genome Engineering with Targetable Nucleases. Annu. Rev. Biochem. 2014, 83, 409–439. [Google Scholar] [CrossRef]
- Urnov, F.D. Ctrl-Alt-inDel: Genome editing to reprogram a cell in the clinic. Curr. Opin. Genet. Dev. 2018, 52, 48–56. [Google Scholar] [CrossRef]
- Capecchi, M.R. Altering the Genome by Homologous Recombination. Science 1989, 244, 1288–1292. [Google Scholar] [CrossRef]
- Dow, L.E. Modeling Disease In Vivo With CRISPR/Cas9. Trends Mol. Med. 2015, 21, 609–621. [Google Scholar] [CrossRef]
- Price, B.D.; D’Andrea, A.D. Chromatin Remodeling at DNA Double-Strand Breaks. Cell 2013, 152, 1344–1354. [Google Scholar] [CrossRef]
- Bateman, A.; Birney, E.; Durbin, R.; Eddy, S.R.; Howe, K.L.; Sonnhammer, E.L. The Pfam Protein Families Database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef]
- Miller, J.; McLachlan, A.D.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef]
- Klug, A. The Discovery of Zinc Fingers and Their Applications in Gene Regulation and Genome Manipulation. Annu. Rev. Biochem. 2010, 79, 213–231. [Google Scholar] [CrossRef]
- Pavletich, N.P.; Pabo, C.O. Zinc Finger-DNA Recognition: Crystal Structure of a Zif268-DNA Complex at 2.1 Å. Science 1991, 252, 809–817. [Google Scholar] [CrossRef]
- Jantz, D.; Amann, B.T.; Gatto, G.J.; Berg, J.M. The Design of Functional DNA-Binding Proteins Based on Zinc Finger Domains. Chem. Rev. 2003, 104, 789–800. [Google Scholar] [CrossRef]
- Dion, S.; Demattéi, M.-V.; Renault, S. Zinc finger proteins: Tools for site-specific correction or modification of the genome. Med. Sci. 2007, 23, 834–839. [Google Scholar] [CrossRef]
- Li, L.; Wu, L.P.; Chandrasegaran, S. Functional domains in Fok I restriction endonuclease. Proc. Natl. Acad. Sci. USA 1992, 89, 4275–4279. [Google Scholar] [CrossRef]
- Bitinaite, J.; Wah, D.A.; Aggarwal, A.K.; Schildkraut, I. Fok I dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. USA 1998, 95, 10570–10575. [Google Scholar] [CrossRef]
- Vanamee, S.; Santagata, S.; Aggarwal, A.K. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001, 309, 69–78. [Google Scholar] [CrossRef]
- Catto, L.E.; Ganguly, S.; Milsom, S.E.; Welsh, A.J.; Halford, S.E. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006, 34, 1711–1720. [Google Scholar] [CrossRef]
- Bibikova, M.; Golic, M.; Golic, K.G.; Carroll, D. Targeted Chromosomal Cleavage and Mutagenesis in Drosophila Using Zinc-Finger Nucleases. Genetics 2002, 161, 1169–1175. [Google Scholar] [CrossRef]
- Bibikova, M.; Beumer, K.; Trautman, J.K.; Carroll, D. Enhancing Gene Targeting with Designed Zinc Finger Nucleases. Science 2003, 300, 764. [Google Scholar] [CrossRef]
- Urnov, F.D.; Miller, J.C.; Lee, Y.-L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef]
- Beumer, K.; Bhattacharyya, G.; Bibikova, M.; Trautman, J.K.; Carroll, D. Efficient Gene Targeting in Drosophila with Zinc-Finger Nucleases. Genetics 2006, 172, 2391–2403. [Google Scholar] [CrossRef]
- Beumer, K.J.; Trautman, J.K.; Bozas, A.; Liu, J.-L.; Rutter, J.; Gall, J.G.; Carroll, D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 2008, 105, 19821–19826. [Google Scholar] [CrossRef]
- Morton, J.; Davis, M.W.; Jorgensen, E.M.; Carroll, D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc. Natl. Acad. Sci. USA 2006, 103, 16370–16375. [Google Scholar] [CrossRef]
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708. [Google Scholar] [CrossRef]
- Meng, X.; Noyes, M.B.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701. [Google Scholar] [CrossRef]
- Foley, J.E.; Yeh, J.-R.J.; Maeder, M.L.; Reyon, D.; Sander, J.D.; Peterson, R.T.; Joung, J.K. Rapid Mutation of Endogenous Zebrafish Genes Using Zinc Finger Nucleases Made by Oligomerized Pool ENgineering (OPEN). PLoS ONE 2009, 4, e4348. [Google Scholar] [CrossRef]
- Ochiai, H.; Fujita, K.; Suzuki, K.-I.; Nishikawa, M.; Shibata, T.; Sakamoto, N.; Yamamoto, T. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 2010, 15, 875–885. [Google Scholar] [CrossRef]
- Carbery, I.D.; Ji, D.; Harrington, A.; Brown, V.; Weinstein, E.J.; Liaw, L.; Cui, X. Targeted Genome Modification in Mice Using Zinc-Finger Nucleases. Genetics 2010, 186, 451–459. [Google Scholar] [CrossRef]
- Meyer, M.; de Angelis, M.H.; Wurst, W.; Kühn, R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 15022–15026. [Google Scholar] [CrossRef]
- Geurts, A.M.; Cost, G.; Freyvert, Y.; Zeitler, B.; Miller, J.C.; Choi, V.M.; Jenkins, S.S.; Wood, A.; Cui, X.; Meng, X.; et al. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science 2009, 325, 433. [Google Scholar] [CrossRef]
- Mashimo, T.; Takizawa, A.; Voigt, B.; Yoshimi, K.; Hiai, H.; Kuramoto, T.; Serikawa, T. Generation of Knockout Rats with X-Linked Severe Combined Immunodeficiency (X-SCID) Using Zinc-Finger Nucleases. PLoS ONE 2010, 5, e8870. [Google Scholar] [CrossRef]
- Maeder, M.L.; Thibodeau-Beganny, S.; Osiak, A.; Wright, D.A.; Anthony, R.M.; Eichtinger, M.; Jiang, T.; Foley, J.E.; Winfrey, R.J.; Townsend, J.A.; et al. Rapid “Open-Source” Engineering of Customized Zinc-Finger Nucleases for Highly Efficient Gene Modification. Mol. Cell 2008, 31, 294–301. [Google Scholar] [CrossRef]
- Lombardo, A.L.; Genovese, P.; Beausejour, C.M.; Colleoni, S.; Lee, Y.-L.; Kim, K.A.; Ando, D.; Urnov, F.D.; Galli, C.; Gregory, P.; et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007, 25, 1298–1306. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, H.J.; Kim, H.; Cho, S.W.; Kim, J.-S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009, 19, 1279–1288. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, E.; Kim, J.-S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2009, 20, 81–89. [Google Scholar] [CrossRef]
- Perez, E.E.; Wang, J.; Miller, J.C.; Jouvenot, Y.; Kim, K.A.; Liu, O.; Wang, N.; Lee, G.; Bartsevich, V.V.; Lee, Y.-L.; et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 808–816. [Google Scholar] [CrossRef]
- Ryan, R.; Vorhölter, F.-J.; Potnis, N.; Jones, J.B.; Van Sluys, M.-A.; Bogdanove, A.J.; Dow, J.M. Pathogenomics of Xanthomonas: Understanding bacterium–plant interactions. Nat. Rev. Genet. 2011, 9, 344–355. [Google Scholar] [CrossRef]
- Szurek, B.; Marois, E.; Bonas, U.; Ackerveken, G.V.D. Eukaryotic features of the Xanthomonas type III effector AvrBs3: Protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 2001, 26, 523–534. [Google Scholar] [CrossRef]
- Scott, J.N.F.; Kupinski, A.P.; Boyes, J. Targeted genome regulation and modification using transcription activator-like effectors. FEBS J. 2014, 281, 4583–4597. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2010, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Xiao, A.; Huang, P.; Wang, W.-Y.; Zhu, Z.-Y.; Zhang, B. TALE nuclease engineering and targeted genome modification. Hereditas (Beijing) 2013, 35, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Carlson, D.F.; Tan, W.; Lillico, S.G.; Stverakova, D.; Proudfoot, C.; Christian, M.; Voytas, D.F.; Long, C.R.; Whitelaw, C.B.A.; Fahrenkrug, S.C. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA 2012, 109, 17382–17387. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.; Annaluru, N.; Kandavelou, K.; Chandrasegaran, S. TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr. Gene Ther. 2014, 14, 461–472. [Google Scholar] [CrossRef]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef]
- Mussolino, C.; Morbitzer, R.; Lütge, F.; Dannemann, N.; Lahaye, T.; Cathomen, T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011, 39, 9283–9293. [Google Scholar] [CrossRef]
- Hockemeyer, D.; Wang, H.; Kiani, S.; Lai, C.S.; Gao, Q.; Cassady, J.P.; Cost, G.; Zhang, L.; Santiago, Y.; Miller, J.C.; et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011, 29, 731–734. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, C.; Cerbini, T.; San, H.; Lin, Y.; Chen, G.; Rao, M.S.; Zou, J. Stable Enhanced Green Fluorescent Protein Expression After Differentiation and Transplantation of Reporter Human Induced Pluripotent Stem Cells Generated by AAVS1 Transcription Activator-Like Effector Nucleases. STEM CELLS Transl. Med. 2014, 3, 821–835. [Google Scholar] [CrossRef]
- Tesson, L.; Usal, C.; Ménoret, S.; Leung, E.; Niles, B.J.; Remy, S.; Santiago, Y.; Vincent, A.I.; Meng, X.; Zhang, L.; et al. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 2011, 29, 695–696. [Google Scholar] [CrossRef]
- Wood, A.J.; Lo, T.-W.; Zeitler, B.; Pickle, C.S.; Ralston, E.J.; Lee, A.H.; Amora, R.; Miller, J.C.; Leung, E.; Meng, X.; et al. Targeted Genome Editing Across Species Using ZFNs and TALENs. Science 2011, 333, 307. [Google Scholar] [CrossRef]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011, 29, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Cade, L.; Khayter, C.; Reyon, D.; Peterson, R.T.; Joung, J.K.; Yeh, J.-R.J. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011, 29, 697–698. [Google Scholar] [CrossRef] [PubMed]
- Bloom, K.; Ely, A.; Mussolino, C.; Cathomen, T.; Arbuthnot, P. Inactivation of Hepatitis B Virus Replication in Cultured Cells and In Vivo with Engineered Transcription Activator-Like Effector Nucleases. Mol. Ther. 2013, 21, 1889–1897. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, W.; Lin, J.; Wang, F.; Wu, M.; Chen, C.; Zheng, Y.; Peng, X.; Li, J.; Yuan, Z. An Efficient Antiviral Strategy for Targeting Hepatitis B Virus Genome Using Transcription Activator-Like Effector Nucleases. Mol. Ther. 2014, 22, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Noh, K.-W.; Park, J.; Kang, M.-S. Targeted disruption of EBNA1 in EBV-infected cells attenuated cell growth. BMB Rep. 2016, 49, 226–231. [Google Scholar] [CrossRef]
- Ernst, M.P.; Broeders, M.; Herrero-Hernandez, P.; Oussoren, E.; van der Ploeg, A.T.; Pijnappel, W.P. Ready for Repair? Gene Editing Enters the Clinic for the Treatment of Human Disease. Mol. Ther.–Methods Clin. Dev. 2020, 18, 532–557. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.; Zhang, Y.; Cao, H.; Li, J.; Duan, R.; Hu, J. TALEN-Mediated Gene Targeting for Cystic Fibrosis-Gene Therapy. Genes 2019, 10, 39. [Google Scholar] [CrossRef]
- Fang, Y.; Cheng, Y.; Lu, D.; Gong, X.; Yang, G.; Gong, Z.; Zhu, Y.; Sang, X.; Fan, S.; Zhang, J.; et al. Treatment of β654-thalassaemia by TALENs in a mouse model. Cell Prolif. 2018, 51, e12491. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Mojica, F.J.; Juez, G.; Rodriguez-Valera, F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993, 9, 613–621. [Google Scholar] [CrossRef]
- Makarova, K.S.; Aravind, L.; Grishin, N.V.; Rogozin, I.; Koonin, E.V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002, 30, 482–496. [Google Scholar] [CrossRef]
- Jansen, R.; Van Embden, J.D.A.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Genet. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef]
- Li, J.-F.; Norville, J.E.; Aach, J.E.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.Y.; Shen, B.; Cui, Y.Q.; Chen, Y.C.; Wang, J.Y.; Wang, L.; Kang, Y.; Zhao, X.Y.; Si, W.; Li, W.; et al. Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics 2013, 194, 1029–1035. [Google Scholar] [CrossRef]
- Liu, P.; Long, L.; Xiong, K.; Yu, B.; Chang, N.; Xiong, J.-W.; Zhu, Z.; Liu, D. Heritable/conditional genome editing in C. elegans using a CRISPR-Cas9 feeding system. Cell Res. 2014, 24, 886–889. [Google Scholar] [CrossRef]
- Friedland, A.E.; Tzur, Y.B.; Esvelt, K.M.; Colaiácovo, M.P.; Church, G.M.; A. Calarco, J. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 2013, 10, 741–743. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Blasco, R.B.; Karaca, E.; Ambrogio, C.; Cheong, T.-C.; Karayol, E.; Minero, V.G.; Voena, C.; Chiarle, R. Simple and Rapid In Vivo Generation of Chromosomal Rearrangements using CRISPR/Cas9 Technology. Cell Rep. 2014, 9, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Canver, M.C.; Bauer, D.E.; Dass, A.; Yien, Y.Y.; Chung, J.; Masuda, T.; Maeda, T.; Paw, B.H.; Orkin, S.H. Characterization of Genomic Deletion Efficiency Mediated by Clustered Regularly Interspaced Palindromic Repeats (CRISPR)/Cas9 Nuclease System in Mammalian Cells*. J. Biol. Chem. 2014, 289, 21312–21324. [Google Scholar] [CrossRef]
- He, Z.; Proudfoot, C.; Mileham, A.J.; McLaren, D.G.; Whitelaw, C.B.A.; Lillico, S.G. Highly efficient targeted chromosome deletions using CRISPR/Cas9. Biotechnol. Bioeng. 2014, 112, 1060–1064. [Google Scholar] [CrossRef]
- Choi, P.; Meyerson, M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 2014, 5, 1–6. [Google Scholar] [CrossRef]
- Torres, R.; Martin, M.C.; Garcia, A.; Cigudosa, J.C.; Ramirez, J.C.; Rodriguez-Perales, S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR–Cas9 system. Nat. Commun. 2014, 5, 3964. [Google Scholar] [CrossRef]
- Maddalo, D.; Manchado, E.; Concepcion, C.P.; Bonetti, C.; Vidigal, J.A.; Han, Y.-C.; Ogrodowski, P.; Crippa, A.; Rekhtman, N.; De Stanchina, E.; et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014, 516, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Xu, Y.; Zhang, X.; Ding, C.; Huang, R.; Zhang, Z.; Lv, J.; Xie, X.; Chen, Y.; Li, Y.; et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015, 6, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. Ethics of embryo editing divides scientists. Nature 2015, 519, 272. [Google Scholar] [CrossRef] [PubMed]
- Lanphier, E.; Urnov, F.; Haecker, S.E.; Werner, M.; Smolenski, J. Don’t edit the human germ line. Nature 2015, 519, 410–411. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Xue, W.; Chen, S.; Bogorad, R.L.; Benedetti, E.; Grompe, M.; Koteliansky, V.; Sharp, P.A.; Jacks, T.E.; Anderson, D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014, 32, 551–553. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Strong, A.; Patel, K.M.; Ng, S.-L.; Gosis, B.S.; Regan, S.N.; Cowan, C.A.; Rader, D.J.; Musunuru, K. Permanent Alteration of PCSK9 With In Vivo CRISPR-Cas9 Genome Editing. Circ. Res. 2014, 115, 488–492. [Google Scholar] [CrossRef]
- Lin, S.R.; Yang, H.C.; Kuo, Y.T.; Liu, C.J.; Yang, T.Y.; Sung, K.C.; Lin, Y.Y.; Wang, H.Y.; Wang, C.C.; Shen, Y.C.; et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol. Ther. Nucleic Acids 2014, 3, e186. [Google Scholar] [CrossRef]
- Shao, Y.; Guan, Y.; Wang, L.; Qiu, Z.; Liu, M.; Chen, Y.; Wu, L.; Li, Y.; Ma, X.; Liu, M.; et al. CRISPR/Cas-mediated genome editing in the rat via direct injection of one-cell embryos. Nat. Protoc. 2014, 9, 2493–2512. [Google Scholar] [CrossRef]
- Shen, B.; Zhang, J.; Wu, H.; Wang, J.; Ma, K.; Li, Z.; Zhang, X.; Zhang, P.; Huang, X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013, 23, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Porteus, M.H. Towards a new era in medicine: Therapeutic genome editing. Genome Biol. 2015, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kotterman, M.A.; Schaffer, D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014, 15, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L., 3rd; Strohl, W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 2017, 31, 317–334. [Google Scholar] [CrossRef]
- Verdera, H.C.; Kuranda, K.; Mingozzi, F. AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Mol. Ther. 2020, 28, 723–746. [Google Scholar] [CrossRef]
- Mengstie, M.A.; Wondimu, B.Z. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biol. Targets Ther. 2021, 15, 353–361. [Google Scholar] [CrossRef]
- Wilson, R.C.; Gilbert, L.A. The Promise and Challenge of In Vivo Delivery for Genome Therapeutics. ACS Chem. Biol. 2018, 13, 376–382. [Google Scholar] [CrossRef]
- Bijlani, S.; Pang, K.M.; Sivanandam, V.; Singh, A.; Chatterjee, S. The Role of Recombinant AAV in Precise Genome Editing. Front. Genome Ed. 2022, 3. [Google Scholar] [CrossRef]
- Vogel, R.; Seyffert, M.; Pereira, B.d.A.; Fraefel, C. Viral and Cellular Components of AAV2 Replication Compartments. Open Virol. J. 2013, 7, 98–120. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Y.; He, L.; Pu, W.; Yu, W.; Li, Y.; Wu, Y.-T.; Xu, C.; Wei, Y.; Ding, Q.; et al. In Vivo AAV-CRISPR/Cas9–Mediated Gene Editing Ameliorates Atherosclerosis in Familial Hypercholesterolemia. Circulation 2020, 141, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Al-Zaidy, S.A.; Rodino-Klapac, L.R.; Goodspeed, K.; Gray, S.J.; Kay, C.N.; Boye, S.L.; Boye, S.E.; George, L.A.; Salabarria, S.; et al. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Mol. Ther. 2020, 29, 464–488. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Rivera, R.M.C.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, C.M.; Schaffer, D.V. Adeno-associated virus-mediated delivery of CRISPR-Cas9 for genome editing in the central nervous system. Curr. Opin. Biomed. Eng. 2018, 7, 33–41. [Google Scholar] [CrossRef]
- Park, S.H.; Bao, G. CRISPR/Cas9 gene editing for curing sickle cell disease. Transfus. Apher. Sci. 2021, 60. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Ma, L.; Shao, T.; Zhang, M.; Guan, Y.; Wang, L.; Hu, Y.; Chen, X.; Han, H.; Shen, N.; et al. Enhanced genome editing to ameliorate a genetic metabolic liver disease through co-delivery of adeno-associated virus receptor. Sci. China Life Sci. 2020, 65, 718–730. [Google Scholar] [CrossRef]
- Torres-Ruíz, R.; Rodriguez-Perales, S. CRISPR-Cas9: A Revolutionary Tool for Cancer Modelling. Int. J. Mol. Sci. 2015, 16, 22151–22168. [Google Scholar] [CrossRef]
- Maresch, R.; Mueller, S.; Veltkamp, C.; Öllinger, R.; Friedrich, M.; Heid, I.; Steiger, K.; Weber, J.; Engleitner, T.; Barenboim, M.; et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 2016, 7, 10770. [Google Scholar] [CrossRef]
- Sánchez-Rivera, F.J.; Papagiannakopoulos, T.; Romero, R.; Tammela, T.; Bauer, M.R.; Bhutkar, A.; Joshi, N.; Subbaraj, L.; Bronson, R.T.; Xue, W.; et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 2014, 516, 428–431. [Google Scholar] [CrossRef]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Roper, J.; Tammela, T.; Akkad, A.; Almeqdadi, M.; Santos, S.B.; Jacks, T.; Yilmaz, H. Colonoscopy-based colorectal cancer modeling in mice with CRISPR–Cas9 genome editing and organoid transplantation. Nat. Protoc. 2018, 13, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Heckl, D.; Kowalczyk, M.S.; Yudovich, D.; Belizaire, R.; Puram, R.V.; McConkey, M.E.; Thielke, A.; Aster, J.C.; Regev, A.; Ebert, B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014, 32, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Zuckermann, M.; Hovestadt, V.; Knobbe-Thomsen, C.B.; Zapatka, M.; Northcott, P.A.; Schramm, K.; Belic, J.; Jones, D.T.W.; Tschida, B.R.; Moriarity, B.S.; et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat. Commun. 2015, 6, 7391. [Google Scholar] [CrossRef]
- Sánchez-Rivera, F.J.; Jacks, T. Applications of the CRISPR–Cas9 system in cancer biology. Nat. Cancer 2015, 15, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Sayin, V.I.; Papagiannakopoulos, T. Application of CRISPR-mediated genome engineering in cancer research. Cancer Lett. 2017, 387, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Li, A.H.; Morrison, A.; Kovar, C.L.; Cupples, L.A.; Brody, J.A.; Polfus, L.M.; Yu, B.; Metcalf, G.; Muzny, D.M.; Veeraraghavan, N.; et al. Analysis of loss-of-function variants and 20 risk factor phenotypes in 8554 individuals identifies loci influencing chronic disease. Nat. Genet. 2015, 47, 640–642. [Google Scholar] [CrossRef]
- Carroll, K.J.; Makarewich, C.A.; McAnally, J.; Anderson, D.M.; Zentilin, L.; Liu, N.; Giacca, M.; Bassel-Duby, R.; Olson, E.N. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc. Natl. Acad. Sci. USA 2015, 113, 338–343. [Google Scholar] [CrossRef]
- Lagace, T.A.; Curtis, D.E.; Garuti, R.; McNutt, M.C.; Park, S.W.; Prather, H.B.; Anderson, N.N.; Ho, Y.K.; Hammer, R.E.; Horton, J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and inlivers of parabiotic mice. J. Clin. Investig. 2006, 116, 2995–3005. [Google Scholar] [CrossRef]
- Abrahimi, P.; Chang, W.G.; Kluger, M.S.; Qyang, Y.; Tellides, G.; Saltzman, W.M.; Pober, J.S. Efficient Gene Disruption in Cultured Primary Human Endothelial Cells by CRISPR/Cas9. Circ. Res. 2015, 117, 121–128. [Google Scholar] [CrossRef]
- Ang, Y.-S.; Rivas, R.N.; Ribeiro, A.J.; Srivas, R.; Rivera, J.; Stone, N.R.; Pratt, K.; Mohamed, T.M.; Fu, J.-D.; Spencer, C.I.; et al. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 2016, 167, 1734–1749.e22. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; McCain, M.L.; Yang, L.; He, A.; Pasqualini, F.S.; Agarwal, A.; Yuan, H.; Jiang, D.; Zhang, D.; Zangi, L.; et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014, 20, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Nance, M.A. Genetics of Huntington disease. Handb. Clin. Neurol. 2017, 144, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.B.; Winslow, A.R.; Strasser, S.D. Systems biology of neurodegenerative diseases. Integr. Biol. 2015, 7, 758–775. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.; Pritzkow, S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1332–1340. [Google Scholar] [CrossRef]
- Niedzielska, E.; Smaga, I.; Gawlik, M.; Moniczewski, A.; Stankowicz, P.; Pera, J.; Filip, M. Oxidative Stress in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 4094–4125. [Google Scholar] [CrossRef]
- Hu, Z.; Yang, B.; Mo, X.; Xiao, H. Mechanism and Regulation of Autophagy and Its Role in Neuronal Diseases. Mol. Neurobiol. 2014, 52, 1190–1209. [Google Scholar] [CrossRef]
- Yan, S.; Tu, Z.C.; Liu, Z.M.; Fan, N.N.; Yang, H.M.; Yang, S.; Yang, W.L.; Zhao, Y.; Ouyang, Z.; Lai, C.D.; et al. A Huntingtin Knockin Pig Model Recapitulates Features of Selective Neurodegeneration in Huntington’s Disease. Cell 2018, 173, 989–1002.e13. [Google Scholar] [CrossRef]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef]
- Liu, J.; Gao, C.; Chen, W.; Ma, W.; Li, X.; Shi, Y.; Zhang, H.; Zhang, L.; Long, Y.; Xu, H.; et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: Mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl. Psychiatry 2016, 6, e703. [Google Scholar] [CrossRef]
- Paquet, D.; Kwart, D.; Chen, A.; Sproul, A.; Jacob, S.; Teo, S.; Olsen, K.M.; Gregg, A.; Noggle, S.; Tessier-Lavigne, M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 2016, 533, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Lubbe, S.; Morris, H. Recent advances in Parkinson’s disease genetics. J. Neurol. 2013, 261, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Soldner, F.; Stelzer, Y.; Shivalila, C.S.; Abraham, B.J.; Latourelle, J.C.; Barrasa, M.I.; Goldmann, J.; Myers, R.H.; Young, R.A.; Jaenisch, R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 2016, 533, 95–99. [Google Scholar] [CrossRef]
- Cai, S.W.; Zhang, Y.; Hou, M.Z.; Liu, Y.; Li, X.R. The research advances and applications of genome editing in hereditary eye diseases. Zhonghua Yan Ke Za Zhi 2017, 53, 386–391. [Google Scholar] [CrossRef]
- Björk, S.; Hurt, C.M.; Ho, V.K.; Angelotti, T. REEPs Are Membrane Shaping Adapter Proteins That Modulate Specific G Protein-Coupled Receptor Trafficking by Affecting ER Cargo Capacity. PLoS ONE 2013, 8, e76366. [Google Scholar] [CrossRef] [PubMed]
- Arno, G.; Agrawal, S.A.; Eblimit, A.; Bellingham, J.; Xu, M.; Wang, F.; Chakarova, C.; Parfitt, D.A.; Lane, A.; Burgoyne, T.; et al. Mutations in REEP6 Cause Autosomal-Recessive Retinitis Pigmentosa. Am. J. Hum. Genet. 2016, 99, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Bowes, C.; Li, T.; Danciger, M.; Baxter, L.C.; Applebury, M.L.; Farber, D.B. Retinal degeneration in the rd mouse is caused by a defect in the β subunit of rod cGMP-phosphodiesterase. Nature 1990, 347, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Keeler, C.E. THE GEOTROPIC REACTION OF RODLESS MICE IN LIGHT AND IN DARKNESS. J. Gen. Physiol. 1928, 11, 361–368. [Google Scholar] [CrossRef]
- Wu, W.-H.; Tsai, Y.-T.; Justus, S.; Lee, T.-T.; Zhang, L.; Lin, C.-S.; Bassuk, A.G.; Mahajan, V.B.; Tsang, S.H. CRISPR Repair Reveals Causative Mutation in a Preclinical Model of Retinitis Pigmentosa. Mol. Ther. 2016, 24, 1388–1394. [Google Scholar] [CrossRef]
- Bassuk, A.; Zheng, A.; Li, Y.; Tsang, S.H.; Mahajan, V.B. Precision Medicine: Genetic Repair of Retinitis Pigmentosa in Patient-Derived Stem Cells. Sci. Rep. 2016, 6, 19969. [Google Scholar] [CrossRef]
- Liao, C.; Zhang, D.; Mungo, C.; Tompkins, D.A.; Zeidan, A.M. Is diabetes mellitus associated with increased incidence and disease-specific mortality in endometrial cancer? A systematic review and meta-analysis of cohort studies. Gynecol. Oncol. 2014, 135, 163–171. [Google Scholar] [CrossRef]
- Zhong, H.; Chen, Y.; Li, Y.; Chen, R.; Mardon, G. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci. Rep. 2015, 5, srep08366. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-H.; Murphree, A.L.; Benedict, W.F. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature 1984, 309, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Naert, T.; Colpaert, R.; Van Nieuwenhuysen, T.; Dimitrakopoulou, D.; Leoen, J.; Haustraete, J.; Boel, A.; Steyaert, W.; Lepez, T.; Deforce, D.; et al. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci. Rep. 2016, 6, 35264. [Google Scholar] [CrossRef]
- Tu, J.; Huo, Z.; Liu, M.; Wang, D.; Xu, A.; Zhou, R.; Zhu, D.; Gingold, J.; Shen, J.; Zhao, R.; et al. Generation of human embryonic stem cell line with heterozygous RB1 deletion by CRIPSR/Cas9 nickase. Stem Cell Res. 2018, 28, 29–32. [Google Scholar] [CrossRef]
- Hollands, H.; Johnson, D.; Hollands, S.; Simel, D.L.; Jinapriya, D.; Sharma, S. Do findings on routine examination identify patients at risk for primary open-angle glaucoma? The rational clinical examination systematic review. JAMA 2013, 309, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Alward, W.L.; Fingert, J.H.; Coote, M.A.; Johnson, A.T.; Lerner, S.F.; Junqua, D.; Durcan, F.J.; McCartney, P.J.; Mackey, D.A.; Sheffield, V.C.; et al. Clinical Features Associated with Mutations in the Chromosome 1 Open-Angle Glaucoma Gene (GLC1A). N. Engl. J. Med. 1998, 338, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Stone, E.M.; Fingert, J.H.; Alward, W.L.M.; Nguyen, T.D.; Polansky, J.R.; Sunden, S.L.F.; Nishimura, D.; Clark, A.F.; Nystuen, A.; Nichols, B.E.; et al. Identification of a Gene That Causes Primary Open Angle Glaucoma. Science 1997, 275, 668–670. [Google Scholar] [CrossRef]
- Kim, B.S.; Savinova, O.V.; Reedy, M.V.; Martin, J.; Lun, Y.; Gan, L.; Smith, R.S.; Tomarev, S.I.; John, S.W.M.; Johnson, R.L. Targeted Disruption of the Myocilin Gene (Myoc) Suggests that Human Glaucoma-Causing Mutations Are Gain of Function. Mol. Cell. Biol. 2001, 21, 7707–7713. [Google Scholar] [CrossRef]
- Carbone, M.A.; Ayroles, J.F.; Yamamoto, A.; Morozova, T.V.; West, S.A.; Magwire, M.M.; Mackay, T.F.C.; Anholt, R.R.H. Overexpression of Myocilin in the Drosophila Eye Activates the Unfolded Protein Response: Implications for Glaucoma. PLoS ONE 2009, 4, e4216. [Google Scholar] [CrossRef]
- Joe, M.K.; Sohn, S.; Hur, W.; Moon, Y.; Choi, Y.R.; Kee, C. Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem. Biophys. Res. Commun. 2003, 312, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Vollrath, D. Reversal of mutant myocilin non-secretion and cell killing: Implications for glaucoma. Hum. Mol. Genet. 2004, 13, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Yam, G.H.-F.; Gaplovska-Kysela, K.; Zuber, C.; Roth, J. Aggregated Myocilin Induces Russell Bodies and Causes Apoptosis: Implications for the Pathogenesis of Myocilin-Caused Primary Open-Angle Glaucoma. Am. J. Pathol. 2007, 170, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Zode, G.; Kasetti, R.B.; Ran, F.A.; Yan, W.; Sharma, T.P.; Bugge, K.; Searby, C.C.; Fingert, J.H.; Zhang, F.; et al. CRISPR-Cas9–based treatment of myocilin-associated glaucoma. Proc. Natl. Acad. Sci. USA 2017, 114, 11199–11204. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-K.; Gu, Y.; Diaz, A.; Marlett, J.M.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.J.; et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015, 6, 6413. [Google Scholar] [CrossRef]
- Ebina, H.; Misawa, N.; Kanemura, Y.; Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013, 3, srep02510. [Google Scholar] [CrossRef]
- Ye, L.; Wang, J.; Beyer, A.I.; Teque, F.; Cradick, T.; Qi, Z.; Chang, J.C.; Bao, G.; Muench, M.; Yu, J.; et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA 2014, 111, 9591–9596. [Google Scholar] [CrossRef]
- Zhen, S.; Hua, L.; Liu, Y.-H.; Gao, L.-C.; Fu, J.; Wan, D.-Y.; Dong, L.-H.; Song, H.-F.; Gao, X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015, 22, 404–412. [Google Scholar] [CrossRef]
- Dong, C.; Qu, L.; Wang, H.; Wei, L.; Dong, Y.; Xiong, S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antivir. Res. 2015, 118, 110–117. [Google Scholar] [CrossRef]
- Ramanan, V.; Shlomai, A.; Cox, D.B.T.; Schwartz, R.E.; Michailidis, E.; Bhatta, A.; Scott, D.A.; Zhang, F.; Rice, C.M.; Bhatia, S.N. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep. 2015, 5, srep10833. [Google Scholar] [CrossRef]
- Zhou, J.; Shen, B.; Zhang, W.; Wang, J.; Yang, J.; Chen, L.; Zhang, N.; Zhu, K.; Xu, J.; Hu, B.; et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int. J. Biochem. Cell Biol. 2014, 46, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Hashikawa, Y.; Hayashi, R.; Tajima, M.; Okubo, T.; Azuma, S.; Kuwamura, M.; Takai, N.; Osada, Y.; Kunihiro, Y.; Mashimo, T.; et al. Generation of knockout rabbits with X-linked severe combined immunodeficiency (X-SCID) using CRISPR/Cas9. Sci. Rep. 2020, 10, 9957. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Yu, D.; Fu, R.; An, P.; Sun, R.; Wang, Z.; Guo, R.; Li, H.; Zhang, Y.; Li, Z.; et al. IL2RG-deficient minipigs generated via CRISPR/Cas9 technology support the growth of human melanoma-derived tumours. Cell Prolif. 2020, 53. [Google Scholar] [CrossRef] [PubMed]
- Meisel, R. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 2021, 384, e91. [Google Scholar] [CrossRef]
- Lu, Y.; Xue, J.; Deng, T.; Zhou, X.; Yu, K.; Deng, L.; Huang, M.; Yi, X.; Liang, M.; Wang, Y.; et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020, 26, 732–740. [Google Scholar] [CrossRef]
- Lacey, S.F.; Fraietta, J.A. First Trial of CRISPR-Edited T cells in Lung Cancer. Trends Mol. Med. 2020, 26, 713–715. [Google Scholar] [CrossRef]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef]
- Wu, Y.; Liang, D.; Wang, Y.; Bai, M.; Tang, W.; Bao, S.; Yan, Z.; Li, D.; Li, J. Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell Stem Cell 2013, 13, 659–662. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, H.; Fan, X.; Zhang, Y.; Zhang, M.; Wang, Y.; Xie, Z.; Bai, M.; Yin, Q.; Liang, D.; et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2014, 25, 67–79. [Google Scholar] [CrossRef]
- Schwank, G.; Koo, B.-K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef]
- Long, C.; McAnally, J.R.; Shelton, J.M.; Mireault, A.A.; Bassel-Duby, R.; Olson, E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science 2014, 345, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Antal, C.E.; Hudson, A.M.; Kang, E.; Zanca, C.; Wirth, C.; Stephenson, N.L.; Trotter, E.W.; Gallegos, L.L.; Miller, C.J.; Furnari, F.B.; et al. Cancer-Associated Protein Kinase C Mutations Reveal Kinase’s Role as Tumor Suppressor. Cell 2015, 160, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, Y.; Liu, L.; Zhuang, C.; Fu, X.; Huang, W.; Cai, Z. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun. 2014, 5, 5393. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Xiao, A.; Cheng, Z.; Kong, L.; Zhu, Z.; Lin, S.; Gao, G.; Zhang, B. CasOT: A genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 2014, 30, 1180–1182. [Google Scholar] [CrossRef]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013, 31, 839–843. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.; Esvelt, K.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-Mediated Base Editing: Technical Considerations and Practical Applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Yang, B.; Yang, L.; Chen, J. Development and Application of Base Editors. CRISPR J. 2019, 2, 91–104. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Sürün, D.; Schneider, A.; Mircetic, J.; Neumann, K.; Lansing, F.; Paszkowski-Rogacz, M.; Hänchen, V.; Lee-Kirsch, M.A.; Buchholz, F. Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors. Genes 2020, 11, 511. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.; He, S.; Huang, S.; Li, C.; Chen, Y.; Liu, Z.; Huang, X.; Wang, X. Efficient generation of mouse models with the prime editing system. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef]
- Gilbert, L.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Li, Y.; Van Nostrand, J.D.; He, Z.; Zhou, J. Cas9-Based Tools for Targeted Genome Editing and Transcriptional Control. Appl. Environ. Microbiol. 2014, 80, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, K.; Unit, P.; et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J.-L. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Rep. 2013, 4, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Ren, M.; Wang, Z.; Zhang, B.; Rong, Y.S.; Jiao, R.; Gao, G. Highly Efficient Genome Modifications Mediated by CRISPR/Cas9 in Drosophila. Genetics 2013, 195, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Zhang, Y.; Heidrich, N.; Ampattu, B.J.; Gunderson, C.W.; Seifert, H.S.; Schoen, C.; Vogel, J.; Sontheimer, E.J. Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Mol. Cell 2013, 50, 488–503. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Gasiunas, G.; Miksys, A.; Barrangou, R.; Horvath, P.; Siksnys, V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013, 10, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Qin, R.; Li, J.; Liu, X.; Xu, R.; Yang, J.; Wei, P. SpCas9-NG self-targets the sgRNA sequence in plant genome editing. Nat. Plants 2020, 6, 197–201. [Google Scholar] [CrossRef]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Liang, F.; Zhang, Y.; Li, L.; Yang, Y.; Fei, J.-F.; Liu, Y.; Qin, W. SpG and SpRY variants expand the CRISPR toolbox for genome editing in zebrafish. Nat. Commun. 2022, 13, 1–10. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014, 56, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Crudele, J.M.; Chamberlain, J.S. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 2018, 9, 1–3. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasrallah, A.; Sulpice, E.; Kobaisi, F.; Gidrol, X.; Rachidi, W. CRISPR-Cas9 Technology for the Creation of Biological Avatars Capable of Modeling and Treating Pathologies: From Discovery to the Latest Improvements. Cells 2022, 11, 3615. https://doi.org/10.3390/cells11223615
Nasrallah A, Sulpice E, Kobaisi F, Gidrol X, Rachidi W. CRISPR-Cas9 Technology for the Creation of Biological Avatars Capable of Modeling and Treating Pathologies: From Discovery to the Latest Improvements. Cells. 2022; 11(22):3615. https://doi.org/10.3390/cells11223615
Chicago/Turabian StyleNasrallah, Ali, Eric Sulpice, Farah Kobaisi, Xavier Gidrol, and Walid Rachidi. 2022. "CRISPR-Cas9 Technology for the Creation of Biological Avatars Capable of Modeling and Treating Pathologies: From Discovery to the Latest Improvements" Cells 11, no. 22: 3615. https://doi.org/10.3390/cells11223615
APA StyleNasrallah, A., Sulpice, E., Kobaisi, F., Gidrol, X., & Rachidi, W. (2022). CRISPR-Cas9 Technology for the Creation of Biological Avatars Capable of Modeling and Treating Pathologies: From Discovery to the Latest Improvements. Cells, 11(22), 3615. https://doi.org/10.3390/cells11223615






