Lycopene Scavenges Cellular ROS, Modulates Autophagy and Improves Survival through 7SK snRNA Interaction in Smooth Muscle Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Design of the Study
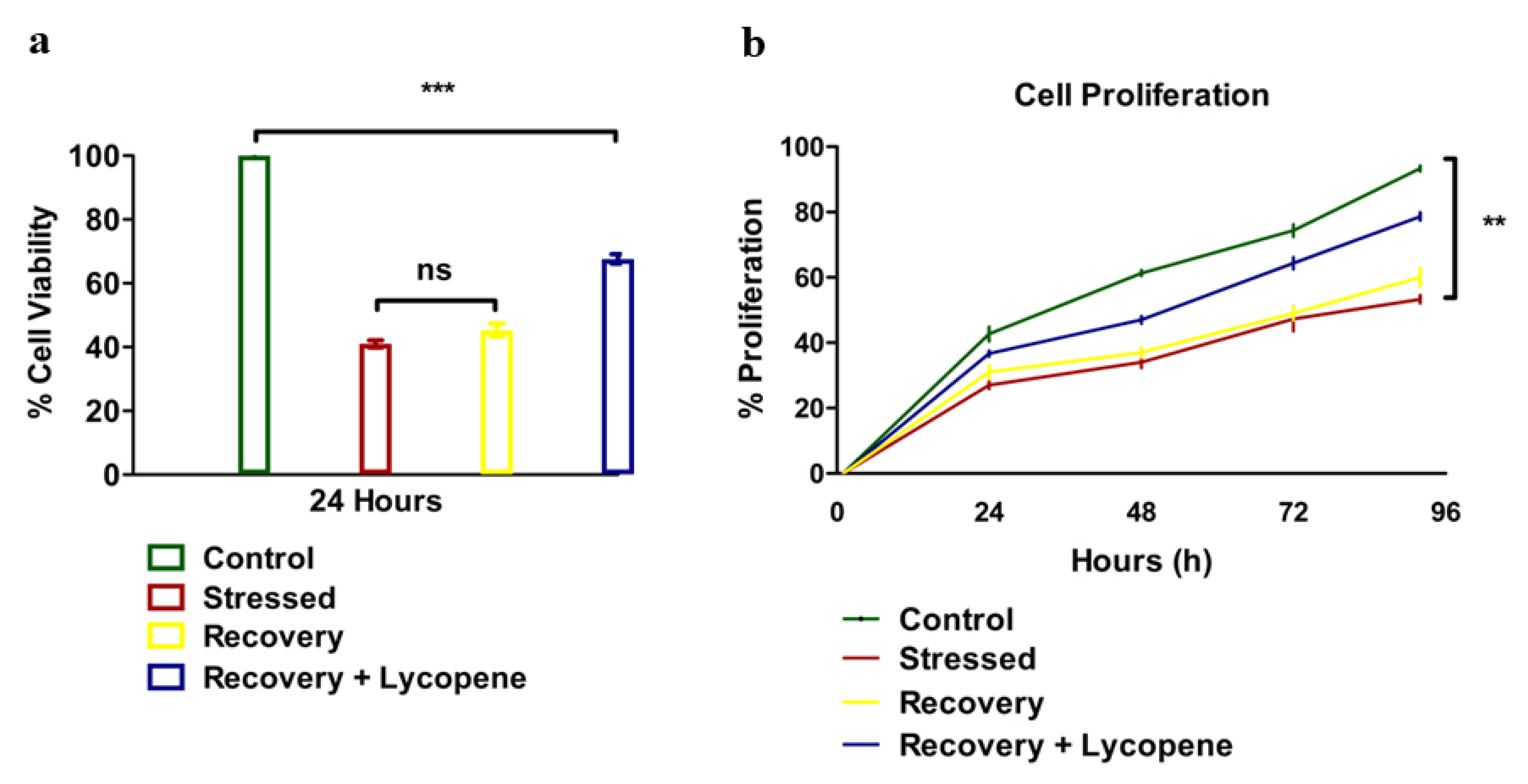
2.2. Viability and Cell Proliferation
2.3. PCR for Autophagic Markers, TNF-α and GPx
2.4. Cytosolic ROS Determination
2.5. Transmission Electron Microscopy (TEM)
2.6. Non-Coding RNA Interaction with Lycopene
2.7. Statistics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Cell Biology of Ischemia/Reperfusion Injury. Int. Rev. Cell Mol. Biol. 2012, 298, 229–317. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Vassiliadis, E.; Barascuk, N.; Karsdal, M.A. Atherofibrosis—A unique and common process of the disease pathogenesis of athero-sclerosis and fibrosis—Lessons for biomarker development. Am. J. Transl. Res. 2013, 5, 1–14. [Google Scholar] [PubMed]
- Miguel, V.; Lamas, S.; Espinosa-Diez, C. Role of non-coding-RNAs in response to environmental stressors and consequences on human health. Redox Biol. 2020, 37, 101580. [Google Scholar] [CrossRef]
- Osonoi, Y.; Mita, T.; Azuma, K.; Nakajima, K.; Masuyama, A.; Goto, H.; Nishida, Y.; Miyatsuka, T.; Fujitani, Y.; Koike, M.; et al. Defective autophagy in vascular smooth muscle cells enhances cell death and atherosclerosis. Autophagy 2018, 14, 1991–2006. [Google Scholar] [CrossRef]
- Klionsky, D.J. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007, 8, 931–937. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, C.; Wang, X.; Sun, Y.; Zhang, J.; Chen, J.; Shi, Y. An Epigenetic Role of Mitochondria in Cancer. Cells 2022, 11, 2518. [Google Scholar] [CrossRef]
- Shankman, L.S.; Gomez, D.; Cherepanova, O.A.; Salmon, M.; Alencar, G.F.; Haskins, R.M.; Swiatlowska, P.; Newman, A.A.C.; Greene, E.S.; Straub, S.C.; et al. KLF4-dependent phenotypic modu-lation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 2015, 21, 628–637. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2017, 34, 142–157. [Google Scholar] [CrossRef]
- Irrinki, K.M.; Mallilankaraman, K.; Thapa, R.J.; Chandramoorthy, H.C.; Smith, F.J.; Jog, N.R.; Gandhirajan, R.; Kelsen, S.G.; Houser, S.R.; May, M.; et al. Requirement of FADD, NEMO, and BAX/BAK for Aberrant Mitochondrial Function in Tumor Necrosis Factor Alpha-Induced Necrosis. Mol. Cell. Biol. 2011, 31, 3745–3758. [Google Scholar] [CrossRef] [PubMed]
- Mallilankaraman, K.; Doonan, P.; Cárdenas, C.; Chandramoorthy, H.C.; Müller, M.; Miller, R.; Hoffman, N.E.; Gandhirajan, R.K.; Molgó, J.; Birnbaum, M.J.; et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell 2012, 151, 630–644. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, J.; Beeraka, N.M.; Tang, C.; Babayeva, Y.V.; Sinelnikov, M.Y.; Zhang, X.; Zhang, J.; Liu, J.; Reshetov, I.V.; et al. Advances in the Prevention and Treatment of Obesity-Driven Effects in Breast Cancers. Front. Oncol. 2022, 12, 2663. [Google Scholar] [CrossRef]
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M.; et al. Corrigendum to Mitochondrial mutations and mitoepigenetics: Focus on regulation of oxidative stress-induced responses in breast cancers. Semin. Cancer Biol. 2022, 83, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Mallilankaraman, K.; Cárdenas, C.; Doonan, P.J.; Chandramoorthy, H.C.; Irrinki, K.M.; Golenár, T.; Csordás, G.; Madireddi, P.; Yang, J.; Müller, M.; et al. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat. Cell Biol. 2012, 14, 1336–1343. [Google Scholar] [CrossRef]
- Imran, M.; Ghorat, F.; Ul-Haq, I.; Ur-Rehman, H.; Aslam, F.; Heydari, M.; Shariati, M.A.; Okuskhanova, E.; Yessimbekov, Z.; Thiruvengadam, M.; et al. Lycopene as a Natural Antioxidant Used to Prevent Human Health Disorders. Antioxidants 2020, 9, 706. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Liu, H.; Wang, J.; Liu, P.; Tan, X.; Ren, B.; Liu, Z.; Liu, X. Lycopene Supplementation Attenuates Oxidative Stress, Neuroinflammation, and Cognitive Impairment in Aged CD-1 Mice. J. Agric. Food Chem. 2018, 66, 3127–3136. [Google Scholar] [CrossRef]
- Li, C.-W.; Li, L.-L.; Chen, S.; Zhang, J.-X.; Lu, W.-L. Antioxidant Nanotherapies for the Treatment of Inflammatory Diseases. Front. Bioeng. Biotechnol. 2020, 8, 200. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Stratos, I.; Li, Z.; Herlyn, P.; Rotter, R.; Behrendt, A.-K.; Mittlmeier, T.; Vollmar, B. Vitamin D Increases Cellular Turnover and Functionally Restores the Skeletal Muscle after Crush Injury in Rats. Am. J. Pathol. 2012, 182, 895–904. [Google Scholar] [CrossRef]
- Cheng, H.M.; Koutsidis, G.; Lodge, J.K.; Ashor, A.; Siervo, M.; Lara, J. Tomato and lycopene supplementation and cardiovascular risk factors: A systematic review and meta-analysis. Atherosclerosis 2017, 257, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Uçar, S.; Pandir, D. Furan induced ovarian damage in non-diabetic and diabetic rats and cellular protective role of lycopene. Arch. Gynecol. Obstet. 2017, 296, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Morgia, G.; Voce, S.; Palmieri, F.; Gentile, M.; Iapicca, G.; Giannantoni, A.; Blefari, F.; Carini, M.; Vespasiani, G.; Santelli, G.; et al. Association between selenium and lycopene sup-plementation and incidence of prostate cancer: Results from the post-hoc analysis of the procomb trial. Phytomedicine 2017, 34, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bánhegyi, G. Lycopene—A natural antioxidant. Orvosi Hetil. 2005, 146, 1621–1624. [Google Scholar]
- El-Ashmawy, N.E.; Khedr, N.F.; El-Bahrawy, H.A.; Hamada, O.B. Suppression of inducible nitric oxide synthase and tumor necrosis factor-alpha level by lycopene is comparable to methylprednisolone in acute pancreatitis. Dig. Liver Dis. 2018, 50, 601–607. [Google Scholar] [CrossRef]
- Liu, C.B.; Wang, R.; Yi, Y.F.; Gao, Z.; Chen, Y.Z. Lycopene mitigates beta-amyloid induced inflammatory response and inhibits NF-kappaB signaling at the choroid plexus in early stages of Alzheimer’s disease rats. J. Nutr. Biochem. 2018, 53, 66–71. [Google Scholar] [CrossRef]
- Alhazzani, A.; Rajagopalan, P.; Albarqi, Z.; Devaraj, A.; Mohamed, M.H.; Al-Hakami, A.; Chandramoorthy, H.C. Mesenchymal Stem Cells (MSCs) Coculture Protects [Ca2+]i Orchestrated Oxidant Mediated Damage in Differentiated Neurons In Vitro. Cells 2018, 7, 250. [Google Scholar] [CrossRef]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3 B 1–A3 B 3. [Google Scholar] [CrossRef]
- Prasanna, R.; Harish, C.C. Anticancer effect of a novel 2-arylidene-4,7-dimethyl indan-1-one against human breast adenocar-cinoma cell line by G2/M cell cycle arrest. Oncol. Res. 2010, 18, 461–468. [Google Scholar] [CrossRef]
- Rezazadeh, D.; Norooznezhad, A.H.; Mansouri, K.; Jahani, M.; Mostafaie, A.; Mohammadi, M.H.; Modarressi, M.H. Rapamycin Reduces Cervical Cancer Cells Viability in Hypoxic Condition: Investigation of the Role of Autophagy and Apoptosis. OncoTargets Ther. 2020, ume 13, 4239–4247. [Google Scholar] [CrossRef]
- Al-Hakami, A.; Alqhatani, S.Q.; Shaik, S.; Jalfan, S.M.; Abu Dhammam, M.S.; Asiri, W.; Alkahtani, A.M.; Devaraj, A.; Chandramoorthy, H.C. Cytokine physiognomies of MSCs from varied sources confirm the regenerative commitment post-coculture with activated neutrophils. J. Cell. Physiol. 2020, 235, 8691–8701. [Google Scholar] [CrossRef] [PubMed]
- Eid, R.A.; Bin-Meferij, M.M.; El-Kott, A.F.; Eleawa, S.M.; Zaki, M.S.A.; Al-Shraim, M.; El-Sayed, F.; Eldeen, M.A.; Alkhateeb, M.A.; Alharbi, S.A.; et al. Exendin-4 Protects Against Myocardial Is-chemia-Reperfusion Injury by Upregulation of SIRT1 and SIRT3 and Activation of AMPK. J. Cardiovasc. Transl. Res. 2021, 14, 619–635. [Google Scholar] [CrossRef] [PubMed]
- Mozos, I.; Stoian, D.; Caraba, A.; Malainer, C.; Horbańczuk, J.O.; Atanasov, A.G. Lycopene and Vascular Health. Front. Pharmacol. 2018, 9, 521. [Google Scholar] [CrossRef]
- Szydłowska, I.; Nawrocka-Rutkowska, J.; Brodowska, A.; Marciniak, A.; Starczewski, A.; Szczuko, M. Dietary Natural Compounds and Vitamins as Potential Cofactors in Uterine Fibroids Growth and Development. Nutrients 2022, 14, 734. [Google Scholar] [CrossRef] [PubMed]
- Castelo-Branco, G.; Amaral, P.P.; Engström, P.G.; Robson, S.C.; Marques, S.C.; Bertone, P.; Kouzarides, T. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013, 14, R98. [Google Scholar] [CrossRef]
- Guiro, J.; Murphy, S. Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, H. The Remedial Potential of Lycopene in Pancreatitis through Regulation of Autophagy. Int. J. Mol. Sci. 2020, 21, 5775. [Google Scholar] [CrossRef]
- Gottlieb, R.A.; Mentzer, R.M. Autophagy During Cardiac Stress: Joys and Frustrations of Autophagy. Annu. Rev. Physiol. 2010, 72, 45–59. [Google Scholar] [CrossRef]
- Ornatowski, W.; Lu, Q.; Yegambaram, M.; Garcia, A.E.; Zemskov, E.A.; Maltepe, E.; Fineman, J.R.; Wang, T.; Black, S.M. Complex interplay between autophagy and oxidative stress in the development of pulmonary disease. Redox Biol. 2020, 36, 101679. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, R.; Zhang, B.; Hu, C.; Zhang, H. Protective effects of lycopene on oxidative stress, proliferation and autophagy in iron supplementation rats. Biol. Res. 2013, 46, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Xiang, Y.; Fujinaga, K.; Bartholomeeusen, K.; Nilson, K.A.; Price, D.H.; Peterlin, B.M. Release of Positive Transcription Elongation Factor b (P-TEFb) from 7SK Small Nuclear Ribonucleoprotein (snRNP) Activates Hexamethylene Bisacetamide-inducible Protein (HEXIM1) Transcription. J. Biol. Chem. 2014, 289, 9918–9925. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Elner, S.G.; Bian, Z.-M.; Till, G.O.; Petty, H.R.; Elner, V.M. Pro-inflammatory cytokines increase reactive oxygen species through mitochondria and NADPH oxidase in cultured RPE cells. Exp. Eye Res. 2007, 85, 462–472. [Google Scholar] [CrossRef]
- Snezhkina, A.V.; Kudryavtseva, A.V.; Kardymon, O.L.; Savvateeva, M.V.; Melnikova, N.V.; Krasnov, G.S.; Dmitriev, A.A. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. Oxidative Med. Cell. Longev. 2019, 2019, 6175804. [Google Scholar] [CrossRef] [PubMed]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive Oxygen Species in Inflammation and Tissue Injury. Antioxid. Redox Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef]
- Golbidi, S.; Li, H.; Laher, I. Oxidative Stress: A Unifying Mechanism for Cell Damage Induced by Noise, (Water-Pipe) Smoking, and Emotional Stress—Therapeutic Strategies Targeting Redox Imbalance. Antioxid. Redox Signal. 2018, 28, 741–759. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular Stress Responses: Cell Survival and Cell Death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef]
- Martindale, J.L.; Holbrook, N.J. Cellular response to oxidative stress: Signaling for suicide and survival. J. Cell. Physiol. 2002, 192, 1–15. [Google Scholar] [CrossRef]
- Kugel, J.F.; Goodrich, J.A. Non-coding RNAs: Key regulators of mammalian transcription. Trends Biochem. Sci. 2012, 37, 144–151. [Google Scholar] [CrossRef]
- Leeper, N.J.; Maegdefessel, L. Non-coding RNAs: Key regulators of smooth muscle cell fate in vascular disease. Cardiovasc. Res. 2017, 114, 611–621. [Google Scholar] [CrossRef]
- Brozovich, F.V.; Nicholson, C.J.; Degen, C.V.; Gao, Y.Z.; Aggarwal, M.; Morgan, K.G. Mechanisms of Vascular Smooth Muscle Con-traction and the Basis for Pharmacologic Treatment of Smooth Muscle Disorders. Pharmacol Rev. 2016, 68, 476–532. [Google Scholar] [CrossRef] [PubMed]
- Arballo, J.; Amengual, J.; Erdman, J.W., Jr. Lycopene: A Critical Review of Digestion, Absorption, Metabolism, and Excretion. Antioxidants 2021, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Lekka, E.; Hall, J. Noncoding RNA s in disease. FEBS Lett. 2018, 592, 2884–2900. [Google Scholar] [CrossRef] [PubMed]




| Target Gene | Primer Sequence F- Forward, R-Reverse | Amplicon Size (bp) |
|---|---|---|
| LC3A | F: 5ʹ-CGTCCTGGACAAGACCAAGT-3′ R: 5ʹ-CTCGTCTTTCTCCTGCTCGT-3′ | 181 |
| LC3B | F: 5ʹ-AGCAGCATCCAACCAAAATC-3′ R: 5ʹ-CTGTGTCCGTTCACCAACAG-3′ | 187 |
| β-Actin | F: 5ʹCAACTGGGACGACATGGAGAAAAT-3′ R: 5ʹ-CCAGAGGCGTACAGGGATAGCAC-3′ | 207 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shati, A.A.; Eid, R.A.; Zaki, M.S.A.; Alqahtani, Y.A.; Al-Qahtani, S.M.; Chandramoorthy, H.C. Lycopene Scavenges Cellular ROS, Modulates Autophagy and Improves Survival through 7SK snRNA Interaction in Smooth Muscle Cells. Cells 2022, 11, 3617. https://doi.org/10.3390/cells11223617
Shati AA, Eid RA, Zaki MSA, Alqahtani YA, Al-Qahtani SM, Chandramoorthy HC. Lycopene Scavenges Cellular ROS, Modulates Autophagy and Improves Survival through 7SK snRNA Interaction in Smooth Muscle Cells. Cells. 2022; 11(22):3617. https://doi.org/10.3390/cells11223617
Chicago/Turabian StyleShati, Ayed A., Refaat A. Eid, Mohamed Samir A. Zaki, Youssef A. Alqahtani, Saleh M. Al-Qahtani, and Harish C. Chandramoorthy. 2022. "Lycopene Scavenges Cellular ROS, Modulates Autophagy and Improves Survival through 7SK snRNA Interaction in Smooth Muscle Cells" Cells 11, no. 22: 3617. https://doi.org/10.3390/cells11223617
APA StyleShati, A. A., Eid, R. A., Zaki, M. S. A., Alqahtani, Y. A., Al-Qahtani, S. M., & Chandramoorthy, H. C. (2022). Lycopene Scavenges Cellular ROS, Modulates Autophagy and Improves Survival through 7SK snRNA Interaction in Smooth Muscle Cells. Cells, 11(22), 3617. https://doi.org/10.3390/cells11223617






