Human 3D Airway Tissue Models for Real-Time Microscopy: Visualizing Respiratory Virus Spreading
Abstract
1. Introduction
2. Materials and Methods
2.1. Primary Cells and Tissue
2.2. Cell Culture
2.3. 3D Tissue Engineering
2.4. Histology and Immunofluorescence Staining
2.5. Determination of Total Cell Numbers of 3D Airway Tissue Models
2.6. Influenza Virus Strains and Infection
2.7. Virus Quantification
2.8. Microscopy, Real-Time Fluorescence Imaging, and Image Processing
2.9. Determination of Epithelial Layer Thickness
3. Results
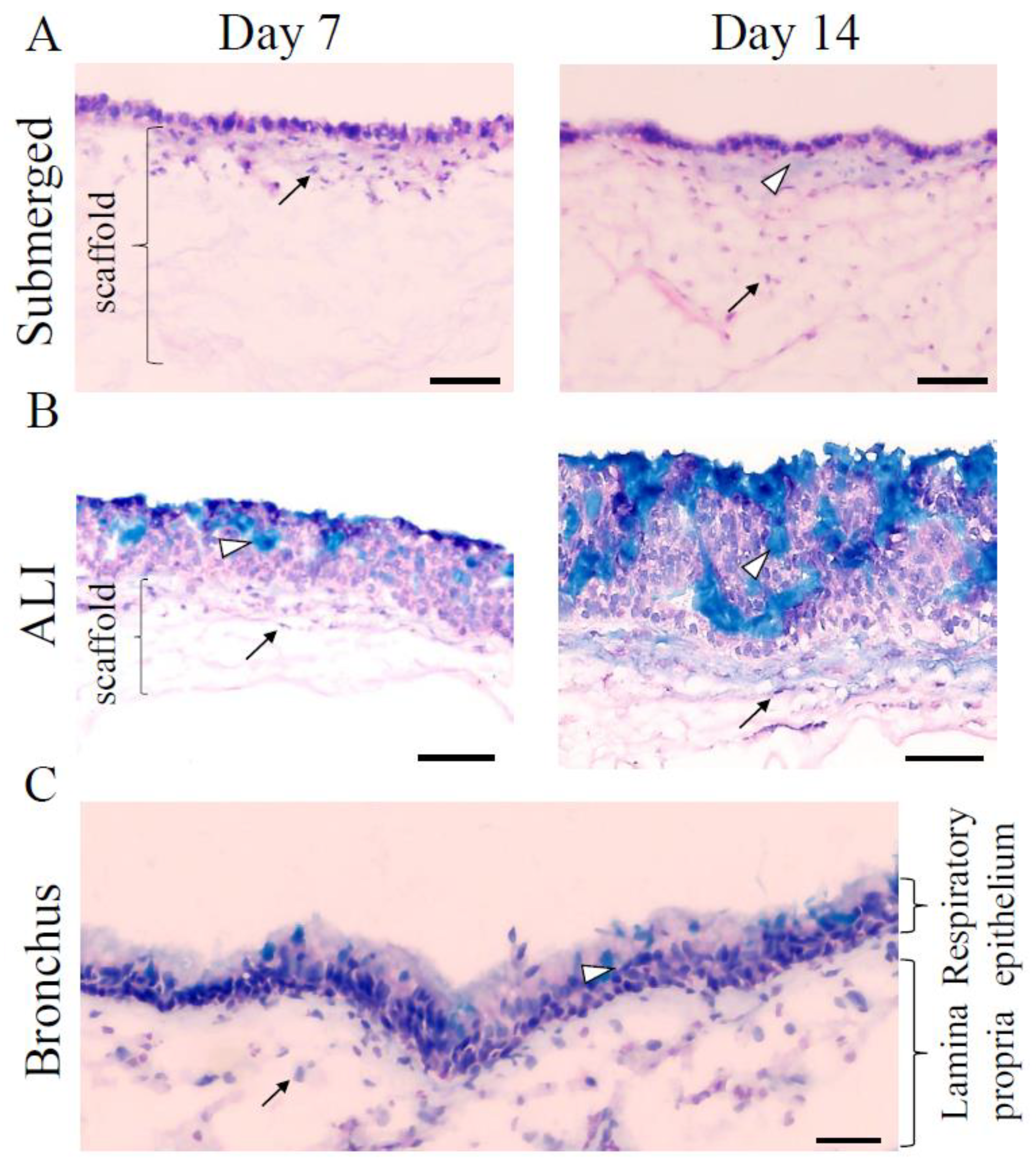
3.1. Human 3D Airway Tissue Models Consisting of Calu-3 Cells and Fibroblasts Resemble Bronchial Epithelial Tissue
3.2. NS1-RFP IAV Replication Is Similar to A/PR/8/34 Wild-Type Virus Strain Replication
3.3. MOI 0.01 and the Presence of FBS Are Suitable to Infect Calu-3 Cells with the NS1-RFP IAV
3.4. Real-Time Fluorescence Imaging of NS1-RFP IAV Infection in 2D Calu-3 Cell Cultures
3.5. Real-Time Fluorescence Imaging of Infection in Submerged 3D Airway Models Allows Monitoring of Infection Progression
3.6. Real-Time Fluorescence Imaging of Infection in ALI Airway Models Showed a Slower Progression of Infection

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manicassamy, B.; Manicassamy, S.; Belicha-Villanueva, A.; Pisanelli, G.; Pulendran, B.; Garcia-Sastre, A. Analysis of in Vivo Dynamics of Influenza Virus Infection in Mice Using a GFP Reporter Virus. Proc. Natl. Acad. Sci. USA 2010, 107, 11531–11536. [Google Scholar] [CrossRef] [PubMed]
- Bottcher-Friebertshauser, E.; Stein, D.A.; Klenk, H.-D.; Garten, W. Inhibition of Influenza Virus Infection in Human Airway Cell Cultures by an Antisense Peptide-Conjugated Morpholino Oligomer Targeting the Hemagglutinin-Activating Protease TMPRSS2. J. Virol. 2011, 85, 1554–1562. [Google Scholar] [CrossRef] [PubMed]
- Zscheppang, K.; Berg, J.; Hedtrich, S.; Verheyen, L.; Wagner, D.E.; Suttorp, N.; Hippenstiel, S.; Hocke, A.C. Human Pulmonary 3D Models For Translational Research. Biotechnol. J. 2018, 13, 1700341. [Google Scholar] [CrossRef] [PubMed]
- Pfenninger, C.; Leinhase, I.; Endres, M.; Rotter, N.; Loch, A.; Ringe, J.; Sittinger, M. Tracheal Remodeling: Comparison of Different Composite Cultures Consisting of Human Respiratory Epithelial Cells and Human Chondrocytes. Vitro Cell. Dev. Biol.-Anim. 2007, 43, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.-Y.; Chu, H.; Poon, V.K.-M.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated Human Airway Organoids to Assess Infectivity of Emerging Influenza Virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Gao, J.; Garcia, I.M.; Chen, H.J.; Castaldi, A.; Chen, Y. Human Pluripotent Stem Cell-derived Lung Organoids: Potential Applications in Development and Disease Modeling. WIREs Dev. Biol. 2021, 10, e399. [Google Scholar] [CrossRef]
- Kreft, M.E.; Jerman, U.D.; Lasič, E.; Hevir-Kene, N.; Rižner, T.L.; Peternel, L.; Kristan, K. The Characterization of the Human Cell Line Calu-3 under Different Culture Conditions and Its Use as an Optimized in Vitro Model to Investigate Bronchial Epithelial Function. Eur. J. Pharm. Sci. 2015, 69, 1–9. [Google Scholar] [CrossRef]
- Grantham, M.L.; Wu, W.-H.; Lalime, E.N.; Lorenzo, M.E.; Klein, S.L.; Pekosz, A. Palmitoylation of the Influenza A Virus M2 Protein Is Not Required for Virus Replication In Vitro but Contributes to Virus Virulence. J. Virol. 2009, 83, 8655–8661. [Google Scholar] [CrossRef]
- Belser, J.A.; Maines, T.R.; Creager, H.M.; Katz, J.M.; Tumpey, T.M. Oseltamivir Inhibits Influenza Virus Replication and Transmission Following Ocular-Only Aerosol Inoculation of Ferrets. Virology 2015, 484, 305–312. [Google Scholar] [CrossRef]
- Fiegel, J.; Ehrhardt, C.; Schaefer, U.F.; Lehr, C.-M.; Hanes, J. Large Porous Particle Impingement on Lung Epithelial Cell Monolayers--toward Improved Particle Characterization in the Lung. Pharm. Res. 2003, 20, 788–796. [Google Scholar] [CrossRef]
- Forbes, B.; Ehrhardt, C. Human Respiratory Epithelial Cell Culture for Drug Delivery Applications. Eur. J. Pharm. Biopharm. 2005, 60, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.E.; Torr, E.E.; Mohd Jamili, N.H.; Bosquillon, C.; Sayers, I. Evaluation of Differentiated Human Bronchial Epithelial Cell Culture Systems for Asthma Research. J. Allergy 2012, 2012, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zanin, M.; Baviskar, P.; Webster, R.; Webby, R. The Interaction between Respiratory Pathogens and Mucus. Cell Host Microbe 2016, 19, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Zmora, P.; Pöhlmann, S. Microscopy as a Useful Tool to Study the Proteolytic Activation of Influenza Viruses. In Microscopy: Advances in Scientific Research and Education; Mendez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2014; pp. 725–731. [Google Scholar]
- Vester, D.; Rapp, E.; Gade, D.; Genzel, Y.; Reichl, U. Quantitative Analysis of Cellular Proteome Alterations in Human Influenza A Virus-Infected Mammalian Cell Lines. PROTEOMICS 2009, 9, 3316–3327. [Google Scholar] [CrossRef]
- Ramos, J.R.C.; Bissinger, T.; Genzel, Y.; Reichl, U. Impact of Influenza A Virus Infection on Growth and Metabolism of Suspension MDCK Cells Using a Dynamic Model. Metabolites 2022, 12, 239. [Google Scholar] [CrossRef]
- Shen, L.-W.; Qian, M.-Q.; Yu, K.; Narva, S.; Yu, F.; Wu, Y.-L.; Zhang, W. Inhibition of Influenza A Virus Propagation by Benzoselenoxanthenes Stabilizing TMPRSS2 Gene G-Quadruplex and Hence down-Regulating TMPRSS2 Expression. Sci. Rep. 2020, 10, 7635. [Google Scholar] [CrossRef]
- Schweinlin, M.; Rossi, A.; Lodes, N.; Lotz, C.; Hackenberg, S.; Steinke, M.; Walles, H.; Groeber, F. Human Barrier Models for the in Vitro Assessment of Drug Delivery. Drug Deliv. Transl. Res. 2017, 7, 217–227. [Google Scholar] [CrossRef]
- Kalbfuss, B.; Knöchlein, A.; Kröber, T.; Reichl, U. Monitoring Influenza Virus Content in Vaccine Production: Precise Assays for the Quantitation of Hemagglutination and Neuraminidase Activity. Biologicals 2008, 36, 145–161. [Google Scholar] [CrossRef]
- Isken, B.; Genzel, Y.; Reichl, U. Productivity, Apoptosis, and Infection Dynamics of Influenza A/PR/8 Strains and A/PR/8-Based Reassortants. Vaccine 2012, 30, 5253–5261. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Killington, R.A. Virus Isolation and Quantitation. In Virology Methods Manual; Academic Press: Cambridge, MA, USA, 1996; pp. 25–46. ISBN 978-0-12-465330-6. [Google Scholar]
- Kupke, S.Y.; Riedel, D.; Frensing, T.; Zmora, P.; Reichl, U. A Novel Type of Influenza A Virus-Derived Defective Interfering Particle with Nucleotide Substitutions in Its Genome. J. Virol. 2018, 93, e01786-18. [Google Scholar] [CrossRef]
- Frensing, T.; Pflugmacher, A.; Bachmann, M.; Peschel, B.; Reichl, U. Impact of Defective Interfering Particles on Virus Replication and Antiviral Host Response in Cell Culture-Based Influenza Vaccine Production. Appl. Microbiol. Biotechnol. 2014, 98, 8999–9008. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Guzman, D.; Boland, S.; Brookes, O.; Mc Cord, C.; Lai Kuen, R.; Sirri, V.; Baeza Squiban, A.; Devineau, S. Long-Term Evolution of the Epithelial Cell Secretome in Preclinical 3D Models of the Human Bronchial Epithelium. Sci. Rep. 2021, 11, 6621. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.S.; Chertow, D.S.; Moyer, J.E.; Suzich, J.; Sandouk, A.; Dorward, D.W.; Logun, C.; Shelhamer, J.H.; Taubenberger, J.K. Validation of Normal Human Bronchial Epithelial Cells as a Model for Influenza A Infections in Human Distal Trachea. J. Histochem. Cytochem. 2015, 63, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Grainger, C.I.; Greenwell, L.L.; Lockley, D.J.; Martin, G.P.; Forbes, B. Culture of Calu-3 Cells at the Air Interface Provides a Representative Model of the Airway Epithelial Barrier. Pharm. Res. 2006, 23, 1482–1490. [Google Scholar] [CrossRef]
- Pasman, T.; Baptista, D.; van Riet, S.; Truckenmüller, R.K.; Hiemstra, P.S.; Rottier, R.J.; Hamelmann, N.M.; Paulusse, J.M.J.; Stamatialis, D.; Poot, A.A. Development of an In Vitro Airway Epithelial–Endothelial Cell Culture Model on a Flexible Porous Poly(Trimethylene Carbonate) Membrane Based on Calu-3 Airway Epithelial Cells and Lung Microvascular Endothelial Cells. Membranes 2021, 11, 197. [Google Scholar] [CrossRef]
- O’Leary, C.; Cavanagh, B.; Unger, R.E.; Kirkpatrick, C.J.; O’Dea, S.; O’Brien, F.J.; Cryan, S.-A. The Development of a Tissue-Engineered Tracheobronchial Epithelial Model Using a Bilayered Collagen-Hyaluronate Scaffold. Biomaterials 2016, 85, 111–127. [Google Scholar] [CrossRef]
- Denney, L.; Ho, L.-P. The Role of Respiratory Epithelium in Host Defence against Influenza Virus Infection. Biomed. J. 2018, 41, 218–233. [Google Scholar] [CrossRef]
- Shen, B.Q.; Finkbeiner, W.E.; Wine, J.J.; Mrsny, R.J.; Widdicombe, J.H. Calu-3: A Human Airway Epithelial Cell Line That Shows CAMP-Dependent Cl- Secretion. Am. J. Physiol.-Lung Cell. Mol. Physiol. 1994, 266, L493–L501. [Google Scholar] [CrossRef]
- Lange, J.; Weil, F.; Riegler, C.; Groeber, F.; Rebhan, S.; Kurdyn, S.; Alb, M.; Kneitz, H.; Gelbrich, G.; Walles, H.; et al. Interactions of Donor Sources and Media Influence the Histo-Morphological Quality of Full-Thickness Skin Models. Biotechnol. J. 2016, 11, 1352–1361. [Google Scholar] [CrossRef]
- Salgueiro, L.; Kummer, S.; Sonntag-Buck, V.; Weiß, A.; Schneider, M.A.; Kräusslich, H.-G.; Sotillo, R. Generation of Human Lung Organoid Cultures from Healthy and Tumor Tissue to Study Infectious Diseases. J. Virol. 2022, 96, e00098-22. [Google Scholar] [CrossRef]
- Gallagher, M.; Brooke, C.; Ke, R.; Koelle, K. Causes and Consequences of Spatial Within-Host Viral Spread. Viruses 2018, 10, 627. [Google Scholar] [CrossRef] [PubMed]
- Lodes, N.; Seidensticker, K.; Perniss, A.; Nietzer, S.; Oberwinkler, H.; May, T.; Walles, T.; Hebestreit, H.; Hackenberg, S.; Steinke, M. Investigation on Ciliary Functionality of Different Airway Epithelial Cell Lines in Three-Dimensional Cell Culture. Tissue Eng. Part A 2020, 26, 432–440. [Google Scholar] [CrossRef] [PubMed]





| Hours Post Infection (hpi) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Age of Model (Days) | MOI | N | 4 | 9 | 12 | 24 | 28 | 37 | 49 |
| 13 | 0.01 | 1 | n.p. | n.p. | n.p. | n.p. | ++ (27 hpi) | +++ | n.p. |
| 13 | 1 | 1 | n.p. | n.p. | n.p. | ++ | n.p. | n.p. | n.p. |
| 14 | 0.01 | 2 | - | + | + | + | + | + | + (43 hpi) |
| 14 | 0.01 | 1 | n.p. | n.p. | n.p. | n.p. | n.p. | n.p. | +++ (48 hpi) |
| 18 | 0.01 | 2 | - | - | + | + | + | ++ | +++ (49.75 hpi) |
| 21 | 0.01 | 2 | - | - | + (14 hpi) | n.p. | n.p. | n.p. | ++ |
| 24 | 0.01 | 1 | - | - | + (11.5 hpi) | + (22 hpi) | ++ (32 hpi) | n.p. | n.p. |
| 24 | 1 | 1 | n.p. | n.p. | n.p. | + (22 hpi) | ++ (32 hpi) | ++ | +++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Möckel, M.; Baldok, N.; Walles, T.; Hartig, R.; Müller, A.J.; Reichl, U.; Genzel, Y.; Walles, H.; Wiese-Rischke, C. Human 3D Airway Tissue Models for Real-Time Microscopy: Visualizing Respiratory Virus Spreading. Cells 2022, 11, 3634. https://doi.org/10.3390/cells11223634
Möckel M, Baldok N, Walles T, Hartig R, Müller AJ, Reichl U, Genzel Y, Walles H, Wiese-Rischke C. Human 3D Airway Tissue Models for Real-Time Microscopy: Visualizing Respiratory Virus Spreading. Cells. 2022; 11(22):3634. https://doi.org/10.3390/cells11223634
Chicago/Turabian StyleMöckel, Marion, Nino Baldok, Thorsten Walles, Roland Hartig, Andreas J. Müller, Udo Reichl, Yvonne Genzel, Heike Walles, and Cornelia Wiese-Rischke. 2022. "Human 3D Airway Tissue Models for Real-Time Microscopy: Visualizing Respiratory Virus Spreading" Cells 11, no. 22: 3634. https://doi.org/10.3390/cells11223634
APA StyleMöckel, M., Baldok, N., Walles, T., Hartig, R., Müller, A. J., Reichl, U., Genzel, Y., Walles, H., & Wiese-Rischke, C. (2022). Human 3D Airway Tissue Models for Real-Time Microscopy: Visualizing Respiratory Virus Spreading. Cells, 11(22), 3634. https://doi.org/10.3390/cells11223634








