JMJD1C Regulates Megakaryopoiesis in In Vitro Models through the Actin Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents, Human Cell Lines, and Primary Samples
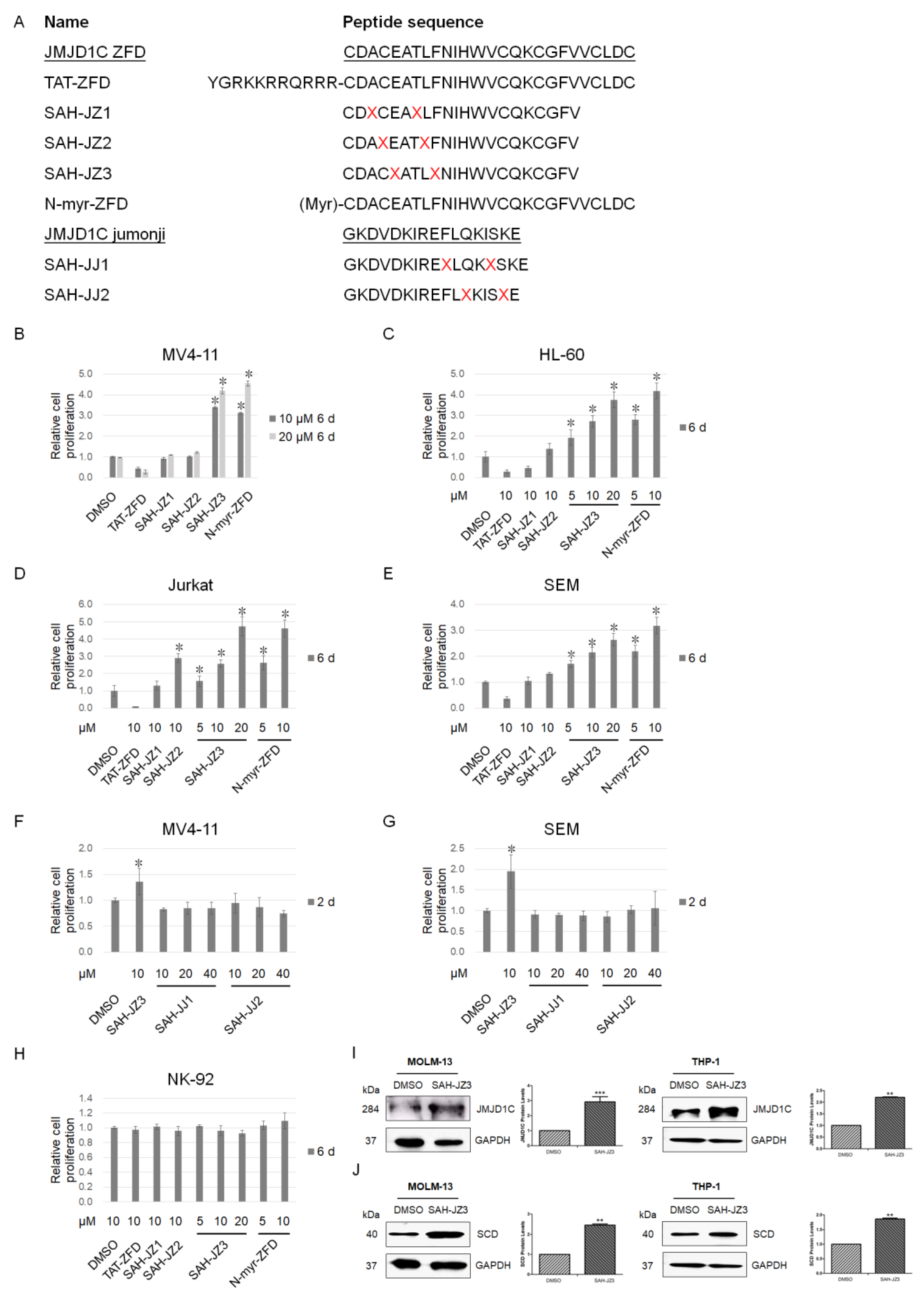
2.2. Design of the Stabilized Alpha Helix of the JMJD1C Peptides
2.3. Peptide Incubation
2.4. Transfection
2.5. Quantitative PCR (qPCR)
2.6. Western Blotting
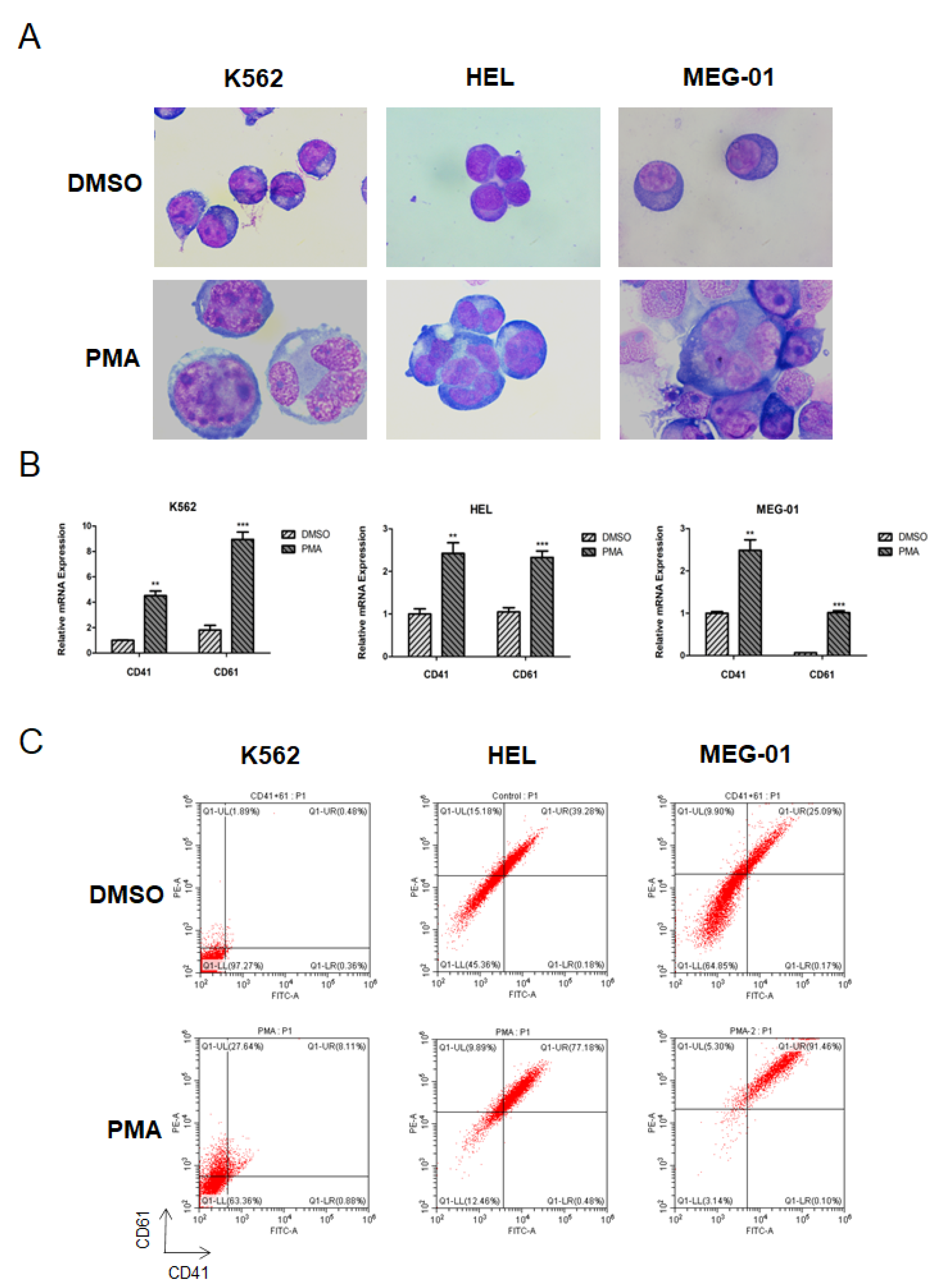
2.7. Wright–Giemsa Staining
2.8. Flow Cytometry
2.9. Cell Proliferation Assay
2.10. Assessment of the Actin Polymerization
2.11. Mass Spectrometry
2.12. Immunoprecipitation
2.13. Statistical Analysis
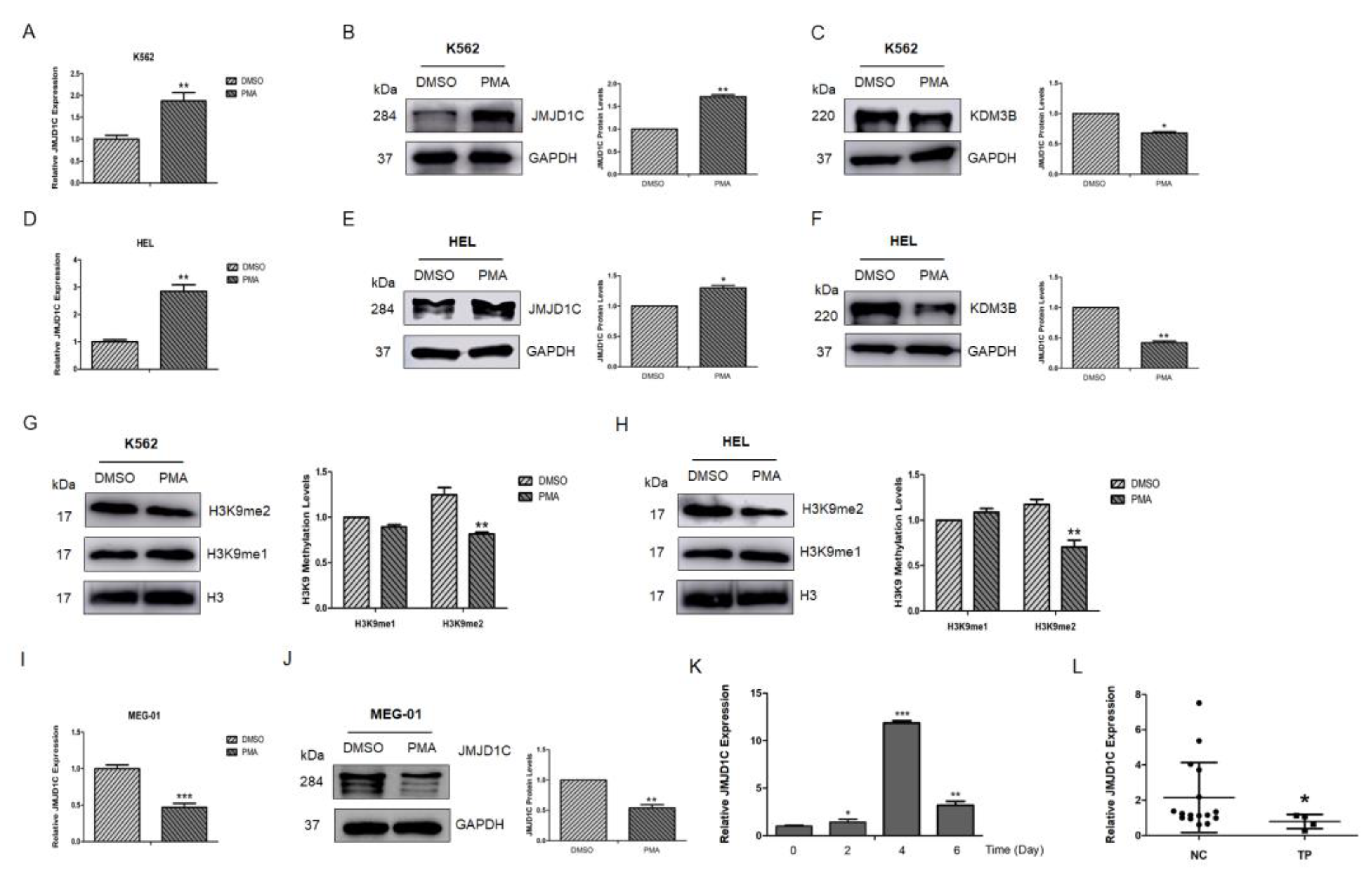
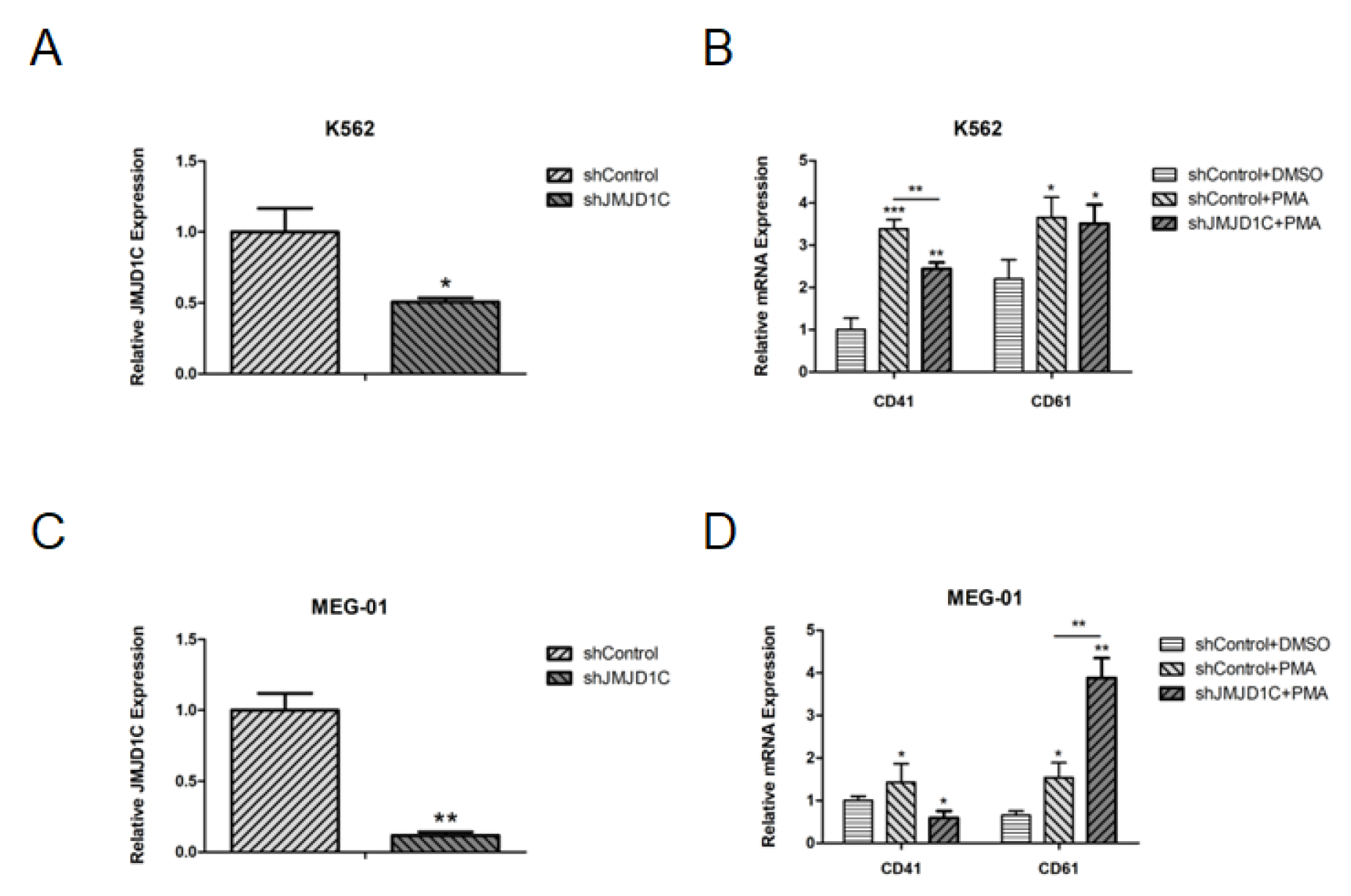
3. Results
3.1. Expression of JMJD1C in Megakaryopoiesis and Thrombocytopenia Patients
3.2. Knockdown of JMJD1C Shows Different Influences on Megakaryopoiesis in the Cell Line Models
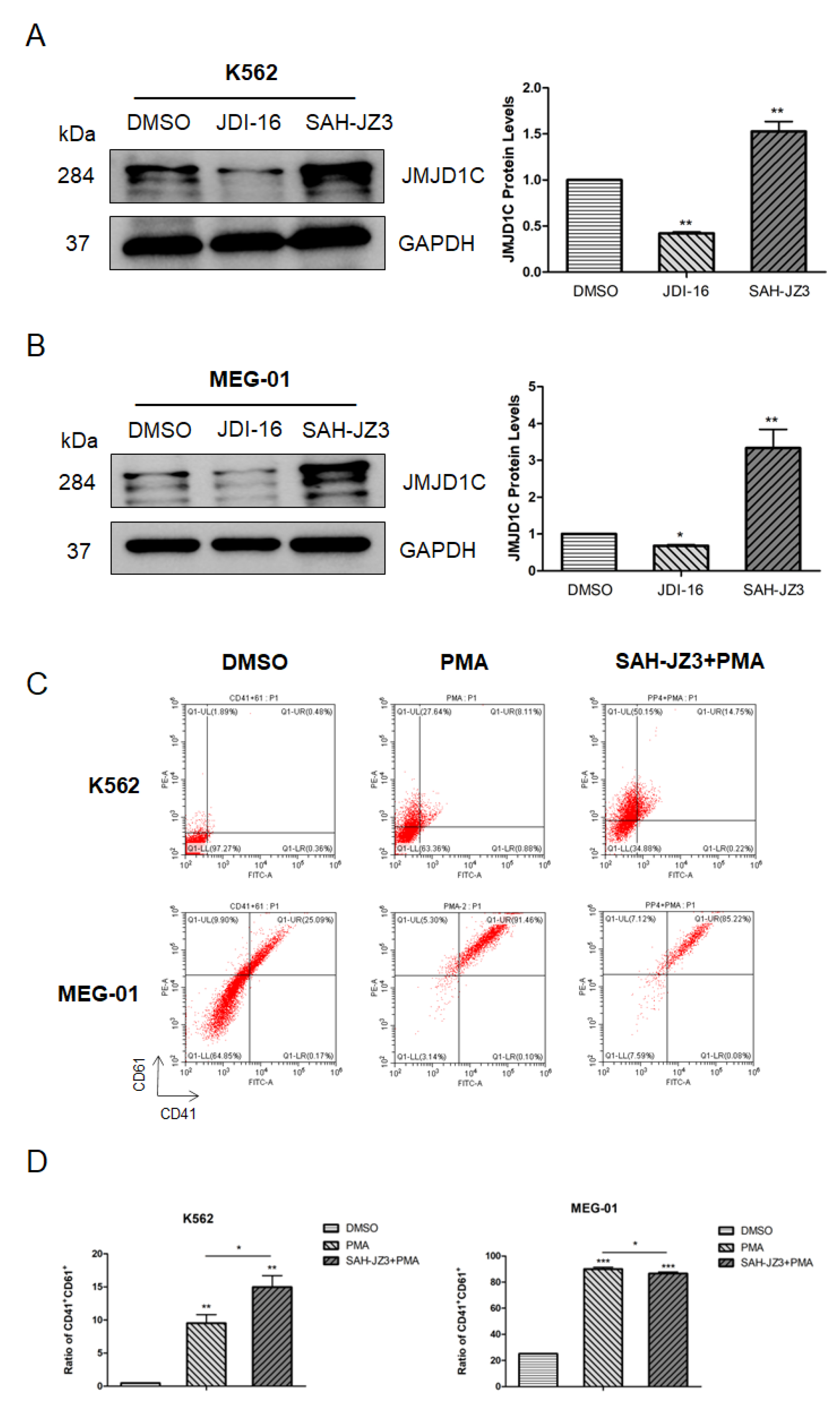
3.3. Identification of a Stapled Peptide as a JMJD1C Agonist
3.4. The Peptide Agonist of JMJD1C Shows a Differential Regulation of K562 and MEG-01 Cells
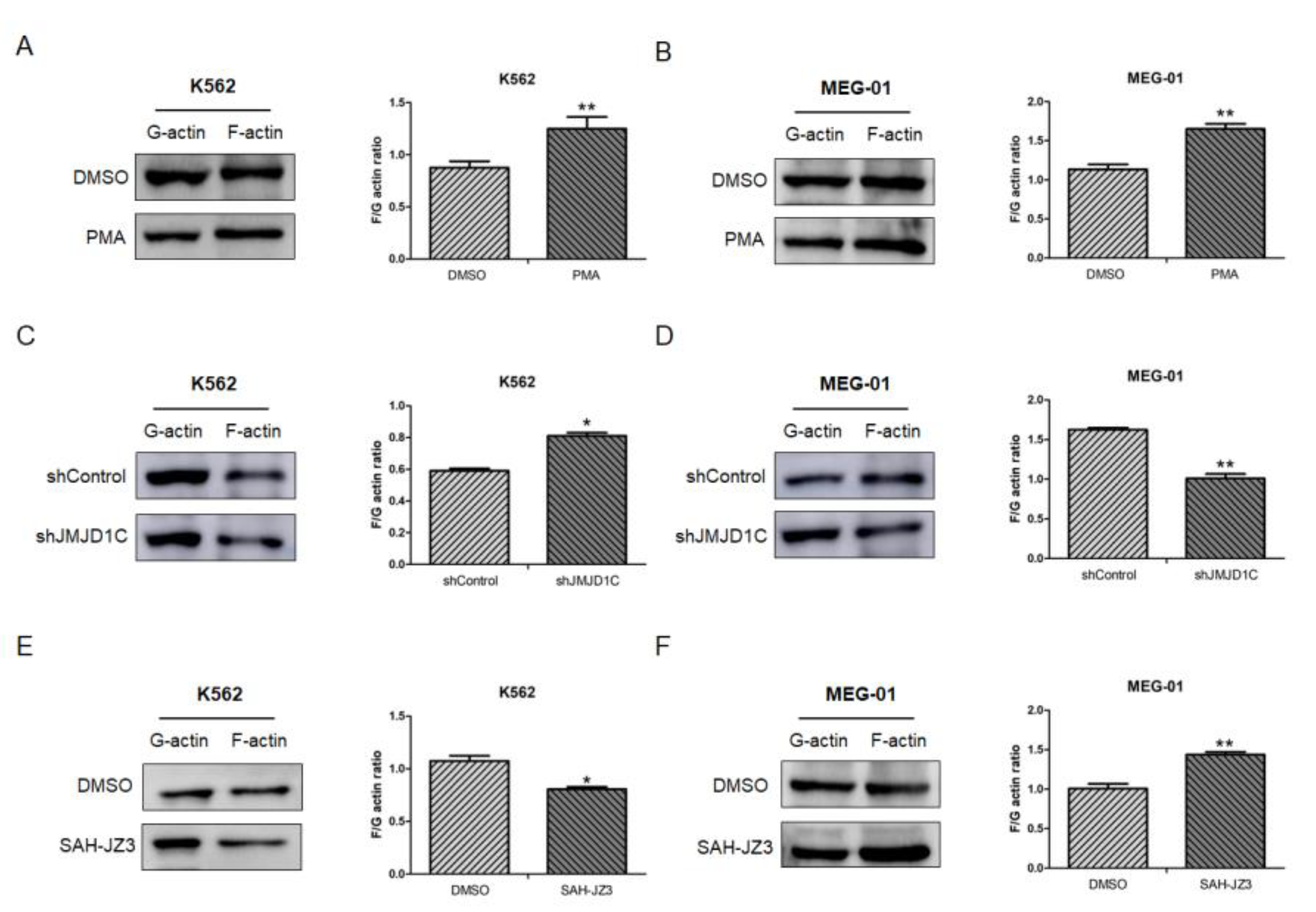
3.5. The Influence of JMJD1C on the Cytoskeleton Network
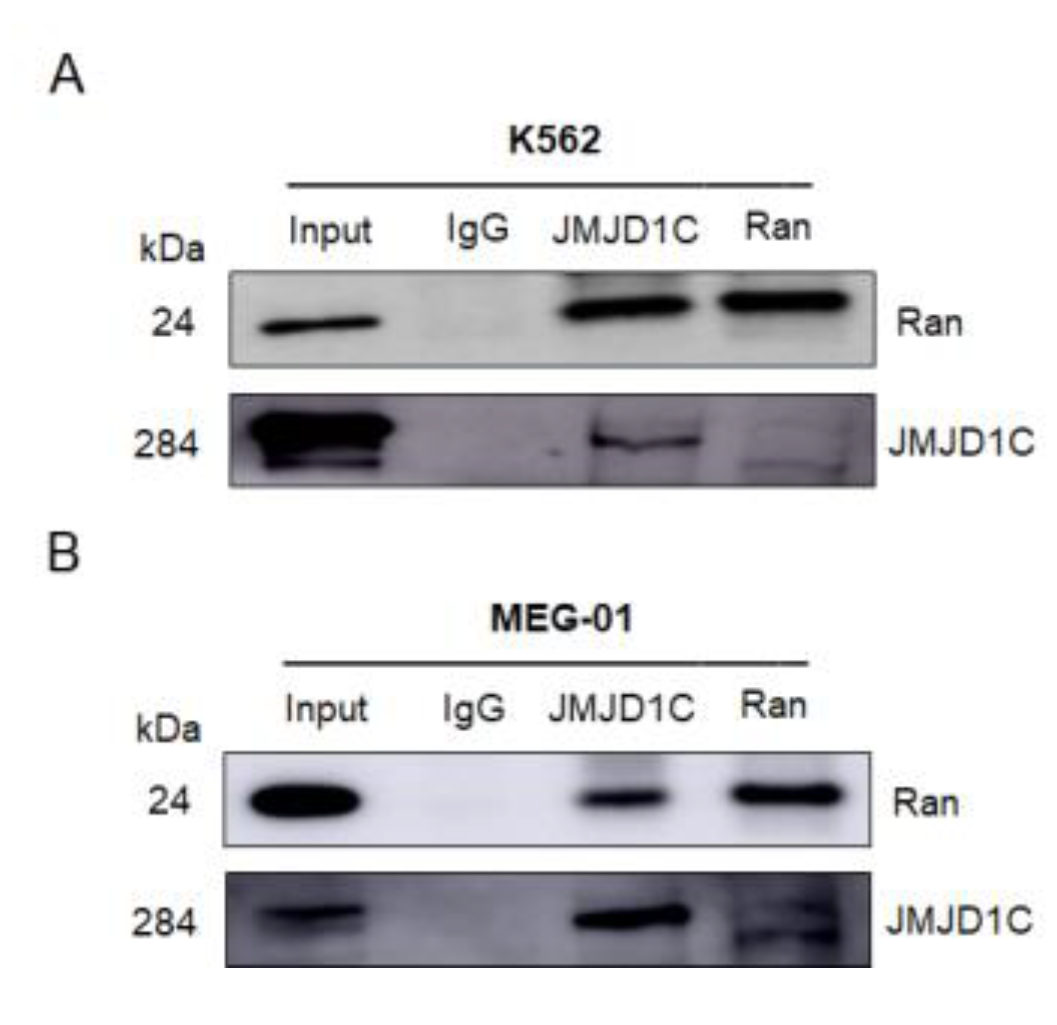
3.6. JMJD1C Interacts with Ran GTPase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soranzo, N.; Spector, T.D.; Mangino, M.; Kühnel, B.; Rendon, A.; Teumer, A.; Willenborg, C.; Wright, B.; Chen, L.; Li, M. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009, 41, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Yanek, L.R.; Chen, M.-H.; Faraday, N.; Larson, M.G.; Tofler, G.; Lin, S.J.; Kraja, A.T.; Province, M.A.; Yang, Q.; et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 2010, 42, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Gieger, C.; Radhakrishnan, A.; Cvejic, A.; Tang, W.; Porcu, E.; Pistis, G.; Serbanovic-Canic, J.; Elling, U.; Goodall, A.H.; Labrune, Y.; et al. New gene functions in megakaryopoiesis and platelet formation. Nature 2011, 480, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, R.; Snively, B.M.; Ziv, E.; Nalls, M.A.; Liu, Y.; Tang, W.; Yanek, L.R.; Lange, L.; Evans, M.K.; Ganesh, S.; et al. A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans. PLoS Genet. 2012, 8, e1002491. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, Z.; Su, L.; Chen, F.; Tejera, P.; Bajwa, E.K.; Wurfel, M.M.; Lin, X.; Christiani, D.C. Platelet count mediates the contribution of a genetic variant in LRRC16A to ARDS risk. Chest 2015, 147, 607–617. [Google Scholar] [CrossRef]
- Eicher, J.D.; Xue, L.; Ben-Shlomo, Y.; Beswick, A.D.; Johnson, A.D. Replication and hematological characterization of human platelet reactivity genetic associations in men from the Caerphilly Prospective Study (CaPS). J. Thromb. Thrombolysis 2016, 41, 343–350. [Google Scholar] [CrossRef]
- Björn, N.; Sigurgeirsson, B.; Svedberg, A.; Pradhananga, S.; Brandén, E.; Koyi, H.; Lewensohn, R.; de Petris, L.; Apellániz-Ruiz, M.; Rodríguez-Antona, C.; et al. Genes and variants in hematopoiesis-related pathways are associated with gemcitabine/carboplatin-induced thrombocytopenia. Pharm. J. 2020, 20, 179–191. [Google Scholar] [CrossRef]
- Choi, S.H.; Ruggiero, D.; Sorice, R.; Song, C.; Nutile, T.; Vernon Smith, A.; Concas, M.P.; Traglia, M.; Barbieri, C.; Ndiaye, N.C.; et al. Six Novel Loci Associated with Circulating VEGF Levels Identified by a Meta-analysis of Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005874. [Google Scholar] [CrossRef]
- Yeung, S.L.A.; Lam, H.S.H.S.; Schooling, C.M. Vascular endothelial growth factor and ischemic heart disease risk: A mendelian randomization study. J. Am. Heart Assoc. 2017, 6, e005619. [Google Scholar] [CrossRef]
- Nethander, M.; Quester, J.; Vandenput, L.; Ohlsson, C. Association of Genetically Predicted Serum Estradiol with Risk of Thromboembolism in Men: A Mendelian Randomization Study. J. Clin. Endocrinol. Metab. 2021, 106, e3078–e3086. [Google Scholar] [CrossRef]
- Noh, J.Y. Megakaryopoiesis and platelet biology: Roles of transcription factors and emerging clinical implications. Int. J. Mol. Sci. 2021, 22, 9615. [Google Scholar] [CrossRef] [PubMed]
- Ogura, M.; Morishima, Y.; Ohno, R.; Kato, Y.; Hirabayashi, N.; Nagura, H.; Saito, H. Establishment of a novel human megakaryoblastic leukemia cell line, MEG-01, with positive Philadelphia chromosome. Blood 1985, 66, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Ogura, M.; Saito, H.; Satoh, M.; Takeuchi, M. Production of platelet-like particles by a human megakaryoblastic leukemia cell line (MEG-01). Exp. Cell Res. 1991, 193, 223–226. [Google Scholar] [CrossRef]
- Trinh, B.Q.; Barengo, N.; Kim, S.B.; Lee, J.S.; Zweidler-McKay, P.A.; Naora, H. The homeobox gene DLX4 regulates erythro-megakaryocytic differentiation by stimulating IL-1β and NF-κB signaling. J. Cell Sci. 2015, 128, 3055–3067. [Google Scholar] [CrossRef]
- Martin, P.; Papayannopoulou, T. HEL cells: A new human erythroleukemia cell line with spontaneous and induced globin expression. Science 1982, 216, 1233–1235. [Google Scholar] [CrossRef]
- Xu, X.; Wang, L.; Hu, L.; Dirks, W.G.; Zhao, Y.; Wei, Z.; Chen, D.; Li, Z.; Wang, Z.; Han, Y.; et al. Small molecular modulators of JMJD1C preferentially inhibit growth of leukemia cells. Int. J. Cancer 2019, 146, 400–412. [Google Scholar] [CrossRef]
- Dirks, W.; MacLeod, R.A.; Jäger, K.; Milch, H.; Drexler, H.G. First searchable database for DNA profiles of human cell lines: Sequential use of fingerprint techniques for authentication. Cell. Mol. Biol. 1999, 45, 841–853. [Google Scholar]
- Messaoudi, K.; Ali, A.; Ishaq, R.; Palazzo, A.; Sliwa, D.; Bluteau, O.; Souquère, S.; Muller, D.; Diop, K.M.; Rameau, P.; et al. Critical role of the HDAC6-cortactin axis in human megakaryocyte maturation leading to a proplatelet-formation defect. Nat. Commun. 2017, 8, 1786. [Google Scholar] [CrossRef]
- Dhenge, A.; Kuhikar, R.; Kale, V.; Limaye, L. Regulation of differentiation of MEG01 to megakaryocytes and platelet-like particles by Valproic acid through Notch3 mediated actin polymerization. Platelets 2019, 30, 780–795. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Zhang, X.; Wang, Y.; Wang, X.; Hu, L.; Zhao, Y.; Wang, H.; Wang, Z.; Wang, H.; et al. Modulators of histone demethylase JMJD1C selectively target leukemic stem cells. FEBS Open Bio. 2020, 11, 265–277. [Google Scholar] [CrossRef]
- Qi, D.; Wang, J.; Zhao, Y.; Yang, Y.; Wang, Y.; Wang, H.; Wang, L.; Wang, Z.; Xu, X.; Hu, Z. JMJD1C-regulated lipid synthesis contributes to the maintenance of MLL-rearranged acute myeloid leukemia. Leuk Lymphoma 2022, 63, 2149–2160. [Google Scholar] [CrossRef] [PubMed]
- Rezaei Araghi, R.; Bird, G.H.; Ryan, J.A.; Jenson, J.M.; Godes, M.; Pritz, J.R.; Grant, R.A.; Letai, A.; Walensky, L.D.; Keating, A.E. Iterative optimization yields Mcl-1–targeting stapled peptides with selective cytotoxicity to Mcl-1–dependent cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, E886–E895. [Google Scholar] [CrossRef] [PubMed]
- Brauchle, M.; Yao, Z.; Arora, R.; Thigale, S.; Clay, I.; Inverardi, B.; Fletcher, J.; Taslimi, P.; Acker, M.G.; Gerrits, B.; et al. Protein Complex Interactor Analysis and Differential Activity of KDM3 Subfamily Members Towards H3K9 Methylation. PLoS ONE 2013, 8, e60549. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.S.; Patchev, V.K.; Obendorf, M. A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch. Biochem. Biophys. 2007, 460, 56–66. [Google Scholar] [CrossRef]
- Watanabe, S.; Watanabe, K.; Akimov, V.; Bartkova, J.; Blagoev, B.; Lukas, J.; Bartek, J. JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1-mediated chromatin response to DNA breaks. Nat. Struct. Mol. Biol. 2013, 20, 1425–1433. [Google Scholar] [CrossRef]
- Izaguirre-Carbonell, J.; Christiansen, L.; Burns, R.; Schmitz, J.; Li, C.; Mokry, R.L.; Bluemn, T.; Zheng, Y.; Shen, J.; Carlson, K.S.; et al. Critical role of Jumonji domain of JMJD1C in MLL-rearranged leukemia. Blood Adv. 2019, 3, 1499–1511. [Google Scholar] [CrossRef]
- Sroczynska, P.; Cruickshank, V.A.; Bukowski, J.P.; Miyagi, S.; Bagger, F.O.; Walfridsson, J.; Schuster, M.B.; Porse, B.; Helin, K. ShRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood 2014, 123, 1870–1882. [Google Scholar] [CrossRef]
- Zhu, N.; Chen, M.; Eng, R.; DeJong, J.; Sinha, A.U.; Rahnamay, N.F.; Koche, R.; Al-Shahrour, F.; Minehart, J.C.; Chen, C.-W.; et al. MLL-AF9- and HOXA9-mediated acute myeloid leukemia stem cell self-renewal requires JMJD1C. J. Clin. Investig. 2016, 126, 997–1011. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, N.; Liu, X.; Laurent, B.; Tang, Z.; Eng, R.; Shi, Y.; Armstrong, S.A.; Roeder, R.G. JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors. Genes Dev. 2015, 29, 2123–2139. [Google Scholar] [CrossRef]
- Kitajima, K.; Kojima, M.; Kondo, S.; Takeuchi, T. A role of jumonji gene in proliferation but not differentiation of megakaryocyte lineage cells. Exp. Hematol. 2001, 29, 507–514. [Google Scholar] [CrossRef]
- Yang, J.; Ma, J.; Xiong, Y.; Wang, Y.; Jin, K.; Xia, W.; Chen, Q.; Huang, J.; Zhang, J.; Jiang, N.; et al. Epigenetic regulation of megakaryocytic and erythroid differentiation by PHF2 histone demethylase. J Cell Physiol. 2018, 233, 6841–6852. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Kim, J.Y.; Choe, N.W.; Cho, I.H.; Kim, J.R.; Kim, D.W.; Seol, J.E.; Lee, S.E.; Kook, H.; Nam, K.I.; et al. Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res. 2010, 38, 6389–6403. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Toumazou, C.; Tsukada, Y.I.; Erdjument-Bromage, H.; Tempst, P.; Wong, J.; Zhang, Y. JHDM2A, a JmjC-Containing H3K9 Demethylase, Facilitates Transcription Activation by Androgen Receptor. Cell 2006, 125, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, Y.I.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Luo, D.; de Morree, A.; Boutet, S.; Quach, N.; Natu, V.; Rustagi, A.; Rando, T.A. Deltex2 represses MyoD expression and inhibits myogenic differentiation by acting as a negative regulator of Jmjd1c. Proc. Natl. Acad. Sci. USA 2017, 114, E3071–E3080. [Google Scholar] [CrossRef]
- Huang, J.; Huang, S.; Ma, Z.; Lin, X.; Li, X.; Huang, X.; Wang, J.; Ye, W.; Li, Y.; He, D.; et al. Ibrutinib Suppresses Early Megakaryopoiesis but Enhances Proplatelet Formation. Thromb. Haemost. 2021, 121, 192–205. [Google Scholar] [CrossRef]
- Roy, A.; Basak, N.P.; Banerjee, S. Notch1 intracellular domain increases cytoplasmic EZH2 levels during early megakaryopoiesis. Cell Death Dis. 2012, 3, e380. [Google Scholar] [CrossRef]
- Mazzi, S.; Dessen, P.; Vieira, M.; Dufour, V.; Cambot, M.; El Khoury, M.; Antony-Debré, I.; Arkoun, B.; Basso-Valentina, F.; BenAbdoulahab, S.; et al. Dual role of EZH2 in megakaryocyte differentiation. Blood 2019, 138, 1603–1614. [Google Scholar] [CrossRef]
- Su, I.H.; Dobenecker, M.W.; Dickinson, E.; Oser, M.; Basavaraj, A.; Marqueron, R.; Viale, A.; Reinberg, D.; Wülfing, C.; Tarakhovsky, A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 2005, 121, 425–436. [Google Scholar] [CrossRef]
- Salus, S.S.; Demeter, J.; Sazer, S. The Ran GTPase system in fission yeast affects microtubules and cytokinesis in cells that are competent for nucleocytoplasmic protein transport. Mol. Cell. Biol. 2002, 22, 8491–8505. [Google Scholar] [CrossRef][Green Version]
- Deng, M.; Suraneni, P.; Schultz, R.M.; Li, R. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev. Cell. 2006, 12, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Yao, T.; Mishina, T.; Endoh, H.; Tanaka, M.; Yonezawa, N.; Shimamoto, Y.; Yonemura, S.; Yamagata, K.; Kitajima, T.S.; et al. RanGTP and the actin cytoskeleton keep paternal and maternal chromosomes apart during fertilization. J. Cell Biol. 2021, 220, e202012001. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Okano, Y.; Nozawa, Y. Differential expression of low Mr GTP-binding proteins in human megakaryoblastic leukemia cell line, MEG-01, and their possible involvement in the differentiation process. Thromb. Haemost. 1997, 77, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Okano, Y.; Nozawa, Y. Expression of GTP-binding proteins and protein kinase C isozymes in platelet-like particles derived from megakaryoblastic leukemia cells (MEG-01). Platelets 1998, 9, 291–296. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, X.; Wang, H.; Qin, L.; Feng, A.; Qi, D.; Wang, H.; Zhao, Y.; Kong, L.; Wang, H.; et al. JMJD1C Regulates Megakaryopoiesis in In Vitro Models through the Actin Network. Cells 2022, 11, 3660. https://doi.org/10.3390/cells11223660
Wang J, Liu X, Wang H, Qin L, Feng A, Qi D, Wang H, Zhao Y, Kong L, Wang H, et al. JMJD1C Regulates Megakaryopoiesis in In Vitro Models through the Actin Network. Cells. 2022; 11(22):3660. https://doi.org/10.3390/cells11223660
Chicago/Turabian StyleWang, Jialing, Xiaodan Liu, Haixia Wang, Lili Qin, Anhua Feng, Daoxin Qi, Haihua Wang, Yao Zhao, Lihua Kong, Haiying Wang, and et al. 2022. "JMJD1C Regulates Megakaryopoiesis in In Vitro Models through the Actin Network" Cells 11, no. 22: 3660. https://doi.org/10.3390/cells11223660
APA StyleWang, J., Liu, X., Wang, H., Qin, L., Feng, A., Qi, D., Wang, H., Zhao, Y., Kong, L., Wang, H., Wang, L., Hu, Z., & Xu, X. (2022). JMJD1C Regulates Megakaryopoiesis in In Vitro Models through the Actin Network. Cells, 11(22), 3660. https://doi.org/10.3390/cells11223660





