The Oocyte-Specific Linker Histone H1FOO Is Not Essential for Mouse Oogenesis and Fertility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Fertility Assessment
2.3. Histology
2.4. Immunocytology and Antibodies
2.5. Unfertilized Oocyte Collection for Metaphase II Analysis
2.6. Cell Culture
2.7. iPSCs Generation from MEFs
2.8. Statistics
2.9. Ethics Statement
3. Results
3.1. Generation of H1FOO Knockout Mice
3.2. H1FOO-Deficient Mice Are Fertile
3.3. H1FOO-Deficient Meiocytes Show Normal Synapsis, Double-Strand Breaks (DSBs) Generation, and Telomere Dynamics
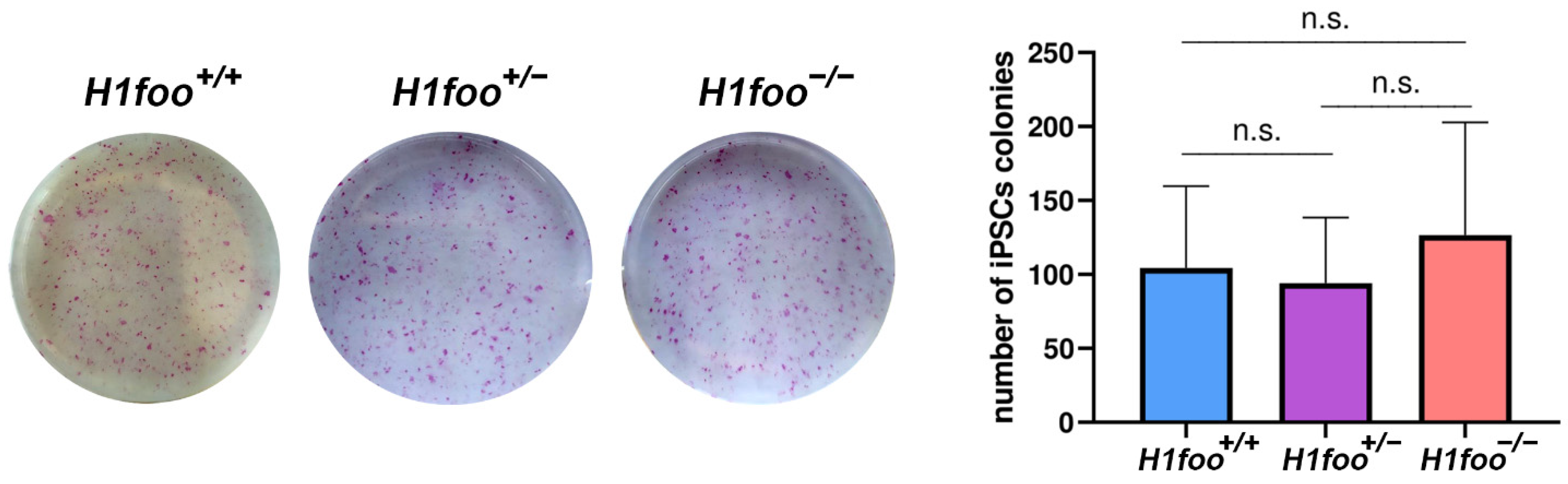
3.4. The Absence of H1FOO Does Not Impair MEFs Reprogramming
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hillers, K.J.; Jantsch, V.; Martinez-Perez, E.; Yanowitz, J.L. Meiosis. WormBook 2017, 2017, 433–434. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, 1–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Zhang, T.; Yang, Y.; Wang, C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Front. Cell Dev. Biol. 2021, 9, 654028. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, Z.; Khawar, M.B.; Liu, C.; Li, W. The Histone Codes for Meiosis. Reproduction 2017, 154, R65–R79. [Google Scholar] [CrossRef]
- Workman, J.L.; Kingston, R.E. Alteration of Nucleosome Structure as a Mechanism of Transcriptional Regulation. Annu. Rev. Biochem. 2003, 67, 545–579. [Google Scholar] [CrossRef] [Green Version]
- Hergeth, S.P.; Schneider, R. The H1 Linker Histones: Multifunctional Proteins beyond the Nucleosomal Core Particle. EMBO Rep. 2015, 16, 1439. [Google Scholar] [CrossRef] [Green Version]
- Pan, C.; Fan, Y. Role of H1 Linker Histones in Mammalian Development and Stem Cell Differentiation. Biochim. Biophys. Acta 2016, 1859, 496. [Google Scholar] [CrossRef] [Green Version]
- Prendergast, L.; Reinberg, D. The Missing Linker: Emerging Trends for H1 Variant-Specific Functions. Genes Dev. 2021, 35, 40–58. [Google Scholar] [CrossRef]
- Fyodorov, D.V.; Zhou, B.R.; Skoultchi, A.I.; Bai, Y. Emerging Roles of Linker Histones in Regulating Chromatin Structure and Function. Nat. Rev. Mol. Cell Biol. 2018, 19, 192. [Google Scholar] [CrossRef] [Green Version]
- Doenecke, D.; Albig, W.; Bode, C.; Drabent, B.; Franke, K.; Gavenis, K.; Witt, O. Histones: Genetic Diversity and Tissue-Specific Gene Expression. Histochem. Cell Biol. 1997, 107, 1–10. [Google Scholar] [CrossRef]
- Tanaka, M.; Hennebold, J.D.; Macfarlane, J.; Adashi, E.Y. A Mammalian Oocyte-Specific Linker Histone Gene H1oo: Homology with the Genes for the Oocyte-Specific Cleavage Stage Histone (cs-H1) of Sea Urchin and the B4/H1M Histone of the Frog. Development 2001, 128, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Kihara, M.; Hennebold, J.D.; Eppig, J.J.; Viveiros, M.M.; Emery, B.R.; Carrell, D.T.; Kirkman, N.J.; Meczekalski, B.; Zhou, J.; et al. H1FOO Is Coupled to the Initiation of Oocytic Growth. Biol. Reprod. 2005, 72, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.; Becker, A.; Miyara, F.; Han, Z.; Kihara, M.; Brown, D.T.; Hager, G.L.; Latham, K.; Adashi, E.Y.; Misteli, T. Differential in Vivo Binding Dynamics of Somatic and Oocyte-Specific Linker Histones in Oocytes and during ES Cell Nuclear Transfer. Mol. Biol. Cell 2005, 16, 3887–3895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funaya, S.; Ooga, M.; Suzuki, M.G.; Aoki, F. Linker Histone H1FOO Regulates the Chromatin Structure in Mouse Zygotes. FEBS Lett. 2018, 592, 2414–2424. [Google Scholar] [CrossRef] [Green Version]
- Furuya, M.; Tanaka, M.; Teranishi, T.; Matsumoto, K.; Hosoi, Y.; Saeki, K.; Ishimoto, H.; Minegishi, K.; Iritani, A.; Yoshimura, Y. H1foo Is Indispensable for Meiotic Maturation of the Mouse Oocyte. J. Reprod. Dev. 2007, 53, 895–902. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Shi, Y.; Dang, Y.; Hu, B.; Xiao, L.; Zhao, P.; Wang, S.; Zhang, K. Linker Histone H1FOO Is Required for Bovine Preimplantation Development by Regulating Lineage Specification and Chromatin Structure. Biol. Reprod. 2022, ioac167. [Google Scholar] [CrossRef]
- Yun, Y.; An, P.; Ning, J.; Zhao, G.M.; Yang, W.L.; Lei, A.M. H1foo Is Essential for in Vitro Meiotic Maturation of Bovine Oocytes. Zygote 2015, 23, 416–425. [Google Scholar] [CrossRef]
- Funaya, S.; Kawabata, Y.; Sugie, K.; Abe, K.I.; Suzuki, Y.; Suzuki, M.G.; Aoki, F. Involvement of the Linker Histone H1Foo in the Regulation of Oogenesis. Reproduction 2022, 164, 19–29. [Google Scholar] [CrossRef]
- Gao, S.; Chung, Y.G.; Parseghian, M.H.; King, G.J.; Adashi, E.Y.; Latham, K.E. Rapid H1 Linker Histone Transitions Following Fertilization or Somatic Cell Nuclear Transfer: Evidence for a Uniform Developmental Program in Mice. Dev. Biol. 2004, 266, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Gomes, K.M.S.; Costa, I.C.; dos Santos, J.F.; Dourado, P.M.M.; Forni, M.F.; Ferreira, J.C.B. Induced Pluripotent Stem Cells Reprogramming: Epigenetics and Applications in the Regenerative Medicine. Rev. Assoc. Med. Bras. 2017, 63, 180–189. [Google Scholar] [CrossRef]
- Papp, B.; Plath, K. Epigenetics of Reprogramming to Induced Pluripotency. Cell 2013, 152, 1324–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terme, J.M.; Sesé, B.; Millán-Ariño, L.; Mayor, R.; Belmonte, J.C.I.; Barrero, M.J.; Jordan, A. Histone H1 Variants Are Differentially Expressed and Incorporated into Chromatin during Differentiation and Reprogramming to Pluripotency. J. Biol. Chem. 2011, 286, 35347–35357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, A.H.F.M.; Plug, A.W.; van Vugt, M.J.; de Boer, P.; Peters, A.H.F.M.; Plug, A.W.; van Vugt, M.J.; De, P. A Drying-down Technique for the Spreading of Mammalian Meiocytes from the Male and Female Germline. Chromosome Res. 1997, 5, 66–71. [Google Scholar] [CrossRef]
- da Cruz, I.; Rodríguez-Casuriaga, R.; Santiñaque, F.F.; Farías, J.; Curti, G.; Capoano, C.A.; Folle, G.A.; Benavente, R.; Sotelo-Silveira, J.R.; Geisinger, A. Transcriptome Analysis of Highly Purified Mouse Spermatogenic Cell Populations: Gene Expression Signatures Switch from Meiotic-to Postmeiotic-Related Processes at Pachytene Stage. BMC Genom. 2016, 17, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, A.; Yamada, Y.; Yamanaka, S. Epigenetic Regulation in Pluripotent Stem Cells: A Key to Breaking the Epigenetic Barrier. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awe, J.P.; Byrne, J.A. Identifying Candidate Oocyte Reprogramming Factors Using Cross-Species Global Transcriptional Analysis. Cell Reprogram 2013, 15, 126–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunitomi, A.; Yuasa, S.; Sugiyama, F.; Saito, Y.; Seki, T.; Kusumoto, D.; Kashimura, S.; Takei, M.; Tohyama, S.; Hashimoto, H.; et al. H1foo Has a Pivotal Role in Qualifying Induced Pluripotent Stem Cells. Stem Cell Rep. 2016, 6, 825–833. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Luo, C.; Wang, Z.; Chen, S.; Morris, D.; Ruan, F.; Chen, Z.; Yang, L.; Wei, X.; Wu, C.; et al. Uncoupling Transcription and Translation through MiRNA-Dependent Poly(A) Length Control in Haploid Male Germ Cells. Development 2022, 149, 199573. [Google Scholar] [CrossRef]
- Nothias, J.Y.; Miranda, M.; DePamphilis, M.L. Uncoupling of Transcription and Translation during Zygotic Gene Activation in the Mouse. EMBO J. 1996, 15, 5715–5725. [Google Scholar] [CrossRef]
- Maeda, C.; Sato, S.; Hattori, N.; Tanaka, S.; Yagi, S.; Shiota, K. DNA Hypomethylation Circuit of the Mouse Oocyte-Specific Histone H1foo Gene in Female Germ Cell Lineage. Biol. Reprod. 2008, 78, 816–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ooga, M.; Funaya, S.; Hashioka, Y.; Fujii, W.; Naito, K.; Suzuki, M.G.; Aoki, F. Chd9 Mediates Highly Loosened Chromatin Structure in Growing Mouse Oocytes. Biochem. Biophys. Res. Commun. 2018, 500, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Ooga, M.; Fulka, H.; Hashimoto, S.; Suzuki, M.G.; Aoki, F. Analysis of Chromatin Structure in Mouse Preimplantation Embryos by Fluorescent Recovery after Photobleaching. Epigenetics 2016, 11, 85–94. [Google Scholar] [CrossRef] [Green Version]
- de Macedo, M.P.; Glanzner, W.G.; Gutierrez, K.; Bordignon, V. Chromatin Role in Early Programming of Embryos. Anim. Front. 2021, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Mizusawa, Y.; Kuji, N.; Tanaka, Y.; Tanaka, M.; Ikeda, E.; Komatsu, S.; Kato, S.; Yoshimura, Y. Expression of Human Oocyte-Specific Linker Histone Protein and Its Incorporation into Sperm Chromatin during Fertilization. Fertil. Steril. 2010, 93, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Jullien, J.; Miyamoto, K.; Pasque, V.; Allen, G.E.; Bradshaw, C.R.; Garrett, N.J.; Halley-Stott, R.P.; Kimura, H.; Ohsumi, K.; Gurdon, J.B. Hierarchical Molecular Events Driven by Oocyte-Specific Factors Lead to Rapid and Extensive Reprogramming. Mol. Cell 2014, 55, 524–536. [Google Scholar] [CrossRef] [Green Version]
- Hayakawa, K.; Ohgane, J.; Tanaka, S.; Yagi, S.; Shiota, K. Oocyte-Specific Linker Histone H1foo Is an Epigenomic Modulator That Decondenses Chromatin and Impairs Pluripotency. Epigenetics 2012, 7, 1029. [Google Scholar] [CrossRef] [Green Version]
- Teranishi, T.; Tanaka, M.; Kimoto, S.; Ono, Y.; Miyakoshi, K.; Kono, T.; Yoshimura, Y. Rapid Replacement of Somatic Linker Histones with the Oocyte-Specific Linker Histone H1foo in Nuclear Transfer. Dev. Biol. 2004, 266, 76–86. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Sáez, F.; Sainz-Urruela, R.; Felipe-Medina, N.; Condezo, Y.B.; Sánchez-Martín, M.; Llano, E.; Pendás, A.M. The Oocyte-Specific Linker Histone H1FOO Is Not Essential for Mouse Oogenesis and Fertility. Cells 2022, 11, 3706. https://doi.org/10.3390/cells11223706
Sánchez-Sáez F, Sainz-Urruela R, Felipe-Medina N, Condezo YB, Sánchez-Martín M, Llano E, Pendás AM. The Oocyte-Specific Linker Histone H1FOO Is Not Essential for Mouse Oogenesis and Fertility. Cells. 2022; 11(22):3706. https://doi.org/10.3390/cells11223706
Chicago/Turabian StyleSánchez-Sáez, Fernando, Raquel Sainz-Urruela, Natalia Felipe-Medina, Yazmine B. Condezo, Manuel Sánchez-Martín, Elena Llano, and Alberto M. Pendás. 2022. "The Oocyte-Specific Linker Histone H1FOO Is Not Essential for Mouse Oogenesis and Fertility" Cells 11, no. 22: 3706. https://doi.org/10.3390/cells11223706






