The XPA Protein—Life under Precise Control
Abstract
:1. Introduction
2. XPA’s Structure and DNA-Binding and Protein-Binding Abilities
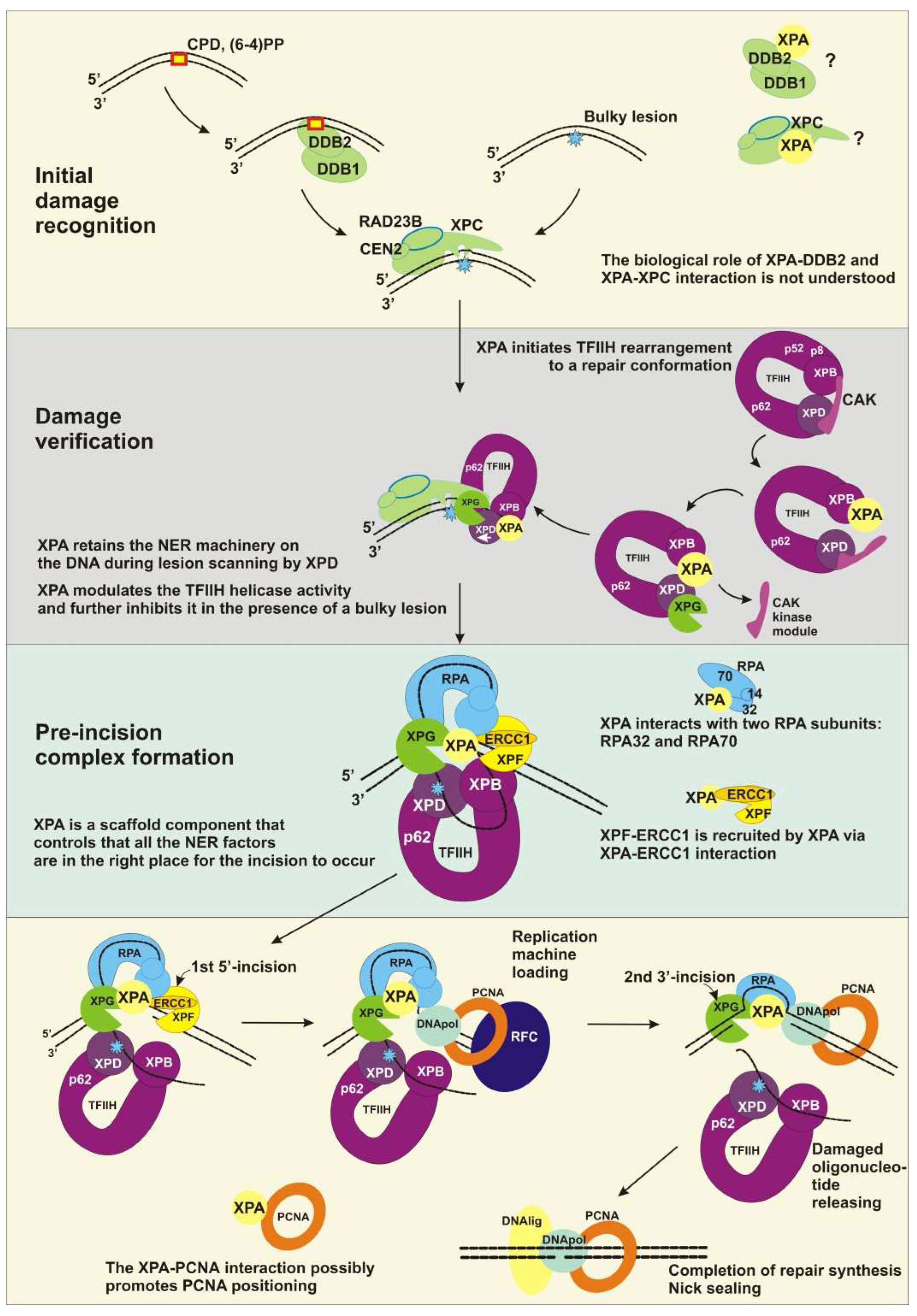
2.1. Initial Damage Recognition
2.2. Damage Verification
2.3. Pre-Incision Complex Formation
2.4. Dual Incision, Resynthesis, and Ligation
2.5. XPA Dimerization
2.6. PTM Proteins
2.7. XABs
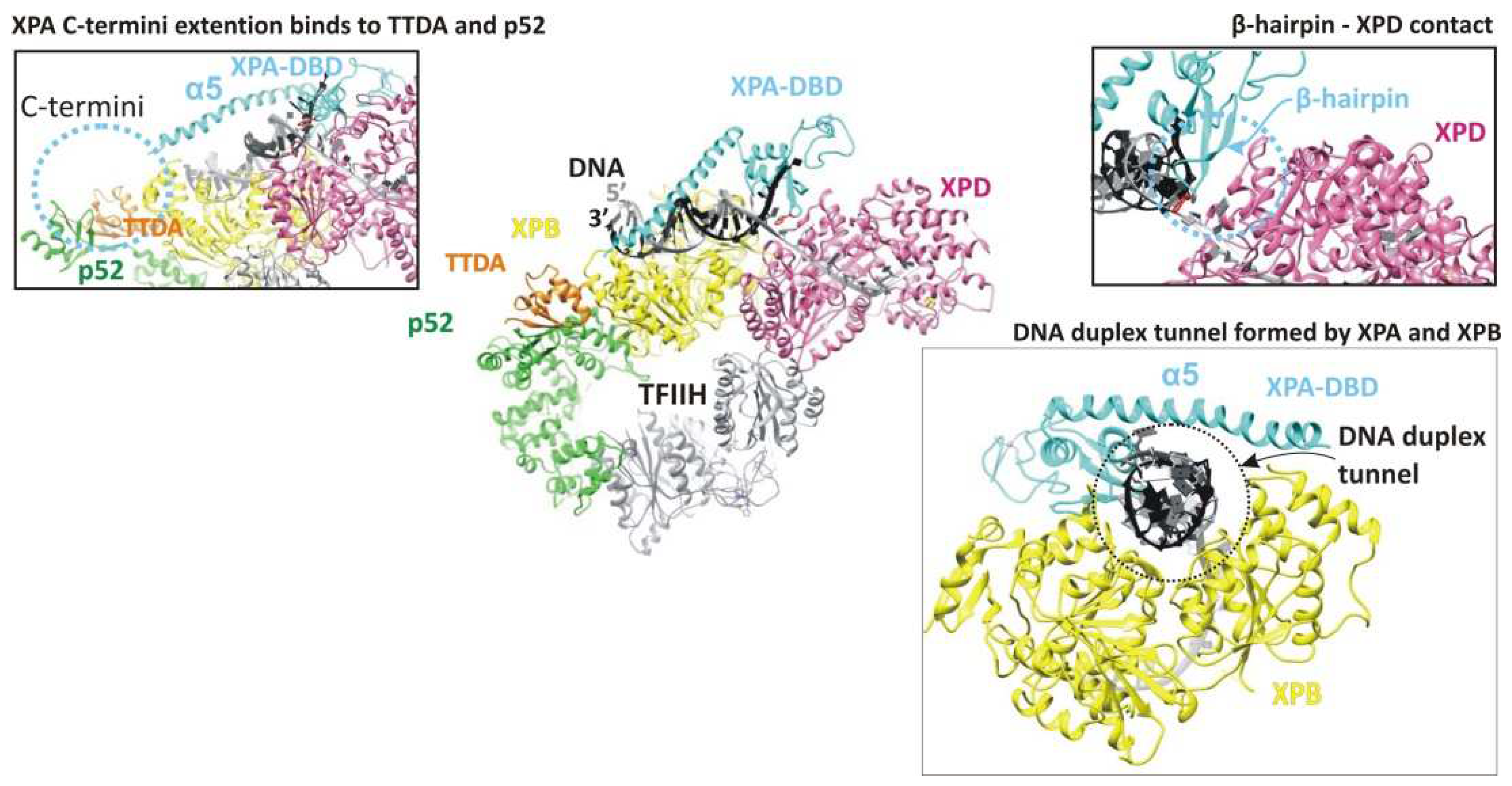
3. The Hinge of the NER Complex
4. The Ways to Control XPA
4.1. The XPA Protein Amount Control—A Balance between Production and Degradation
4.1.1. The Amount of XPA Molecules
4.1.2. The XPA Residence: The Cytoplasm or Nucleus?
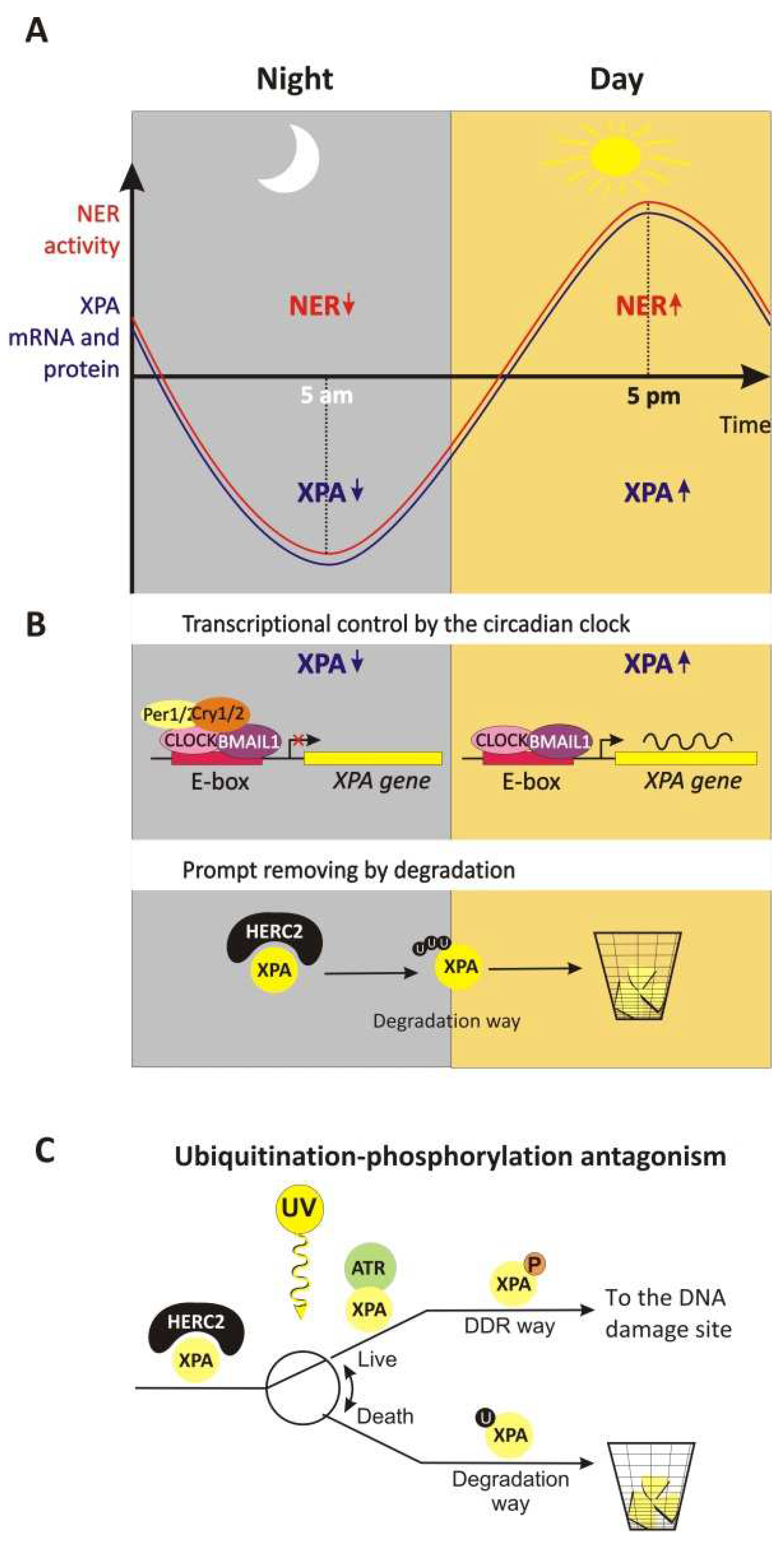
4.1.3. The Circadian Rhythm of XPA’s Life
4.1.4. XPA Transcription Control Inside a Solid Tumor
4.1.5. Other Ways to Control XPA Transcription
4.2. Fine Tuning of XPA by PTMs
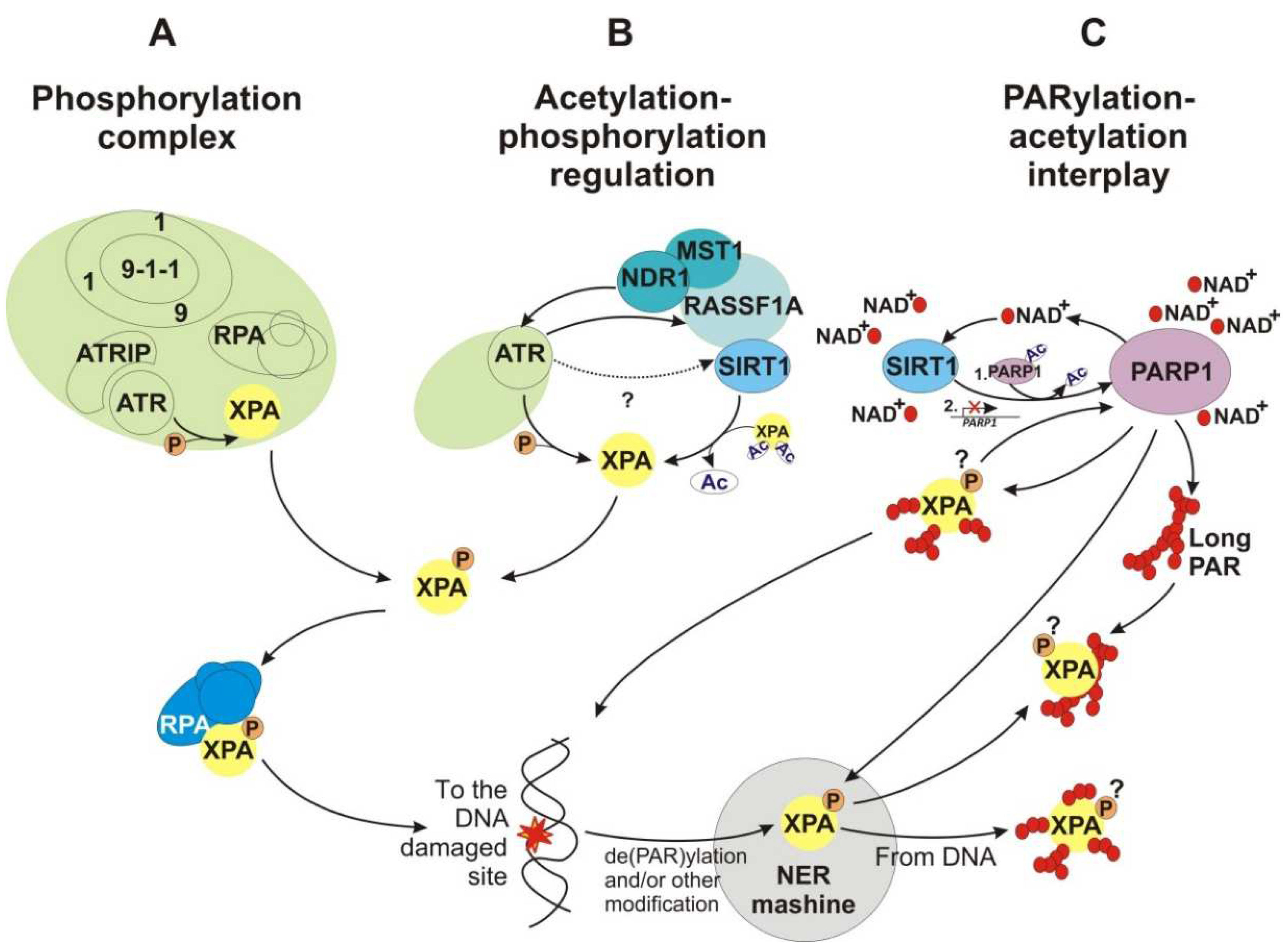
4.2.1. Phosphorylation and Checkpoint/DNA Repair Duties
4.2.2. Dephosphorylation
4.2.3. Acetylation
4.2.4. Deacetylation
4.2.5. PARylation
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gillet, L.C.; Schärer, O.D. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006, 106, 253–276. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Geacintov, N.E.; Broyde, S.; Yeo, J.E.; Schärer, O.D. Molecular basis for damage recognition and verification by XPC-RAD23B and TFIIH in nucleotide excision repair. DNA Repair 2018, 71, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Kusakabe, M.; Onishi, Y.; Tada, H.; Kurihara, F.; Kusao, K.; Furukawa, M.; Iwai, S.; Yokoi, M.; Sakai, W.; Sugasawa, K. Mechanism and regulation of DNA damage recognition in nucleotide excision repair. Genes Environ. 2019, 41, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapin, I.; Lindenbaum, Y.; Dickson, D.W.; Kraemer, K.H.; Robbins, J.H. Cockayne syndrome and xeroderma pigmentosum. Neurology 2000, 55, 1442–1449. [Google Scholar] [CrossRef] [Green Version]
- DiGiovanna, J.J.; Kraemer, K.H. Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 2012, 132, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Fassihi, H.; Sethi, M.; Fawcett, H.; Wing, J.; Chandler, N.; Mohammed, S.; Craythorne, E.; Morley, A.M.; Lim, R.; Turner, S.; et al. Deep phenotyping of 89 xeroderma pigmentosum patients reveals unexpected heterogeneity dependent on the precise molecular defect. Proc. Natl. Acad. Sci. USA 2016, 113, 1236–1245. [Google Scholar] [CrossRef] [Green Version]
- Krasikova, Y.; Rechkunova, N.; Lavrik, O. Nucleotide Excision Repair: From Molecular Defects to Neurological Abnormalities. Int. J. Mol. Sci. 2021, 22, 6220. [Google Scholar] [CrossRef]
- Cleaver, J.E.; States, J.C. The DNA damage-recognition problem in human and other eukaryotic cells: The XPA damage binding protein. Biochem. J. 1997, 328, 1–12. [Google Scholar] [CrossRef]
- Wakasugi, M.; Sancar, A. Order of assembly of human DNA repair excision nuclease. J. Biol. Chem. 1999, 274, 18759–18768. [Google Scholar] [CrossRef] [Green Version]
- Thoma, B.S.; Vasquez, K.M. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003, 38, 1–13. [Google Scholar] [CrossRef]
- Li, C.L.; Golebiowski, F.M.; Onishi, Y.; Samara, N.L.; Sugasawa, K.; Yang, W. Tripartite DNA Lesion Recognition and Verification by XPC, TFIIH, and XPA in Nucleotide Excision Repair. Mol. Cell 2015, 59, 1025–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugitani, N.; Sivley, R.M.; Perry, K.E.; Capra, J.A.; Chazin, W.J. XPA: A key scaffold for human nucleotide excision repair. DNA Repair 2016, 44, 123–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borszéková Pulzová, L.; Ward, T.A.; Chovanec, M. XPA: DNA Repair Protein of Significant Clinical Importance. Int. J. Mol. Sci. 2020, 21, 2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikegami, T.; Kuraoka, I.; Saijo, M.; Kodo, N.; Kyogoku, Y.; Morikawa, K.; Tanaka, K.; Shirakawa, M. Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nat. Struct. Biol. 1998, 5, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Buchko, G.W.; Daughdrill, G.W.; de Lorimier, R.; Rao, B.K.; Isern, N.G.; Lingbeck, J.M.; Taylor, J.S.; Wold, M.S.; Gochin, M.; Spicer, L.D.; et al. Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: Chemical shift mapping and 15N NMR relaxation studies. Biochemistry 1999, 38, 15116–15128. [Google Scholar] [CrossRef]
- Lian, F.M.; Yang, X.; Yang, W.; Jiang, Y.L.; Qian, C. Structural characterization of the redefined DNA-binding domain of human XPA. Biochem. Biophys. Res. Commun. 2019, 514, 985–990. [Google Scholar] [CrossRef]
- Lian, F.M.; Yang, X.; Jiang, Y.L.; Yang, F.; Li, C.; Yang, W.; Qian, C. New structural insights into the recognition of undamaged splayed-arm DNA with a single pair of non-complementary nucleotides by human nucleotide excision repair protein XPA. Int. J. Biol. Macromol. 2020, 148, 466–474. [Google Scholar] [CrossRef]
- Koch, S.C.; Kuper, J.; Gasteiger, K.L.; Simon, N.; Strasser, R.; Eisen, D.; Geiger, S.; Schneider, S.; Kisker, C.; Carell, T. Structural insights into the recognition of cisplatin and AAF-dG lesion by Rad14 (XPA). Proc. Natl. Acad. Sci. USA 2015, 112, 8272–8277. [Google Scholar] [CrossRef] [Green Version]
- Simon, N.; Ebert, C.; Schneider, S. Structural basis for bulky-Adduct DNA-lesion recognition by the nucleotide excision repair protein Rad14. Chemistry 2016, 22, 10782–10785. [Google Scholar] [CrossRef]
- Ebert, C.; Simon, N.; Schneider, S.; Carell, T. Structural insights into the recognition of N(2) -Aryl- and C8-Aryl DNA lesions by the repair protein XPA/ Rad14. Chembiochem 2017, 18, 1379–1382. [Google Scholar] [CrossRef]
- Kokic, G.; Chernev, A.; Tegunov, D.; Dienemann, C.; Urlaub, H.; Cramer, P. Structural basis of TFIIH activation for nucleotide excision repair. Nat. Commun. 2019, 10, 2885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasikova, Y.S.; Rechkunova, N.I.; Maltseva, E.A.; Petruseva, I.O.; Lavrik, O.I. Localization of xeroderma pigmentosum group A protein and replication protein A on damaged DNA in nucleotide excision repair. Nucleic Acids Res. 2010, 38, 8083–8094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.M.; Sboner, A.; Xia, Y.; Gerstein, M. The role of disorder in interaction networks: A structural analysis. Mol. Syst. Biol. 2008, 4, 179. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, I.; Miura, N.; Niwa, H.; Miyazaki, J.; Tanaka, K. Mutational analysis of the structure and function of the xeroderma pigmentosum group A complementing protein. Identification of essential domains for nuclear localization and DNA excision repair. J. Biol. Chem. 1992, 267, 12182–12187. [Google Scholar] [CrossRef]
- Barve, A.; Ghaskadbi, S.; Ghaskadbi, S. Structural and Sequence Similarities of Hydra Xeroderma Pigmentosum A Protein to Human Homolog Suggest Early Evolution and Conservation. BioMed Res. Int. 2013, 2013, 854745. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Miura, N.; Satokata, I.; Miyamoto, I.; Yoshida, M.C.; Satoh, Y.; Kondo, S.; Yasui, A.; Okayama, H.; Okada, Y. Analysis of a human DNA excision repair gene involved in group A xeroderma pigmentosum and containing a zinc-finger domain. Nature 1990, 348, 73–76. [Google Scholar] [CrossRef]
- Buchko, G.W.; Tung, C.S.; McAteer, K.; Isern, N.G.; Spicer, L.D.; Kennedy, M.A. DNA-XPA interactions: A 31P NMP and molecular modeling study of dCCAATTAACC association with the minimal DNA-binding domain (M98-F219) of the Nucleotide Excision Repair protein XPA. Nucleic Acids Res. 2001, 29, 2635–2643. [Google Scholar] [CrossRef] [Green Version]
- Smirnova, J.; Zhukova, L.; Witkiewicz-Kucharczyk, A.; Kopera, E.; Oledzki, J.; Wysłouch-Cieszyńska, A.; Palumaa, P.; Hartwig, A.; Bal, W. Quantitative electrospray ionization mass spectrometry of zinc finger oxidation: The reaction of XPA zinc finger with H(2)O(2). Anal Biochem 2007, 369, 226–231. [Google Scholar] [CrossRef]
- Kuraoka, I.; Morita, E.H.; Saijo, M.; Matsuda, T.; Morikawa, K.; Shirakawa, M.; Tanaka, K. Identification of a damaged-DNA binding domain of the XPA protein. Mutat. Res. 1996, 362, 87–95. [Google Scholar] [CrossRef]
- Buchko, G.W.; Ni, S.; Thrall, B.D.; Kennedy, M.A. Structural features of the minimal DNA binding domain (M98-F219) of human nucleotide excision repair protein XPA. Nucleic Acids Res. 1998, 26, 2779–2788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilton, B.; Shkriabai, N.; Musich, P.R.; Kvaratskhelia, M.; Shell, S.; Zou, Y. A new structural insight into XPA-DNA interactions. Biosci. Rep. 2014, 34, e00162. [Google Scholar] [CrossRef] [PubMed]
- Sugitani, N.; Shell, S.M.; Soss, S.E.; Chazin, W.J. Redefining the DNA-binding domain of human XPA. J. Am. Chem. Soc. 2014, 136, 10830–10833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugitani, N.; Voehler, M.W.; Roh, M.S.; Topolska-Wos, A.M.; Chazin, W.J. Analysis of DNA binding by human factor xeroderma pigmentosum complementation group A (XPA) provides insight into its interactions with nucleotide excision repair substrates. J. Biol. Chem. 2017, 292, 16847–16857. [Google Scholar] [CrossRef] [Green Version]
- Missura, M.; Buterin, T.; Hindges, R.; Hubscher, U.; Kasparkova, J.; Brabec, V.; Naegeli, H. Double-check probing of DNA bending and unwinding by XPA-RPA: An architectural function in DNA repair. EMBO J. 2001, 20, 3554–3564. [Google Scholar] [CrossRef]
- Camenisch, U.; Dip, R.; Schumacher, S.B.; Schuler, B.; Naegeli, H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat. Struct. Mol. Biol. 2006, 13, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Roginskaya, M.; Colis, L.C.; Basu, A.K.; Shell, S.M.; Liu, Y.; Musich, P.R.; Harris, C.M.; Harris, T.M.; Zou, Y. Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3'- and/or 5'-ssDNA branches. Biochemistry 2006, 45, 15921–15930. [Google Scholar] [CrossRef] [Green Version]
- Krasikova, Y.S.; Rechkunova, N.I.; Maltseva, E.A.; Lavrik, O.I. RPA and XPA interaction with DNA structures mimicking intermediates of the late stages in nucleotide excision repair. PLoS ONE 2018, 13, e0190782. [Google Scholar] [CrossRef] [Green Version]
- Beckwitt, E.C.; Jang, S.; Carnaval Detweiler, I.; Kuper, J.; Sauer, F.; Simon, N.; Bretzler, J.; Watkins, S.C.; Carell, T.; Kisker, C.; et al. Single molecule analysis reveals monomeric XPA bends DNA and undergoes episodic linear diffusion during damage search. Nat. Commun. 2020, 11, 1356. [Google Scholar] [CrossRef]
- Wakasugi, M.; Kasashima, H.; Fukase, Y.; Imura, M.; Imai, R.; Yamada, S.; Cleaver, J.E.; Matsunaga, T. Physical and functional interaction between DDB and XPA in nucleotide excision repair. Nucleic Acids Res. 2009, 37, 516–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blessing, C.; Apelt, K.; van den Heuvel, D.; Gonzalez-Leal, C.; Rother, M.B.; van der Woude, M.; González-Prieto, R.; Yifrach, A.; Parnas, A.; Shah, R.G.; et al. XPC-PARP complexes engage the chromatin remodeler ALC1 to catalyze global genome DNA damage repair. Nat. Commun. 2022, 13, 4762. [Google Scholar] [CrossRef] [PubMed]
- Schärer, O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunick, C.G.; Miller, M.R.; Fuller, B.E.; Fanning, E.; Chazin, W.J. Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry 2006, 45, 14965–14979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasikova, Y.S.; Rechkunova, N.I.; Maltseva, E.A.; Petruseva, I.O.; Silnikov, V.N.; Zatsepin, T.S.; Oretskaya, T.S.; Schärer, O.D.; Lavrik, O.I. Interaction of nucleotide excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA. Biochemistry 2008, 73, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Tsutakawa, S.E.; Tsai, C.L.; Yan, C.; Bralić, A.; Chazin, W.J.; Hamdan, S.M.; Schärer, O.D.; Ivanov, I.; Tainer, J.A. Envisioning how the prototypic molecular machine TFIIH functions in transcription initiation and DNA repair. DNA Repair 2020, 96, 102972. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Mu, D.; Reardon, J.T.; Sancar, A. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J. Biol. Chem. 1995, 270, 4896–4902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Wold, M.S. Replication protein A: Single-stranded DNA's first responder: Dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 2014, 36, 1156–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wold, M.S. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997, 66, 61–92. [Google Scholar] [CrossRef] [PubMed]
- Krasikova, Y.S.; Rechkunova, N.I.; Lavrik, O.I. Replication protein A as a major eukaryotic single-stranded DNA-binding protein and its role in DNA repair. Mol. Biol. 2016, 50, 735–750. [Google Scholar] [CrossRef]
- Topolska-Woś, A.M.; Sugitani, N.; Cordoba, J.J.; Le Meur, K.V.; Le Meur, R.A.; Kim, H.S.; Yeo, J.E.; Rosenberg, D.; Hammel, M.; Schärer, O.D.; et al. A key interaction with RPA orients XPA in NER complexes. Nucleic Acids Res. 2020, 48, 2173–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Kim, H.S.; D'Souza, A.; Gallagher, K.; Jeong, E.; Topolska-Wós, A.; Ogorodnik Le Meur, K.; Tsai, C.L.; Tsai, M.S.; Kee, M.; et al. Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair. Proc. Natl. Acad. Sci. USA 2022, 119, e2207408119. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Elledge, S.J.; Peterson, C.A.; Bales, E.S.; Legerski, R.J. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. USA 1994, 91, 5012–5016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.H.; Sancar, A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 5017–5021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsodikov, O.V.; Ivanov, D.; Orelli, B.; Staresincic, L.; Shoshani, I.; Oberman, R.; Schärer, O.D.; Wagner, G.; Ellenberger, T. Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J. 2007, 26, 4768–4776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croteau, D.L.; Peng, Y.; Van Houten, B. DNA repair gets physical: Mapping an XPA-binding site on ERCC1. DNA Repair 2008, 7, 819–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadda, E. Conformational determinants for the recruitment of ERCC1 by XPA in the nucleotide excision repair (NER) Pathway: Structure and dynamics of the XPA binding motif. Biophys J. 2013, 104, 2503–2511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilljam, K.M.; Feyzi, E.; Aas, P.A.; Sousa, M.M.; Muller, R.; Vagbo, C.B.; Catterall, T.C.; Liabakk, N.B.; Slupphaug, G.; Drablos, F.; et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J. Cell Biol. 2009, 186, 645–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilljam, K.M.; Müller, R.; Liabakk, N.B.; Otterlei, M. Nucleotide excision repair is associated with the replisome and its efficiency depends on a direct interaction between XPA and PCNA. PLoS ONE 2012, 7, e49199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, K.; Uchida, M.; Tagata, R.; Yokoyama, H.; Ishikawa, Y.; Hishiki, A.; Hashimoto, H. Structure of proliferating cell nuclear antigen (PCNA) bound to an APIM peptide reveals the universality of PCNA interaction. Acta Cryst. F Struct. Biol. Commun. 2018, 74, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.G.; Liu, Y.; Mao, L.Y.; Zhang, J.T.; Zou, Y. Dimerization of human XPA and formation of XPA(2)-RPA protein complex. Biochemistry 2002, 41, 13012–13020. [Google Scholar] [CrossRef]
- Rademakers, S.; Volker, M.; Hoogstraten, D.; Nigg, A.L.; Moné, M.J.; Van Zeeland, A.A.; Hoeijmakers, J.H.; Houtsmuller, A.B.; Vermeulen, W. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 2003, 23, 5755–5767. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Sarma, H.; Mattaparthi, V.S.K. Investigation of the probable homo-dimer model of the Xeroderma pigmentosum complementation group A (XPA) protein to represent the DNA-binding core. J. Biomol. Struct. Dyn. 2019, 37, 3322–3336. [Google Scholar] [CrossRef] [PubMed]
- Muniesa-Vargas, A.; Theil, A.F.; Ribeiro-Silva, C.; Vermeulen, W.; Lans, H. XPG: A multitasking genome caretaker. Cell Mol. Life Sci. 2022, 79, 166. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Kang, T.H. Transcriptional and posttranslational regulation of nucleotide excision repair: The guardian of the genome against ultraviolet radiation. Int. J. Mol. Sci. 2016, 17, 1840. [Google Scholar] [CrossRef] [PubMed]
- Rechkunova, N.I.; Maltseva, E.A.; Lavrik, O.I. Post-translational Modifications of Nucleotide Excision Repair Proteins and Their Role in the DNA Repair. Biochemistry 2019, 84, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.M.; Li, Z.; Shkriabai, N.; Kvaratskhelia, M.; Brosey, C.; Serrano, M.A.; Chazin, W.J.; Musich, P.R.; Zou, Y. Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A. J. Biol. Chem. 2009, 284, 24213–24222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.R.; Lee, E.Y. UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle 2009, 8, 655–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitta, M.; Saijo, M.; Kodo, N.; Matsuda, T.; Nakatsu, Y.; Tamai, H.; Tanaka, K. A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res. 2000, 28, 4212–4218. [Google Scholar] [CrossRef] [Green Version]
- Nakatsu, Y.; Asahina, H.; Citterio, E.; Rademakers, S.; Vermeulen, W.; Kamiuchi, S.; Yeo, J.P.; Khaw, M.C.; Saijo, M.; Kodo, N.; et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J. Biol. Chem. 2000, 275, 34931–34937. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I.; Ito, S.; Wada, T.; Hayashida, M.; Lee, L.; Saijo, M.; Nakatsu, Y.; Matsumoto, M.; Matsunaga, T.; Handa, H.; et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J. Biol. Chem. 2008, 283, 940–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.; Qu, D.; Li, Y.; Zhu, B.; Liang, D.; Wei, X.; Tang, W.; Zhang, Q.; Hao, J.; Guo, W.; et al. XAB2 depletion induces intron retention in POLR2A to impair global transcription and promote cellular senescence. Nucleic Acids Res. 2019, 47, 8239–8254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onyango, D.O.; Howard, S.M.; Neherin, K.; Yanez, D.A.; Stark, J.M. Tetratricopeptide repeat factor XAB2 mediates the end resection step of homologous recombination. Nucleic Acids Res. 2016, 44, 5702–5716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulielmaki, E.; Tsekrekou, M.; Batsiotos, N.; Ascensão-Ferreira, M.; Ledaki, E.; Stratigi, K.; Chatzinikolaou, G.; Topalis, P.; Kosteas, T.; Altmüller, J.; et al. The splicing factor XAB2 interacts with ERCC1-XPF and XPG for R-loop processing. Nat. Commun. 2021, 12, 3153. [Google Scholar] [CrossRef] [PubMed]
- Donnio, L.M.; Cerutti, E.; Magnani, C.; Neuillet, D.; Mari, P.O.; Giglia-Mari, G. XAB2 dynamics during DNA damage-dependent transcription inhibition. Elife 2022, 11, e77094. [Google Scholar] [CrossRef] [PubMed]
- Moné, M.J.; Bernas, T.; Dinant, C.; Goedvree, F.A.; Manders, E.M.; Volker, M.; Houtsmuller, A.B.; Hoeijmakers, J.H.; Vermeulen, W.; van Driel, R. In vivo dynamics of chromatin-associated complex formation in mammalian nucleotide excision repair. Proc. Natl. Acad. Sci. USA 2004, 101, 15933–15937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, J.H.; Pavletich, N.P. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 2007, 449, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Mu, H.; Zhao, H.; Ouerfelli, O.; Jeffrey, P.D.; Broyde, S.; Min, J.H. Structure and mechanism of pyrimidine-pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex. Nucleic Acids Res. 2019, 47, 6015–6028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheon, N.Y.; Kim, H.S.; Yeo, J.E.; Schärer, O.D.; Lee, J.Y. Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 2019, 47, 8337–8347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, D.; Mu, H.; Tavakoli, A.; Dai, Q.; Chen, X.; Chakraborty, S.; He, C.; Ansari, A.; Broyde, S.; Min, J.H. Tethering- facilitated DNA 'opening' and complementary roles of β-hairpin motifs in the Rad4/XPC DNA damage sensor protein. Nucleic Acids Res. 2020, 48, 12348–12364. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Silva, C.; Sabatella, M.; Helfricht, A.; Marteijn, J.A.; Theil, A.F.; Vermeulen, W.; Lans, H. Ubiquitin and TFIIH-stimulated DDB2 dissociation drives DNA damage handover in nucleotide excision repair. Nat. Commun. 2020, 11, 4868. [Google Scholar] [CrossRef]
- Van Eeuwen, T.; Shim, Y.; Kim, H.J.; Zhao, T.; Basu, S.; Garcia, B.A.; Kaplan, C.D.; Min, J.H.; Murakami, K. Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair. Nat. Commun. 2021, 12, 3338. [Google Scholar] [CrossRef] [PubMed]
- Alemasova, E.E.; Lavrik, O.I. Poly(ADP-ribosyl)ation by PARP1: Reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019, 47, 3811–3827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakazawa, Y.; Hara, Y.; Oka, Y.; Komine, O.; van den Heuvel, D.; Guo, C.; Daigaku, Y.; Isono, M.; He, Y.; Shimada, M.; et al. Ubiquitination of DNA Damage-Stalled RNAPII Promotes Transcription-Coupled Repair. Cell 2020, 180, 1228–1244. [Google Scholar] [CrossRef] [PubMed]
- Son, K.; Schärer, O.D. Repair, Removal, and Shutdown: It All Hinges on RNA Polymerase II Ubiquitylation. Cell 2020, 180, 1039–1041. [Google Scholar] [CrossRef] [PubMed]
- Van der Weegen, Y.; Golan-Berman, H.; Mevissen, T.E.T.; Apelt, K.; González-Prieto, R.; Goedhart, J.; Heilbrun, E.E.; Vertegaal, A.C.O.; van den Heuvel, D.; Walter, J.C.; et al. The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II. Nat. Commun. 2020, 11, 2104. [Google Scholar] [CrossRef] [PubMed]
- Tsutakawa, S.E.; Sarker, A.H.; Ng, C.; Arvai, A.S.; Shin, D.S.; Shih, B.; Jiang, S.; Thwin, A.C.; Tsai, M.S.; Willcox, A.; et al. Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. Proc. Natl. Acad. Sci. USA 2020, 117, 14127–14138. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yan, C.; Fang, J.; Inouye, C.; Tjian, R.; Ivanov, I.; Nogales, E. Near-atomic resolution visualization of human transcription promoter opening. Nature 2016, 533, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Schilbach, S.; Hantsche, M.; Tegunov, D.; Dienemann, C.; Wigge, C.; Urlaub, H.; Cramer, P. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 2017, 551, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, W.; Iltis, I.; Radu, L.; Braun, C.; Maglott-Roth, A.; Giraudon, C.; Egly, J.M.; Poterszman, A. ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc. Natl. Acad. Sci. USA 2013, 110, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Greber, B.J.; Nguyen, T.H.D.; Fang, J.; Afonine, P.V.; Adams, P.D.; Nogales, E. The cryo-electron microscopy structure of human transcription factor IIH. Nature 2017, 549, 414–417. [Google Scholar] [CrossRef] [Green Version]
- Greber, B.J.; Toso, D.B.; Fang, J.; Nogales, E. The complete structure of the human TFIIH core complex. Elife 2019, 8, e44771. [Google Scholar] [CrossRef]
- Dueva, R.; Iliakis, G. Replication protein A: A multifunctional protein with roles in DNA replication, repair and beyond. NAR Cancer 2020, 2, zcaa022. [Google Scholar] [CrossRef]
- Hass, C.S.; Lam, K.; Wold, M.S. Repair-specific functions of replication protein A. J. Biol. Chem. 2012, 287, 3908–3918. [Google Scholar] [CrossRef] [Green Version]
- Kolpashchikov, D.M.; Khodyreva, S.N.; Khlimankov, D.Y.; Wold, M.S.; Favre, A.; Lavrik, O.I. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 2001, 29, 373–379. [Google Scholar] [CrossRef]
- Fan, J.; Pavletich, N.P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012, 26, 2337–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, L.A.; Bessho, M.; Wakasugi, M.; Matsunaga, T.; Bessho, T. Role of interaction of XPF with RPA in nucleotide excision repair. J. Mol. Biol. 2011, 413, 337–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagbemi, A.F.; Orelli, B.; Schärer, O.D. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair 2011, 10, 722–729. [Google Scholar] [CrossRef] [Green Version]
- Staresincic, L.; Fagbemi, A.F.; Enzlin, J.H.; Gourdin, A.M.; Wijgers, N.; Dunand-Sauthier, I.; Giglia-Mari, G.; Clarkson, S.G.; Vermeulen, W.; Schärer, O.D. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009, 28, 1111–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, T.; Hanaoka, F.; Egly, J.M. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003, 22, 5293–5303. [Google Scholar] [CrossRef] [PubMed]
- Mocquet, V.; Laine, J.P.; Riedl, T.; Yajin, Z.; Lee, M.Y.; Egly, J.M. Sequential recruitment of the repair factors during NER: The role of XPG in initiating the resynthesis step. EMBO J. 2008, 27, 155–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overmeer, R.M.; Moser, J.; Volker, M.; Kool, H.; Tomkinson, A.E.; van Zeeland, A.A.; Mullenders, L.H.; Fousteri, M. Replication protein A safeguards genome integrity by controlling NER incision events. J. Cell Biol. 2011, 192, 401–415. [Google Scholar] [CrossRef] [Green Version]
- Moser, J.; Kool, H.; Giakzidis, I.; Caldecott, K.; Mullenders, L.H.; Fousteri, M. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol. Cell 2007, 27, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Kemp, M.G.; Gaddameedhi, S.; Choi, J.H.; Hu, J.; Sancar, A. DNA repair synthesis and ligation affect the processing of excised oligonucleotides generated by human nucleotide excision repair. J. Biol. Chem. 2014, 289, 26574–26583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemp, M.G.; Reardon, J.T.; Lindsey-Boltz, L.A.; Sancar, A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J. Biol. Chem. 2012, 287, 22889–22899. [Google Scholar] [CrossRef] [Green Version]
- Köberle, B.; Roginskaya, V.; Wood, R.D. XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair 2006, 5, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Araújo, S.J.; Nigg, E.A.; Wood, R.D. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 2001, 21, 2281–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luijsterburg, M.S.; von Bornstaedt, G.; Gourdin, A.M.; Politi, A.Z.; Moné, M.J.; Warmerdam, D.O.; Goedhart, J.; Vermeulen, W.; van Driel, R.; Höfer, T. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 2010, 189, 445–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seroussi, E.; Lavi, S. Replication protein A is the major single-stranded DNA binding protein detected in mammalian cell extracts by gel retardation assays and UV cross-linking of long and short single-stranded DNA molecules. J. Biol. Chem. 1993, 268, 7147–7154. [Google Scholar] [CrossRef]
- Köberle, B.; Roginskaya, V.; Zima, K.S.; Masters, J.R.; Wood, R.D. Elevation of XPA protein level in testis tumor cells without increasing resistance to cisplatin or UV radiation. Mol. Carcinog. 2008, 47, 580–586. [Google Scholar] [CrossRef]
- Kang, T.-H.; Reardon, J.T.; Kemp, M.; Sancar, A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl. Acad. Sci. USA 2009, 106, 2864–2867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, T.-H.; Lindsey-Boltz, L.A.; Reardon, J.T.; Sancar, A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2010, 107, 4890–4895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaddameedhi, S.; Selby, C.P.; Kaufmann, W.K.; Smart, R.C.; Sancar, A. Control of skin cancer by the circadian rhythm. Proc. Natl. Acad. Sci. USA 2011, 108, 18790–18795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, T.-H.; Reardon, J.T.; Sancar, A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2010, 39, 3176–3187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cleaver, J.E.; Charles, W.C.; McDowell, M.L.; Sadinski, W.J.; Mitchell, D.L. Overexpression of the XPA repair gene increases resistance to ultraviolet radiation in human cells by selective repair of DNA damage. Cancer Res. 1995, 55, 6152–6160. [Google Scholar] [PubMed]
- Muotri, A.R.; Marchetto, M.C.; Suzuki, M.F.; Okazaki, K.; Lotfi, C.F.; Brumatti, G.; Amarante-Mendes, G.P.; Menck, C.F. Low amounts of the DNA repair XPA protein are sufficient to recover UV-resistance. Carcinogenesis 2002, 23, 1039–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musich, P.R.; Li, Z.; Shell, S.M.; Zou, Y. XPA is primarily cytoplasmic but is transported into the nucleus upon UV damage in a cell cycle dependent manner. DNA Repair 2017, 60, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shell, S.M.; Liu, Y.; Zou, Y. ATR-dependent checkpoint modulates XPA nuclear import in response to UV irradiation. Oncogene 2007, 26, 757–764. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Musich, P.R.; Cartwright, B.M.; Wang, H.; Zou, Y. UV-induced nuclear import of XPA is mediated by importin-α4 in an ATR-dependent manner. PLoS ONE 2013, 8, e68297. [Google Scholar] [CrossRef] [Green Version]
- Wood, R.D.; Manandhar, M.; Lowery, M.; Boulware, K.S.; Lin, K.; Lu, Y. Response to “XPA is primarily cytoplasmic but is transported into the nucleus upon UV damage”. DNA Repair 2018, 62, 30–31. [Google Scholar] [CrossRef] [PubMed]
- Auclair, Y.; Rouget, R.; Affar, E.B.; Drobetsky, E.A. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc. Natl. Acad. Sci. USA 2008, 105, 17896–17901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Musich, P.R.; Serrano, M.A.; Dong, Z.; Zou, Y. XPA-mediated regulation of global nucleotide excision repair by ATR Is p53-dependent and occurs primarily in S-phase. PLoS ONE 2011, 6, e28326. [Google Scholar] [CrossRef] [Green Version]
- Musich, P.R.; Li, Z.; Zou, Y. Xeroderma PigmentosaGroup A (XPA), Nucleotide Excision Repair and Regulation by ATR in Response to Ultraviolet Irradiation. Adv. Exp. Med. Biol. 2017, 996, 41–54. [Google Scholar]
- Scott, I.C.; Halila, R.; Jenkins, J.M.; Mehan, S.; Apostolou, S.; Winqvist, R.; Callen, D.F.; Prockop, D.J.; Peltonen, L.; Kadler, K.E. Molecular cloning, expression and chromosomal localization of a human gene encoding a 33 kDa putative metallopeptidase (PRSM1). Gene 1996, 174, 135–143. [Google Scholar] [CrossRef]
- Barr, F.A.; Puype, M.; Vandekerckhove, J.; Warren, G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 1997, 91, 253–262. [Google Scholar] [CrossRef] [Green Version]
- Cox, K.H.; Takahashi, J.S. Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 2019, 63, R93–R102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, T.-H. Circadian Rhythm of NER and ATR Pathways. Biomolecules 2021, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Dakup, P.P.; Porter, K.I.; Little, A.A.; Gajula, R.P.; Zhang, H.; Skornyakov, E.; Kemp, M.G.; Van Dongen, H.P.A.; Gaddameedhi, S. The circadian clock regulates cisplatin-induced toxicity and tumor regression in melanoma mouse and human models. Oncotarget 2018, 9, 14524–14538. [Google Scholar] [CrossRef] [Green Version]
- Dakup, P.; Gaddameedhi, S. Impact of the Circadian Clock on UV-Induced DNA Damage Response and Photocarcinogenesis. Photochem Photobiol. 2017, 93, 296–303. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.H.; Park, J.M.; Leem, S.H.; Kang, T.H. Coordinated regulation of XPA stability by ATR and HERC2 during nucleotide excision repair. Oncogene 2014, 33, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Pruteanu, M.; Baker, T.A. Controlled degradation by ClpXP protease tunes the levels of the excision repair protein UvrA to the extent of DNA damage. Mol. Microbiol. 2009, 71, 912–924. [Google Scholar] [CrossRef]
- Rey, S.; Schito, L.; Koritzinsky, M.; Wouters, B.G. Molecular Targeting of Hypoxia in Radiotherapy. Adv. Drug Deliv. 2017, 109, 45–62. [Google Scholar] [CrossRef]
- Barker, H.E.; Paget, J.T.E.; Khan, A.A.; Harrington, K.J. The Tumour Microenvironment After Radiotherapy: Mechanisms of Resistance and Recurrence. Nat. Rev. Cancer 2015, 15, 409–425. [Google Scholar] [CrossRef] [Green Version]
- Rakotomalala, A.; Escande, A.; Furlan, A.; Meignan, S.; Lartigau, E. Hypoxia in Solid Tumors: How Low Oxygenation Impacts the "Six Rs" of Radiotherapy. Front. Endocrinol. 2021, 12, 742215. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bernauer, A.M.; Yingling, C.M.; Belinsky, S.A. HIF1α regulated expression of XPA contributes to cisplatin resistance in lung cancer. Carcinogenesis 2012, 33, 1187–1192. [Google Scholar] [CrossRef]
- Rezvani, H.R.; Mahfouf, W.; Ali, N.; Chemin, C.; Ged, C.; Kim, A.L.; de Verneuil, H.; Taïeb, A.; Bickers, D.R.; Mazurier, F. Hypoxia-inducible factor-1alpha regulates the expression of nucleotide excision repair proteins in keratinocytes. Nucleic Acids Res. 2010, 38, 797–809. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Gong, L.; Peng, Y.; Li, L.; Liu, G. Enhancer-bound Nrf2 licenses HIF-1α transcription under hypoxia to promote cisplatin resistance in hepatocellular carcinoma cells. Aging 2020, 13, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair 2015, 36, 122–136. [Google Scholar] [CrossRef]
- Liang, G.; He, Z. High Mobility Group Proteins in Sepsis. Front. Immunol. 2022, 13, 911152. [Google Scholar] [CrossRef]
- Adair, J.E.; Kwon, Y.; Dement, G.A.; Smerdon, M.J.; Reeves, R. Inhibition of nucleotide excision repair by high mobility group protein HMGA1. J. Biol. Chem. 2005, 280, 32184–32192. [Google Scholar] [CrossRef] [Green Version]
- Adair, J.E.; Maloney, S.C.; Dement, G.A.; Wertzler, K.J.; Smerdon, M.J.; Reeves, R. High-mobility group A1 proteins inhibit expression of nucleotide excision repair factor xeroderma pigmentosum group A. Cancer Res. 2007, 67, 6044–6052. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, C.; Chen, K.; Chen, Z.; Sun, Z.; Zhang, Z.; Ding, D.; Ren, S.; Zuo, Y. The clinical significance of DC-SIGN and DC-SIGNR, which are novel markers expressed in human colon cancer. PLoS ONE 2014, 9, e114748. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, H.; Su, L.; Yang, P.; Xin, Z.; Zou, J.; Ren, S.; Zuo, Y. Low expression of dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin-related protein in lung cancer and significant correlations with brain metastasis and natural killer cells. Mol. Cell Biochem. 2015, 407, 151–160. [Google Scholar] [CrossRef]
- Na, H.; Liu, X.; Li, X.; Zhang, X.; Wang, Y.; Wang, Z.; Yuan, M.; Zhang, Y.; Ren, S.; Zuo, Y. Novel roles of DC-SIGNR in colon cancer cell adhesion, migration, invasion, and liver metastasis. J. Hematol. Oncol. 2017, 10, 28. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, Q.; Zhang, M.; Yuan, M.; Wang, Z.; Zhang, J.; Zhou, X.; Zhang, Y.; Lin, F.; Na, H.; et al. DC—SIGNR by influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4 in gastric cancer liver metastasis. Mol. Cancer 2017, 16, 78. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wei, H.; Fan, L.; Fang, M.; He, X.; Lu, B.; Pang, Z. CLEC4s as Potential Therapeutic Targets in Hepatocellular Carcinoma Microenvironment. Front. Cell Dev. Biol. 2021, 9, 681372. [Google Scholar] [CrossRef]
- Tan, L.M.; Li, X.; Qiu, C.F.; Zhu, T.; Hu, C.P.; Yin, J.Y.; Zhang, W.; Zhou, H.H.; Liu, Z.Q. CLEC4M is associated with poor prognosis and promotes cisplatin resistance in NSCLC patients. J. Cancer 2019, 10, 6374–6383. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Ren, S.; Zuo, Y. DC-SIGN, DC-SIGNR and LSECtin: C-type lectins for infection. Int. Rev. Immunol. 2014, 33, 54–66. [Google Scholar] [CrossRef]
- Khoo, U.S.; Chan, K.Y.; Chan, V.S.; Lin, C.L. DC-SIGN and L-SIGN: The SIGNs for infection. J. Mol. Med. 2008, 86, 861–874. [Google Scholar] [CrossRef]
- Luo, L.; Chen, L.; Ke, K.; Zhao, B.; Wang, L.; Zhang, C.; Wang, F.; Liao, N.; Zheng, X.; Liu, X.; et al. High expression levels of CLEC4M indicate poor prognosis in patients with hepatocellular carcinoma. Oncol. Lett. 2020, 19, 1711–1720. [Google Scholar] [CrossRef]
- Yu, Q.; Gao, K. CLEC4M overexpression inhibits progression and is associated with a favorable prognosis in hepatocellular carcinoma. Mol. Med. Rep. 2020, 22, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Lindsey-Boltz, L.A. Bringing It All Together: Coupling Excision Repair to the DNA Damage Checkpoint. Photochem. Photobiol. 2016, 93, 238–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maréchal, A.; Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015, 25, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokhrel, N.; Caldwell, C.C.; Corless, E.I.; Tillison, E.A.; Tibbs, J.; Jocic, N.; Tabei, S.M.A.; Wold, M.S.; Spies, M.; Antony, E. Dynamics and selective remodeling of the DNA-binding domains of RPA. Nat. Struct. Mol. Biol. 2019, 26, 129–136. [Google Scholar] [CrossRef]
- Yang, X.H.; Zou, L. Recruitment of ATR-ATRIP, Rad17, and 9-1-1 complexes to DNA damage. Methods Enzymol. 2006, 409, 118–131. [Google Scholar] [CrossRef]
- Wu, X.; Shell, S.M.; Zou, Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene 2005, 24, 4728–4735. [Google Scholar] [CrossRef] [Green Version]
- Ray, A.; Milum, K.; Battu, A.; Wani, G.; Wani, A.A. NER initiation factors, DDB2 and XPC, regulate UV radiation response by recruiting ATR and ATM kinases to DNA damage sites. DNA Repair 2013, 12, 273–283. [Google Scholar] [CrossRef] [Green Version]
- Lindsey-Boltz, L.A.; Kemp, M.G.; Reardon, J.T.; DeRocco, V.; Iyer, R.R.; Modrich, P.; Sancar, A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 2014, 289, 5074–5082. [Google Scholar] [CrossRef] [Green Version]
- Ray, A.; Blevins, C.; Wani, G.; Wani, A.A. ATR- and ATM-Mediated DNA damage response is dependent on excision repair assembly during G1 but bot in S phase of cell cycle. PLoS ONE 2016, 11, e0159344. [Google Scholar] [CrossRef] [Green Version]
- Sivasubramaniam, S.; Sun, X.M.; Pan, Y.R.; Wang, S.H.; Lee, E.Y.H.P. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA and CHK1. Genes Dev. 2008, 22, 587–600. [Google Scholar] [CrossRef]
- Hergovich, A.; Cornils, H.; Hemmings, B.A. Mammalian NDR protein kinases: From regulation to a role in centrosome duplication. Biochim. Biophys. Acta 2008, 1784, 3–15. [Google Scholar] [CrossRef]
- Park, J.M.; Choi, J.Y.; Yi, J.M.; Chung, J.W.; Leem, S.H.; Koh, S.S.; Kang, T.H. NDR1 modulates the UV-induced DNA-damage checkpoint and nucleotide excision repair. Biochem. Biophys. Res. Commun. 2015, 461, 543–548. [Google Scholar] [CrossRef]
- Nishi, R.; Okuda, Y.; Watanabe, E.; Mori, T.; Iwai, S.; Masutani, C.; Sugasawa, K.; Hanaoka, F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 2005, 25, 5664–5674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasikova, Y.S.; Rechkunova, N.I.; Maltseva, E.A.; Craescu, C.T.; Petruseva, I.O.; Lavrik, O.I. Influence of centrin 2 on the interaction of nucleotide excision repair factors with damaged DNA. Biochemistry 2012, 77, 346–353. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, Y.; Yang, B. Phosphorylation promotes the endonuclease-like activity of human centrin 2. RSC Adv. 2022, 12, 21892–21903. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Slattery, S.D.; Moon, S.H.; Darlington, Y.F.; Lu, X.B.; Donehower, L.A. The oncogenic phosphatase WIP1 negatively regulates nucleotide excision repair. DNA Repair 2010, 9, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Luo, J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol. Cell 2010, 39, 247–258. [Google Scholar] [CrossRef]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef] [Green Version]
- Dobbin, M.M.; Madabhushi, R.; Pan, L.; Chen, Y.; Kim, D.; Gao, J.; Ahanonu, B.; Pao, P.-C.; Qiu, Y.; Zhao, Y.; et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 2013, 16, 1008–1015. [Google Scholar] [CrossRef] [Green Version]
- Donninger, H.; Clark, J.; Rinaldo, F.; Nelson, N.; Barnoud, T.; Schmidt, M.L.; Hobbing, K.R.; Vos, M.D.; Sils, B.; Clark, G.J. The RASSF1A tumor suppressor regulates XPA-mediated DNA repair. Mol. Cell. Biol. 2015, 35, 277–287. [Google Scholar] [CrossRef] [PubMed]
- García-Gutiérrez, L.; McKenna, S.; Kolch, W.; Matallanas, D. RASSF1A Tumour Suppressor: Target the Network for Effective Cancer Therapy. Cancers 2020, 12, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, M.; El-Kalla, M.; Baksh, S. RASSF1 Polymorphisms in Cancer. Mol. Biol. Int. 2012, 2012, 365213. [Google Scholar] [CrossRef] [PubMed]
- Pefani, D.E.; Latusek, R.; Pires, I.; Grawenda, A.M.; Yee, K.S.; Hamilton, G.; van der Weyden, L.; Esashi, F.; Hammond, E.M.; O'Neill, E. RASSF1A-LATS1 signalling stabilizes replication forks by restricting CDK2-mediated phosphorylation of BRCA2. Nat. Cell Biol. 2014, 16, 962–971. [Google Scholar] [CrossRef] [Green Version]
- Vichalkovski, A.; Gresko, E.; Cornils, H.; Hergovich, A.; Schmitz, D.; Hemmings, B.A. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr. Biol. 2008, 18, 1889–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Wang, J.; Liu, T. UV-Induced RPA1 Acetylation Promotes Nucleotide Excision Repair. Cell Rep. 2017, 20, 2010–2025. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Geng, R.; Guo, X.; Yuan, R.; Zhou, X.; Zhong, Y.; Huo, Y.; Zhou, M.; Shen, Q.; Li, Y.; et al. PCAF/GCN5-mediated acetylation of RPA1 promotes nucleotide excision repair. Cell Rep. 2017, 20, 1997–2009. [Google Scholar] [CrossRef] [Green Version]
- Ming, M.; Shea, C.R.; Guo, X.; Li, X.; Soltani, K.; Han, W.; He, Y.Y. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc. Natl. Acad. Sci USA 2010, 107, 22623–22628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, M.; Soltani, K.; Shea, C.R.; Li, X.; He, Y.Y. Dual role of SIRT1 in UVB-induced skin tumorigenesis. Oncogene 2015, 34, 357–363. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.Y.; Park, J.M.; Yi, J.M.; Leem, S.H.; Kang, T.H. Enhanced nucleotide excision repair capacity in lung cancer cells by preconditioning with DNA-damaging agents. Oncotarget 2015, 6, 22575–22586. [Google Scholar] [CrossRef] [Green Version]
- Yousafzai, N.A.; Jin, H.; Ullah, M.; Wang, X. Recent advances of SIRT1 and implications in chemotherapeutics resistance in cancer. Am. J. Cancer Res. 2021, 11, 5233–5248. [Google Scholar]
- Mangerich, A.; Burkle, A. Pleiotropic cellular functions of PARP1 in longevity and aging: Genome maintenance meets inflammation. Oxidative Med. Cell. Longev. 2012, 2012, 321653. [Google Scholar] [CrossRef] [PubMed]
- Robu, M.; Shah, R.G.; Petitclerc, N.; Brind’Amour, J.; Kandan-Kulangara, F.; Shah, G.M. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc. Natl. Acad. Sci. USA 2013, 110, 1658–1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khodyreva, S.N.; Lavrik, O.I. Poly(ADP-ribose) polymerase 1 as a key regulator of DNA repair. Mol. Biol. 2016, 50, 580–595. [Google Scholar] [CrossRef]
- Liu, C.; Vyas, A.; Kassab, M.A.; Singh, A.K.; Yu, X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 2017, 45, 8129–8141. [Google Scholar] [CrossRef]
- Rechkunova, N.I.; Krasikova, Y.S.; Lavrik, O.I. Interactome of Base and Nucleotide Excision DNA Repair Systems. Mol. Biol. 2021, 55, 181–193. [Google Scholar] [CrossRef]
- Krietsch, J.; Rouleau, M.; Pic, E.; Ethier, C.; Dawson, T.M.; Dawson, V.L.; Masson, J.Y.; Poirier, G.G.; Gagne, J.P. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Asp. Med. 2013, 34, 1066–1087. [Google Scholar] [CrossRef] [PubMed]
- Alemasova, E.E.; Lavrik, O.I. At the Interface of Three Nucleic Acids: The Role of RNA-Binding Proteins and Poly(ADP-ribose) in DNA Repair. Acta Nat. 2017, 9, 4–16. [Google Scholar] [CrossRef]
- Purohit, N.K.; Robu, M.; Shah, R.G.; Geacintov, N.E.; Shah, G.M. Characterization of the interactions of PARP-1 with UV-damaged DNA in vivo and in vitro. Sci. Rep. 2016, 6, 19020. [Google Scholar] [CrossRef]
- Robu, M.; Shah, R.G.; Purohit, N.K.; Zhou, P.; Naegeli, H.; Shah, G.M. Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, E6847–E6856. [Google Scholar] [CrossRef] [Green Version]
- Robu, M.; Shah, R.G.; Shah, G.M. Methods to Study Intracellular Movement and Localization of the Nucleotide Excision Repair Proteins at the DNA Lesions in Mammalian Cells. Front. Cell Dev. Biol. 2020, 8, 590242. [Google Scholar] [CrossRef]
- King, B.S.; Cooper, K.L.; Liu, K.J.; Hudson, L.G. Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair. J. Biol. Chem. 2012, 287, 39824–39833. [Google Scholar] [CrossRef]
- Fischer, J.M.; Popp, O.; Gebhard, D.; Veith, S.; Fischbach, A.; Beneke, S.; Leitenstorfer, A.; Bergemann, J.; Scheffner, M.; Ferrando-May, E.; et al. Poly(ADP-ribose)-mediated interplay of XPA and PARP1 leads to reciprocal regulation of protein function. FEBS J. 2014, 281, 3625–3641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000, 275, 40974–40980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahrer, J.; Kranaster, R.; Altmeyer, M.; Marx, A.; Burkle, A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007, 35, e143. [Google Scholar] [CrossRef] [PubMed]
- Singatulina, A.S.; Hamon, L.; Sukhanova, M.V.; Desforges, B.; Joshi, V.; Bouhss, A.; Lavrik, O.I.; Pastré, D. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep. 2019, 27, 1809–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolthur-Seetharam, U.; Dantzer, F.; McBurney, M.W.; de Murcia, G.; Sassone-Corsi, P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle 2006, 5, 873–877. [Google Scholar] [CrossRef] [Green Version]
- Pillai, J.B.; Isbatan, A.; Imai, S.; Gupta, M.P. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J. Biol. Chem. 2005, 280, 43121–43130. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Wang, S.Y.; Fleuriel, C.; Leprince, D.; Rocheleau, J.V.; Piston, D.W.; Goodman, R.H. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc. Natl. Acad. Sci. USA 2007, 104, 829–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajamohan, S.B.; Pillai, V.B.; Gupta, M.; Sundaresan, N.R.; Birukov, K.G.; Samant, S.; Hottinger, M.O.; Gupta, M.P. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell. Biol. 2009, 29, 4116–4129. [Google Scholar] [CrossRef] [Green Version]
- Fang, E.F.; Scheibye-Knudsen, M.; Brace, L.E.; Kassahun, H.; SenGupta, T.; Nilsen, H.; Mitchell, J.R.; Croteau, D.L.; Bohr, V.A. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 2014, 157, 882–896. [Google Scholar] [CrossRef] [Green Version]
- Maltseva, E.A.; Krasikova, Y.S.; Sukhanova, M.V.; Rechkunova, N.I.; Lavrik, O.I. Replication protein A as a modulator of the poly(ADP-ribose)polymerase 1 activity. DNA Repair 2018, 72, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Thorslund, T.; von Kobbe, C.; Harrigan, J.A.; Indig, F.E.; Christiansen, M.; Stevnsner, T.; Bohr, V.A. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol. Cell. Biol. 2005, 25, 7625–7636. [Google Scholar] [CrossRef] [Green Version]
- Pines, A.; Vrouwe, M.G.; Marteijn, J.A.; Typas, D.; Luijsterburg, M.S.; Cansoy, M.; Hensbergen, P.; Deelder, A.; de Groot, A.; Matsumoto, S.; et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 2012, 199, 235–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apelt, K.; Lans, H.; Schärer, O.D.; Luijsterburg, M.S. Nucleotide excision repair leaves a mark on chromatin: DNA damage detection in nucleosomes. Cell Mol. Life Sci. 2021, 78, 7925–7942. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef]
- Bowden, N.A. Nucleotide excision repair: Why is it not used to predict response to platinum-based chemotherapy? Cancer Lett. 2014, 346, 163–171. [Google Scholar] [CrossRef]
- Stevens, E.V.; Raffeld, M.; Espina, V.; Kristensen, G.B.; Trope, C.G.; Kohn, E.C.; Davidson, B. Expression of xeroderma pigmentosum A protein predicts improved outcome in metastatic ovarian carcinoma. Cancer 2005, 103, 2313–2319. [Google Scholar] [CrossRef]
- Welsh, C.; Day, R.; McGurk, C.; Masters, J.R.; Wood, R.D.; Koberle, B. Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. Int. J. Cancer 2004, 110, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.; Martinez, J.; Hernandez, C.; Perez-Montiel, D.; Castro, C.; Fabian-Morales, E.; Santibanez, M.; Gonzalez-Barrios, R.; Diaz-Chavez, J.; Andonegui, M.A.; et al. Association between ERCC1 and XPA expression and polymorphisms and the response to cisplatin in testicular germ cell tumours. Br. J. Cancer 2013, 109, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Ballesta, A.; Innominato, P.F.; Dallmann, R.; Rand, D.A.; Lévi, F.A. Systems Chronotherapeutics. Pharmacol. Rev. 2017, 69, 161–199. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krasikova, Y.S.; Lavrik, O.I.; Rechkunova, N.I. The XPA Protein—Life under Precise Control. Cells 2022, 11, 3723. https://doi.org/10.3390/cells11233723
Krasikova YS, Lavrik OI, Rechkunova NI. The XPA Protein—Life under Precise Control. Cells. 2022; 11(23):3723. https://doi.org/10.3390/cells11233723
Chicago/Turabian StyleKrasikova, Yuliya S., Olga I. Lavrik, and Nadejda I. Rechkunova. 2022. "The XPA Protein—Life under Precise Control" Cells 11, no. 23: 3723. https://doi.org/10.3390/cells11233723
APA StyleKrasikova, Y. S., Lavrik, O. I., & Rechkunova, N. I. (2022). The XPA Protein—Life under Precise Control. Cells, 11(23), 3723. https://doi.org/10.3390/cells11233723






