Genome-Wide Association Study Identifies CDKN1A as a Novel Locus Associated with Muscle Fiber Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Participants
2.2.1. The Russian Cohorts
2.2.2. The Japanese Cohorts
2.2.3. The Finnish Cohort
2.3. Evaluation of Muscle Fiber Composition by Immunohistochemistry
2.3.1. Russian Study
2.3.2. Japanese Study
2.3.3. Finnish Study
2.4. Genotyping
2.4.1. Russian Study
2.4.2. Japanese Study
2.4.3. Finnish Study
2.5. Gene Expression Analysis
2.5.1. Total RNA Isolation
2.5.2. RNA Sequencing
2.6. Statistical Analyses
3. Results
3.1. Muscle Fiber Composition and Athlete Status Studies
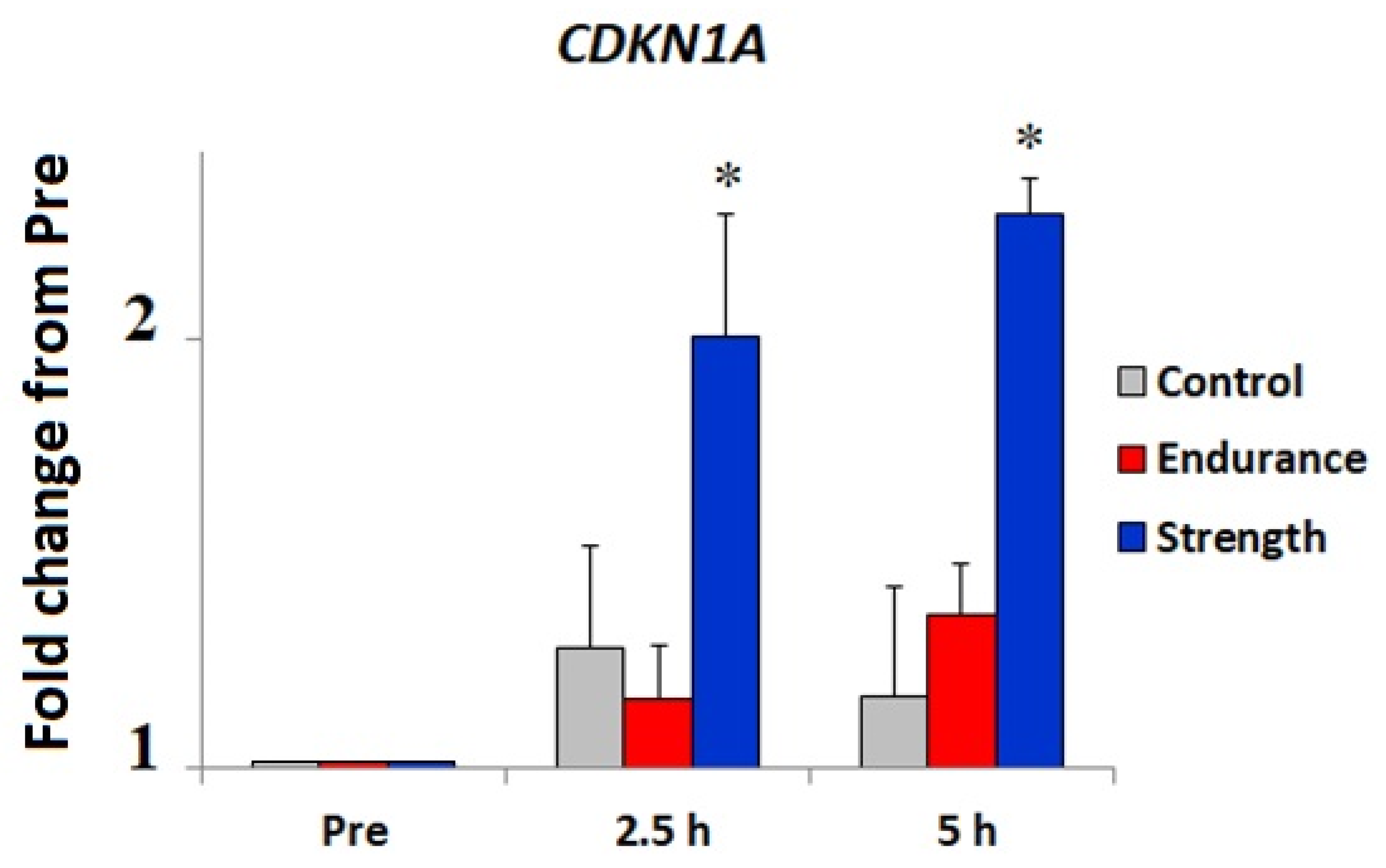
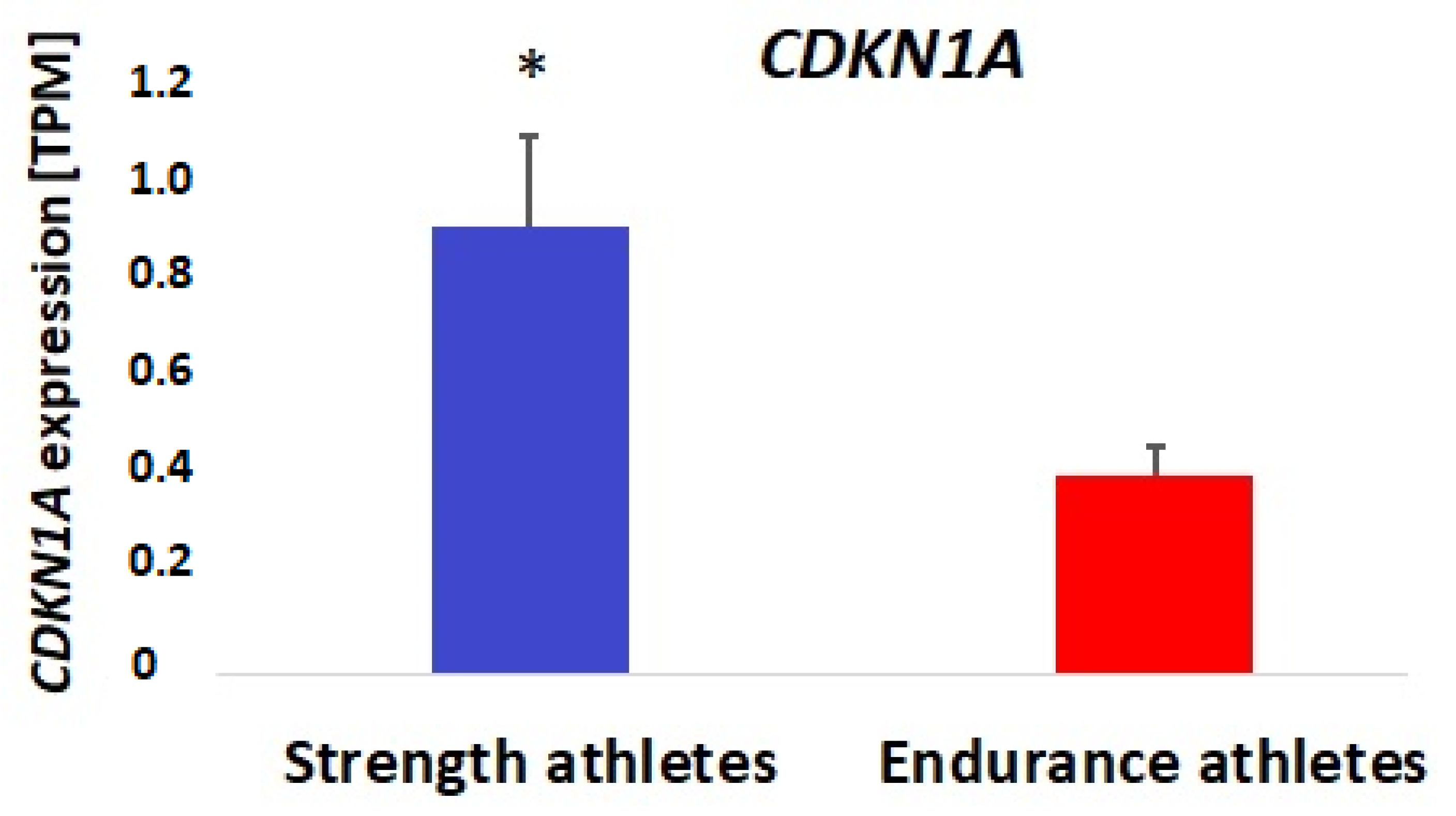
3.2. Bioinformatic and Gene Expression Studies
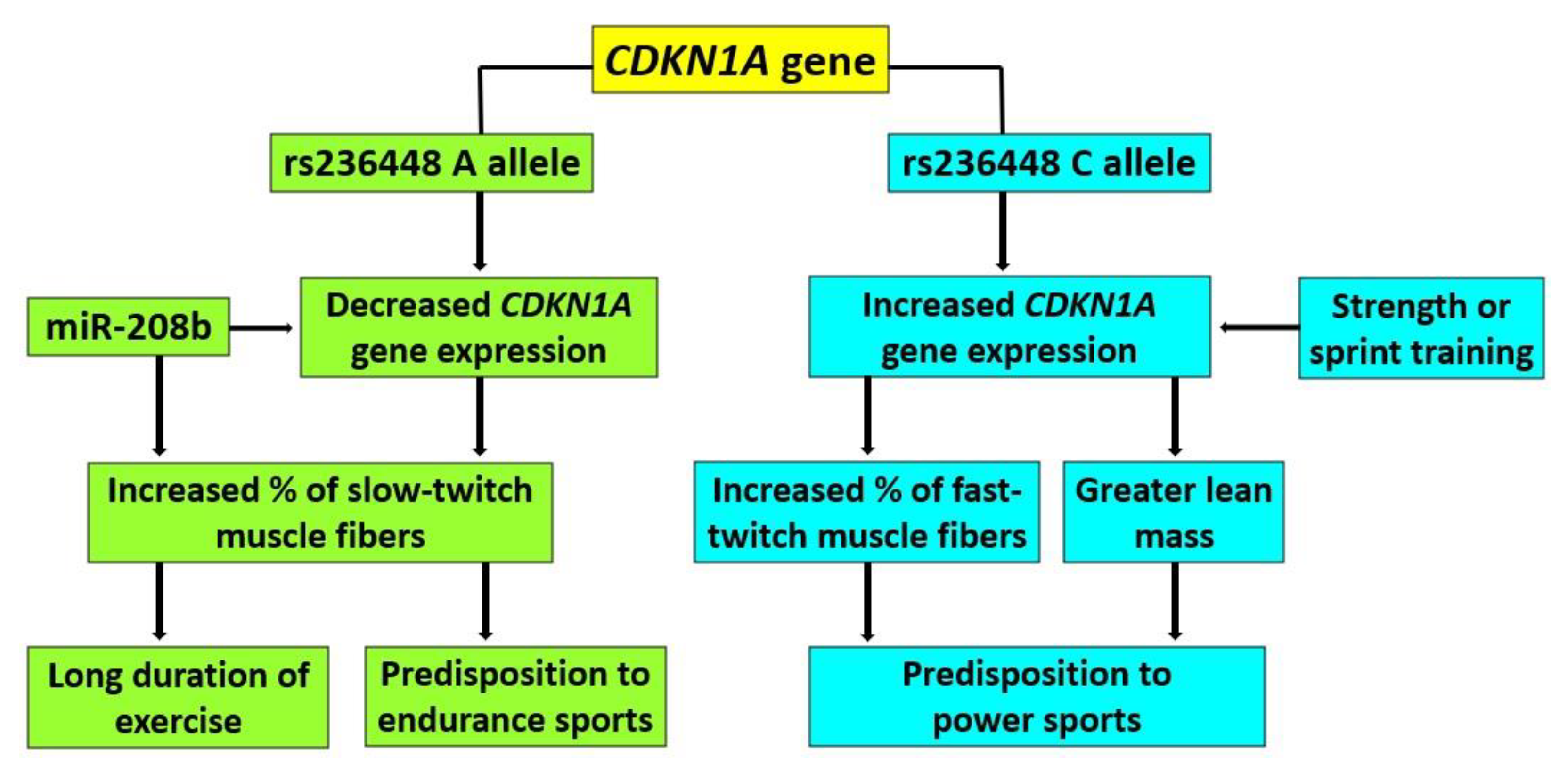
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [Green Version]
- Simoneau, J.A.; Bouchard, C. Human variation in skeletal muscle fiber-type proportion and enzyme activities. Am. J. Physiol. 1989, 257, 567–572. [Google Scholar] [CrossRef]
- Klitgaard, H.; Mantoni, M.; Schiaffino, S.; Ausoni, S.; Gorza, L.; Laurent-Winter, C.; Schnohr, P.; Saltin, B. Function, morphology and protein expression of ageing skeletal muscle, a cross-sectional study of elderly men with different training backgrounds. Acta Physiol. Scand. 1990, 140, 41–54. [Google Scholar] [CrossRef]
- Andersen, J.L.; Schjerling, P.; Saltin, B. Muscle, genes, and athletic performance. Sci. Am. 2000, 283, 48–55. [Google Scholar] [CrossRef]
- Hall, E.C.R.; Lysenko, E.A.; Semenova, E.A.; Borisov, O.V.; Andryushchenko, O.N.; Andryushchenko, L.B.; Vepkhvadze, T.F.; Lednev, E.M.; Zmijewski, P.; Popov, D.V.; et al. Prediction of muscle fiber composition using multiple repetition testing. Biol. Sport 2021, 38, 277–283. [Google Scholar] [CrossRef]
- Hall, E.C.R.; Semenova, E.A.; Borisov, O.V.; Andryushchenko, O.N.; Andryushchenko, L.B.; Zmijewski, P.; Generozov, E.V.; Ahmetov, I.I. Association of muscle fiber composition with health and exercise-related traits in athletes and untrained subjects. Biol. Sport 2021, 38, 659–666. [Google Scholar] [CrossRef]
- Ricoy, J.R.; Encinas, A.R.; Cabello, A.; Madero, S.; Arenas, J. Histochemical study of the vastus lateralis muscle fibre types of athletes. J. Physiol. Biochem. 1998, 54, 41–47. [Google Scholar]
- Zawadowska, B.; Majerczak, J.; Semik, D.; Karasinski, J.; Kolodziejski, L.; Kilarski, W.M.; Duda, K.; Zoladz, J.A. Characteristics of myosin profile in human vastus lateralis muscle in relation to training background. Folia Histochem. Cytobiol. 2004, 42, 181–190. [Google Scholar]
- Andersen, J.L.; Klitgaard, H.; Saltin, B. Myosin heavy chain isoforms in single fibres from m. vastus lateralis of sprinters, influence of training. Acta Physiol. Scand. 1994, 151, 135–142. [Google Scholar] [CrossRef]
- Damer, A.; El Meniawy, S.; McPherson, R.; Wells, G.; Harper, M.E.; Dent, R. Association of muscle fiber type with measures of obesity: A systematic review. Obes. Rev. 2022, 23, e13444. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Vinogradova, O.L.; Williams, A.G. Gene polymorphisms and fiber-type composition of human skeletal muscle. Int. J. Sport Nutr. Exerc. Metab. 2012, 22, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, P.; Trappe, S.; Harber, M.; Creer, A.; Mazzetti, S.; Trappe, T.; Alkner, B.; Tesch, P. Effects of 84-days of bedrest and resistance training on single muscle fibre myosin heavy chain distribution in human vastus lateralis and soleus muscles. Acta Physiol. Scand. 2005, 185, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Bathgate, K.E.; Bagley, J.R.; Jo, E.; Talmadge, R.J.; Tobias, I.S.; Brown, L.E.; Coburn, J.W.; Arevalo, J.A.; Segal, N.L.; Galpin, A.J. Muscle health and performance in monozygotic twins with 30 years of discordant exercise habits. Eur. J. Appl. Physiol. 2018, 118, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, D.L.; Roberts, M.D.; Haun, C.T.; Schoenfeld, B.J. Muscle Fiber Type Transitions with Exercise Training: Shifting Perspectives. Sports 2021, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Simoneau, J.A.; Bouchard, C. Genetic determinism of fiber type proportion in human skeletal muscle. FASEB J. 1995, 9, 1091–1095. [Google Scholar] [CrossRef]
- Zhang, B.; Tanaka, H.; Shono, N.; Miura, S.; Kiyonaga, A.; Shindo, M.; Saku, K. The I allele of the angiotensin-converting enzyme gene is associated with an increased percentage of slow-twitch type I fibers in human skeletal muscle. Clin. Genet. 2003, 63, 139–144. [Google Scholar] [CrossRef]
- Akhmetov, I.I.; Astratenkova, I.V.; Druzhevskaia, A.M.; Komkova, A.I.; Liubaeva, E.V.; Tarakin, P.P.; Shenkman, B.S.; Rogozkin, V.A. The association of gene polymorphisms with the muscle fiber type composition. Ross. Fiziol. Zh. Im. I. M. Sechenova 2006, 92, 883–888. [Google Scholar]
- Kumagai, H.; Tobina, T.; Ichinoseki-Sekine, N.; Kakigi, R.; Tsuzuki, T.; Zempo, H.; Shiose, K.; Yoshimura, E.; Kumahara, H.; Ayabe, M.; et al. Role of selected polymorphisms in determining muscle fiber composition in Japanese men and women. J. Appl. Physiol. 2018, 124, 1377–1384. [Google Scholar] [CrossRef] [Green Version]
- Vincent, B.; De Bock, K.; Ramaekers, M.; Van den Eede, E.; Van Leemputte, M.; Hespel, P.; Thomis, M.A. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol. Genom. 2007, 32, 58–63. [Google Scholar] [CrossRef] [Green Version]
- Ahmetov, I.I.; Druzhevskaya, A.M.; Lyubaeva, E.V.; Popov, D.V.; Vinogradova, O.L.; Williams, A.G. The dependence of preferred competitive racing distance on muscle fibre type composition and ACTN3 genotype in speed skaters. Exp. Physiol. 2011, 96, 1302–1310. [Google Scholar] [CrossRef]
- Mustafina, L.J.; Naumov, V.A.; Cieszczyk, P.; Popov, D.V.; Lyubaeva, E.V.; Kostryukova, E.S.; Fedotovskaya, O.N.; Druzhevskaya, A.M.; Astratenkova, I.V.; Glotov, A.S.; et al. AGTR2 gene polymorphism is associated with muscle fibre composition, athletic status and aerobic performance. Exp. Physiol. 2014, 99, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, J.P.L.F.; Semenova, E.A.; Borisov, O.V.; Kostryukova, E.S.; Vepkhvadze, T.F.; Lysenko, E.A.; Andryushchenko, O.N.; Andryushchenko, L.B.; Lednev, E.M.; Larin, A.K.; et al. The BDNF-Increasing Allele is Associated with Increased Proportion of Fast-Twitch Muscle Fibers, Handgrip Strength, and Power Athlete Status. J. Strength Cond. Res. 2022, 36, 1884–1889. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.; Suraci, B.; Semenova, E.A.; Boulygina, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Borisov, O.V.; Khabibova, S.A.; Larin, A.K.; Pavlenko, A.V.; et al. A genome-wide association study of sprint performance in elite youth football players. J. Strength Cond. Res. 2019, 33, 2344–2351. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, J.; Semenova, E.A.; Zempo, H.; Martins, G.L.; Lancha Junior, A.H.; Miyamoto-Mikami, E.; Kumagai, H.; Tobina, T.; Shiose, K.; Kakigi, R.; et al. Are genome-wide association study identified single-nucleotide polymorphisms associated with sprint athletic status? A replication study with 3 different cohorts. Int. J. Sports Physiol. Perform. 2021, 16, 489–495. [Google Scholar] [CrossRef]
- Guilherme, J.; Egorova, E.S.; Semenova, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Borisov, O.V.; Khabibova, S.A.; Larin, A.K.; Ospanova, E.A.; Pavlenko, A.V.; et al. The A-allele of the FTO gene rs9939609 polymorphism is associated with decreased proportion of slow oxidative muscle fibers and over-represented in heavier athletes. J. Strength Cond. Res. 2019, 33, 691–700. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Hakimullina, A.M.; Lyubaeva, E.V.; Vinogradova, O.L.; Rogozkin, V.A. Effect of HIF1A gene polymorphism on human muscle performance. Bull. Exp. Biol. Med. 2008, 146, 351–353. [Google Scholar] [CrossRef]
- Guilherme, J.P.L.F.; Semenova, E.A.; Larin, A.K.; Yusupov, R.A.; Generozov, E.V.; Ahmetov, I.I. Genomic Predictors of Brisk Walking Are Associated with Elite Sprinter Status. Genes 2022, 13, 1710. [Google Scholar] [CrossRef]
- Kumagai, H.; Natsume, T.; Kim, S.J.; Tobina, T.; Miyamoto-Mikami, E.; Shiose, K.; Ichinoseki-Sekine, N.; Kakigi, R.; Tsuzuki, T.; Miller, B.; et al. The MOTS-c K14Q polymorphism in the mtDNA is associated with muscle fiber composition and muscular performance. Biochim. Biophys. Acta Gen. Subj. 2022, 1866, 130048. [Google Scholar] [CrossRef]
- Yvert, T.; Miyamoto-Mikami, E.; Tobina, T.; Shiose, K.; Kakigi, R.; Tsuzuki, T.; Takaragawa, M.; Ichinoseki-Sekine, N.; Pérez, M.; Kobayashi, H.; et al. PPARGC1A rs8192678 and NRF1 rs6949152 Polymorphisms Are Associated with Muscle Fiber Composition in Women. Genes 2020, 11, 1012. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Mozhayskaya, I.A.; Flavell, D.M.; Astratenkova, I.V.; Komkova, A.I.; Lyubaeva, E.V.; Tarakin, P.P.; Shenkman, B.S.; Vdovina, A.B.; Netreba, A.I.; et al. PPARalpha gene variation and physical performance in Russian athletes. Eur. J. Appl. Physiol. 2006, 97, 103–108. [Google Scholar] [CrossRef]
- Kusić, D.; Connolly, J.; Kainulainen, H.; Semenova, E.A.; Borisov, O.V.; Larin, A.K.; Popov, D.V.; Generozov, E.V.; Ahmetov, I.I.; Britton, S.L.; et al. Striated muscle-specific serine/threonine-protein kinase beta segregates with high versus low responsiveness to endurance exercise training. Physiol. Genom. 2020, 52, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Willems, S.M.; Wright, D.J.; Day, F.R.; Trajanoska, K.; Joshi, P.K.; Morris, J.A.; Matteini, A.M.; Garton, F.C.; Grarup, N.; Oskolkov, N.; et al. Large-scale GWAS identifies multiple loci for hand grip strength providing biological insights into muscular fitness. Nat. Commun. 2017, 8, 16015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmetov, I.I.; Hakimullina, A.M.; Popov, D.V.; Lyubaeva, E.V.; Missina, S.S.; Vinogradova, O.L.; Williams, A.G.; Rogozkin, V.A. Association of the VEGFR2 gene His472Gln polymorphism with endurance-related phenotypes. Eur. J. Appl. Physiol. 2009, 107, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Williams, A.G.; Popov, D.V.; Lyubaeva, E.V.; Hakimullina, A.M.; Fedotovskaya, O.N.; Mozhayskaya, I.A.; Vinogradova, O.L.; Astratenkova, I.V.; Montgomery, H.E.; et al. The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum. Genet. 2009, 126, 751–761. [Google Scholar] [CrossRef]
- Takaragawa, M.; Tobina, T.; Shiose, K.; Kakigi, R.; Tsuzuki, T.; Ichinoseki-Sekine, N.; Kumagai, H.; Zempo, H.; Miyamoto-Mikami, E.; Kobayashi, H.; et al. Genotype Score for Iron Status Is Associated with Muscle Fiber Composition in Women. Genes 2021, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Fuku, N.; Kumagai, H.; Ahmetov, I.I. Genetics of muscle fiber composition. In Sports, Exercise, and Nutritional Genomics; Academic Press: Cambridge, MA, USA, 2019; pp. 295–314. [Google Scholar]
- Al-Khelaifi, F.; Yousri, N.A.; Diboun, I.; Semenova, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Borisov, O.V.; Andryushchenko, L.B.; Larin, A.K.; Generozov, E.V.; et al. Genome-wide association study reveals a novel association between MYBPC3 gene polymorphism, endurance athlete status, aerobic capacity and steroid metabolism. Front. Genet. 2020, 11, 595. [Google Scholar] [CrossRef]
- Boulygina, E.A.; Borisov, O.V.; Valeeva, E.V.; Semenova, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Larin, A.K.; Nabiullina, R.M.; Mavliev, F.A.; Akhatov, A.M.; et al. Whole genome sequencing of elite athletes. Biol. Sport 2020, 37, 295–304. [Google Scholar] [CrossRef]
- Taylor, D.L.; Jackson, A.U.; Narisu, N.; Hemani, G.; Erdos, M.R.; Chines, P.S.; Swift, A.; Idol, J.; Didion, J.P.; Welch, R.P.; et al. Integrative analysis of gene expression, DNA methylation, physiological traits, and genetic variation in human skeletal muscle. Proc. Natl. Acad. Sci. USA 2019, 116, 10883–10888. [Google Scholar] [CrossRef] [Green Version]
- Kakigi, R.; Naito, H.; Ogura, Y.; Kobayashi, H.; Saga, N.; Ichinoseki-Sekine, N.; Yoshihara, T.; Katamoto, S. Heat stress enhances mTOR signaling after resistance exercise in human skeletal muscle. J. Physiol. Sci. 2011, 61, 131–140. [Google Scholar] [CrossRef]
- Kawai, Y.; Mimori, T.; Kojima, K.; Nariai, N.; Danjoh, I.; Saito, R.; Yasuda, J.; Yamamoto, M.; Nagasaki, M. Japonica array: Improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J. Hum. Genet. 2015, 60, 581–587. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Võsa, U.; Claringbould, A.; Westra, H.J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.F.; Liu, Y.Z.; Yang, X.L.; Zhang, H.; Feng, G.J.; Wei, X.T.; Zhang, L. The genetic architecture of appendicular lean mass characterized by association analysis in the UK Biobank study. Commun. Biol. 2020, 3, 608. [Google Scholar] [CrossRef]
- UK Biobank GWAS Round 2 Results [Released 1 August 2018]. Available online: http://www.nealelab.is/uk-biobank/ (accessed on 7 September 2022).
- Vissing, K.; Schjerling, P. Simplified data access on human skeletal muscle transcriptome responses to differentiated exercise. Sci. Data 2014, 1, 140041. [Google Scholar] [CrossRef] [Green Version]
- Ticli, G.; Cazzalini, O.; Stivala, L.A.; Prosperi, E. Revisiting the Function of p21CDKN1A in DNA Repair: The Influence of Protein Interactions and Stability. Int. J. Mol. Sci. 2022, 23, 7058. [Google Scholar] [CrossRef]
- Cazzalini, O.; Scovassi, A.I.; Savio, M.; Stivala, L.A.; Prosperi, E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat. Res. 2010, 704, 12–20. [Google Scholar] [CrossRef]
- Wang, J.; Song, C.; Cao, X.; Li, H.; Cai, H.; Ma, Y.; Huang, Y.; Lan, X.; Lei, C.; Ma, Y.; et al. MiR-208b regulates cell cycle and promotes skeletal muscle cell proliferation by targeting CDKN1A. J. Cell Physiol. 2019, 234, 3720–3729. [Google Scholar] [CrossRef]
- Van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J., Jr.; Olson, E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Wang, H.; Liao, Y.; Zhou, P.; Xu, Y.; Zhao, Y.; Xie, S.; Zhao, S.; Li, X. miR-208b modulating skeletal muscle development and energy homoeostasis through targeting distinct targets. RNA Biol. 2020, 17, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Rundqvist, H.C.; Montelius, A.; Osterlund, T.; Norman, B.; Esbjornsson, M.; Jansson, E. Acute sprint exercise transcriptome in human skeletal muscle. PLoS ONE 2019, 14, e0223024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, M.; Liu, Q.; Yan, C. Skeletal Muscle Transcriptomic Comparison Between Men and Women in Response to Acute Sprint Exercise. Front. Genet. 2022, 13, 860815. [Google Scholar] [CrossRef] [PubMed]
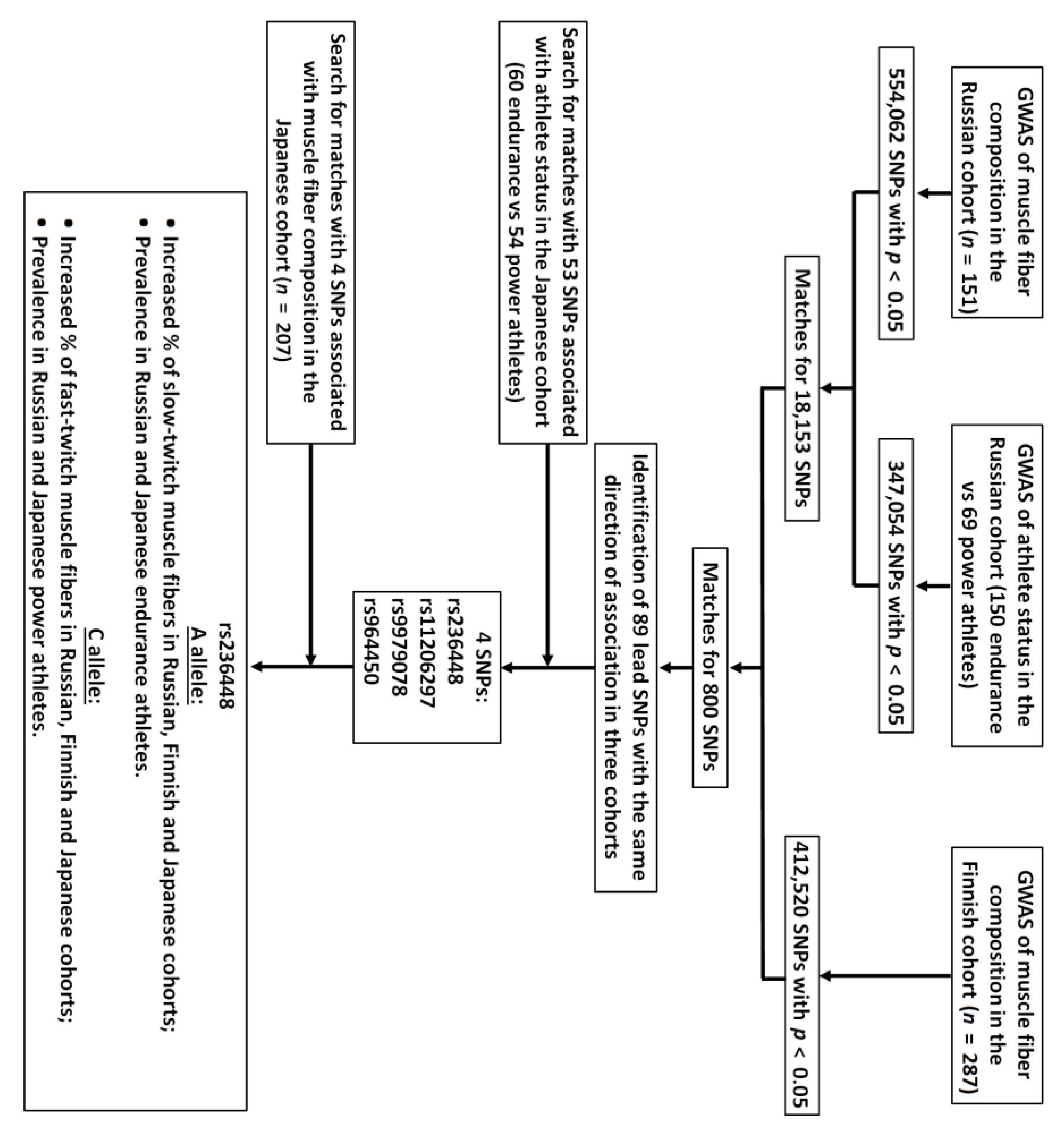
| Group | n | rs236448 Genotypes | A Allele, % | p Value (OR) (Endurance vs. Power) | ||
|---|---|---|---|---|---|---|
| AA | AC | CC | ||||
| Japanese endurance athletes | 60 | 46 | 13 | 1 | 87.5 | 0.038 * (2.1) |
| Japanese power athletes | 54 | 30 | 23 | 1 | 76.9 | |
| Russian endurance athletes | 150 | 89 | 54 | 7 | 77.3 | 0.045 * (1.6) |
| Russian power athletes | 69 | 33 | 28 | 8 | 68.1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semenova, E.A.; Zempo, H.; Miyamoto-Mikami, E.; Kumagai, H.; Larin, A.K.; Sultanov, R.I.; Babalyan, K.A.; Zhelankin, A.V.; Tobina, T.; Shiose, K.; et al. Genome-Wide Association Study Identifies CDKN1A as a Novel Locus Associated with Muscle Fiber Composition. Cells 2022, 11, 3910. https://doi.org/10.3390/cells11233910
Semenova EA, Zempo H, Miyamoto-Mikami E, Kumagai H, Larin AK, Sultanov RI, Babalyan KA, Zhelankin AV, Tobina T, Shiose K, et al. Genome-Wide Association Study Identifies CDKN1A as a Novel Locus Associated with Muscle Fiber Composition. Cells. 2022; 11(23):3910. https://doi.org/10.3390/cells11233910
Chicago/Turabian StyleSemenova, Ekaterina A., Hirofumi Zempo, Eri Miyamoto-Mikami, Hiroshi Kumagai, Andrey K. Larin, Rinat I. Sultanov, Konstantin A. Babalyan, Andrey V. Zhelankin, Takuro Tobina, Keisuke Shiose, and et al. 2022. "Genome-Wide Association Study Identifies CDKN1A as a Novel Locus Associated with Muscle Fiber Composition" Cells 11, no. 23: 3910. https://doi.org/10.3390/cells11233910
APA StyleSemenova, E. A., Zempo, H., Miyamoto-Mikami, E., Kumagai, H., Larin, A. K., Sultanov, R. I., Babalyan, K. A., Zhelankin, A. V., Tobina, T., Shiose, K., Kakigi, R., Tsuzuki, T., Ichinoseki-Sekine, N., Kobayashi, H., Naito, H., Burniston, J., Generozov, E. V., Fuku, N., & Ahmetov, I. I. (2022). Genome-Wide Association Study Identifies CDKN1A as a Novel Locus Associated with Muscle Fiber Composition. Cells, 11(23), 3910. https://doi.org/10.3390/cells11233910










