Activation of OR10A3 by Suberic Acid Promotes Collagen Synthesis in UVB-Irradiated Dermal Fibroblasts via the cAMP-Akt Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Quantitative Assessment of Procollagen Secretion
2.3. Determination of Cell Viability
2.4. Next Generation Sequencing (NGS)
2.5. Molecular Docking Analysis
2.6. Measurement of mRNA Expression
2.7. Small Interfering RNAs (siRNAs) Transfection
2.8. Cyclic Adenosine Monophosphate (cAMP) Measurement
2.9. Western Blot
2.10. Plasmid Construct, Transfection, and Dual Luciferase Reporter Assay
2.11. Statistical Analysis
3. Results
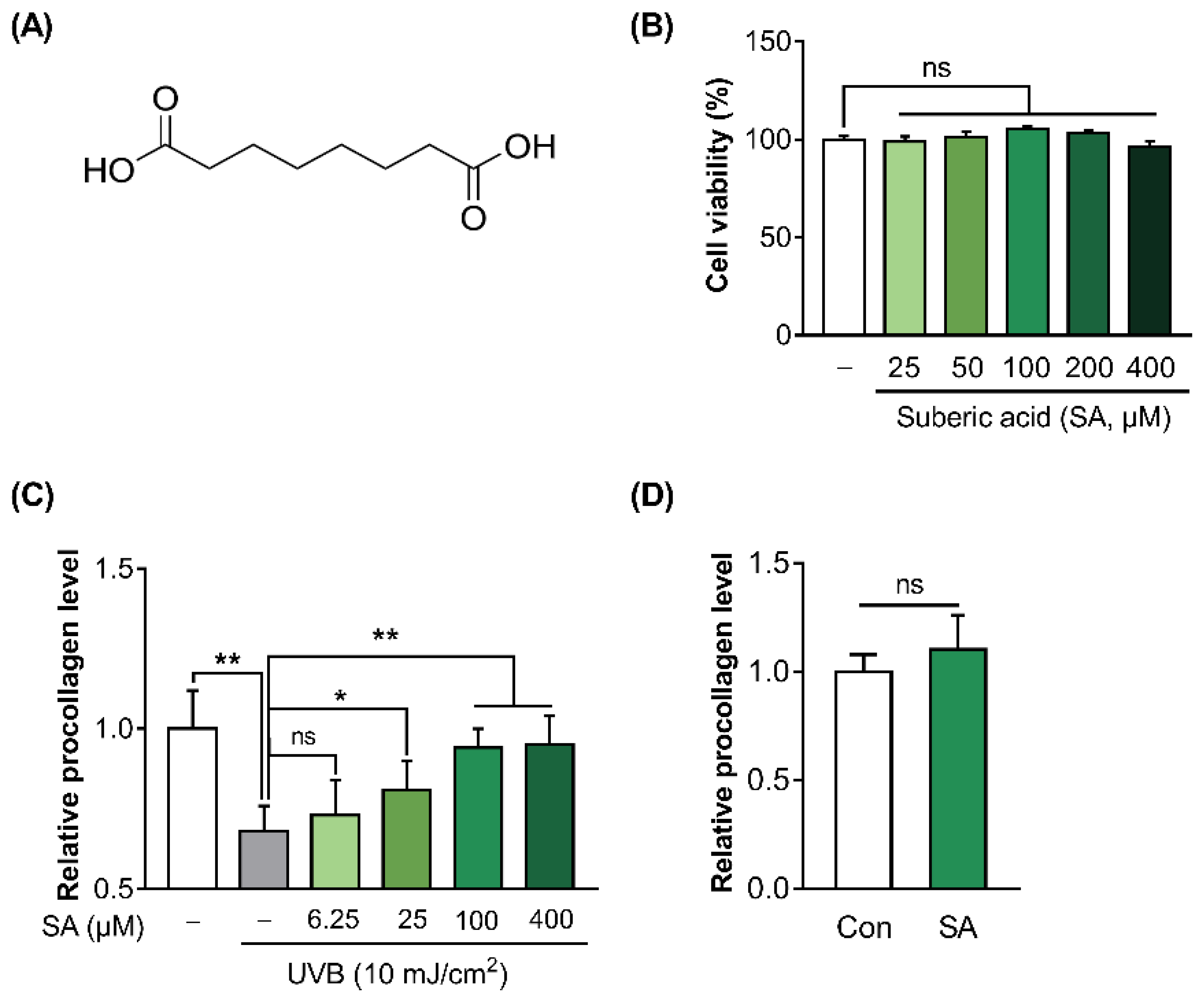
3.1. Ultraviolet B (UVB) Inhibits Procollagen Synthesis in Hs68 Cells
3.2. Suberic Acid Attenuated the Reduction of Collagen Production in UVB-Irradiated Hs68 Cells
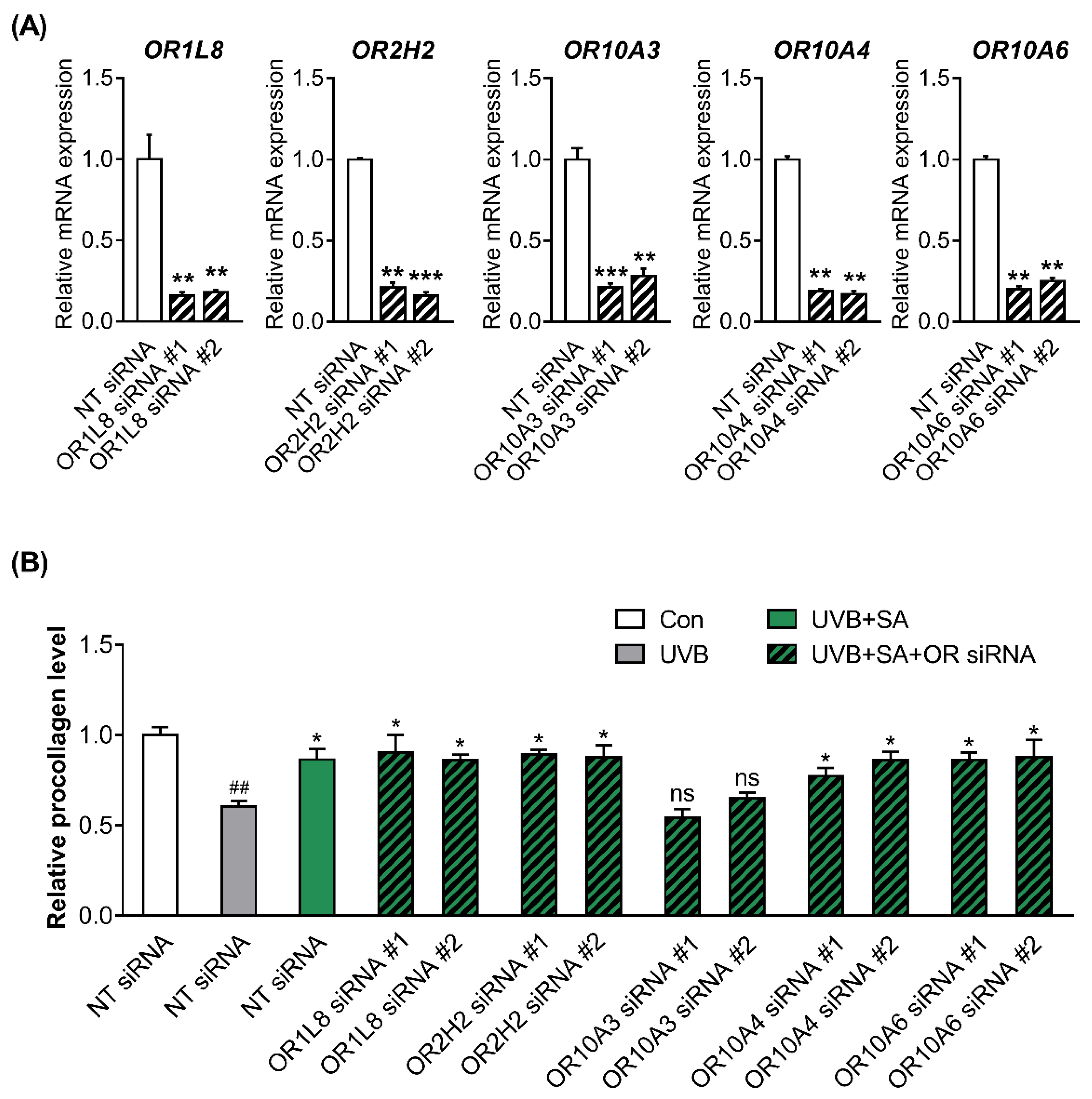
3.3. Suberic Acid Increased the Production of Collagen through OR10A3 in UVB-Exposed Hs68 Cells
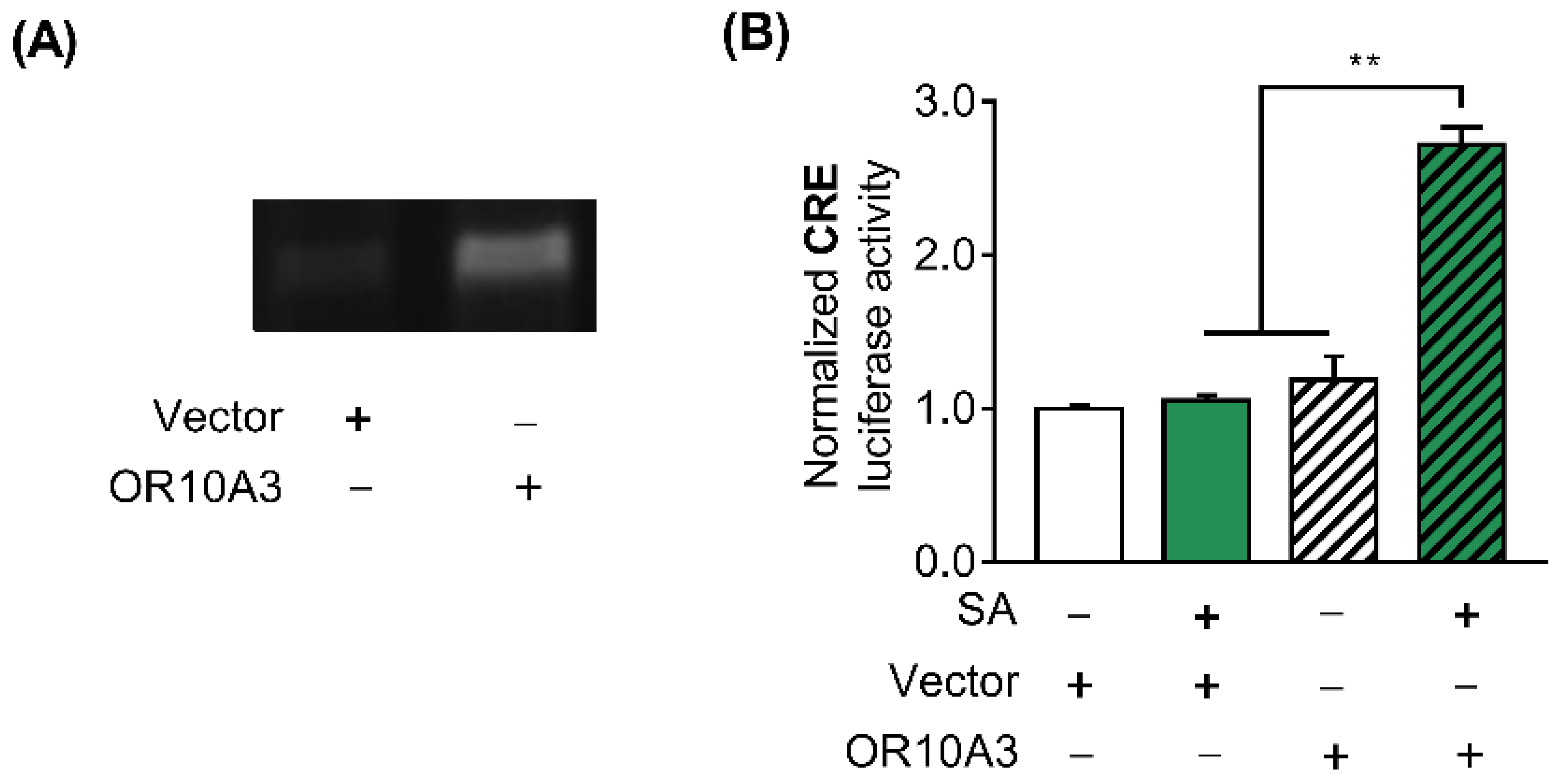
3.4. The Treatment with Suberic Acid Led to the Activation of OR10A3
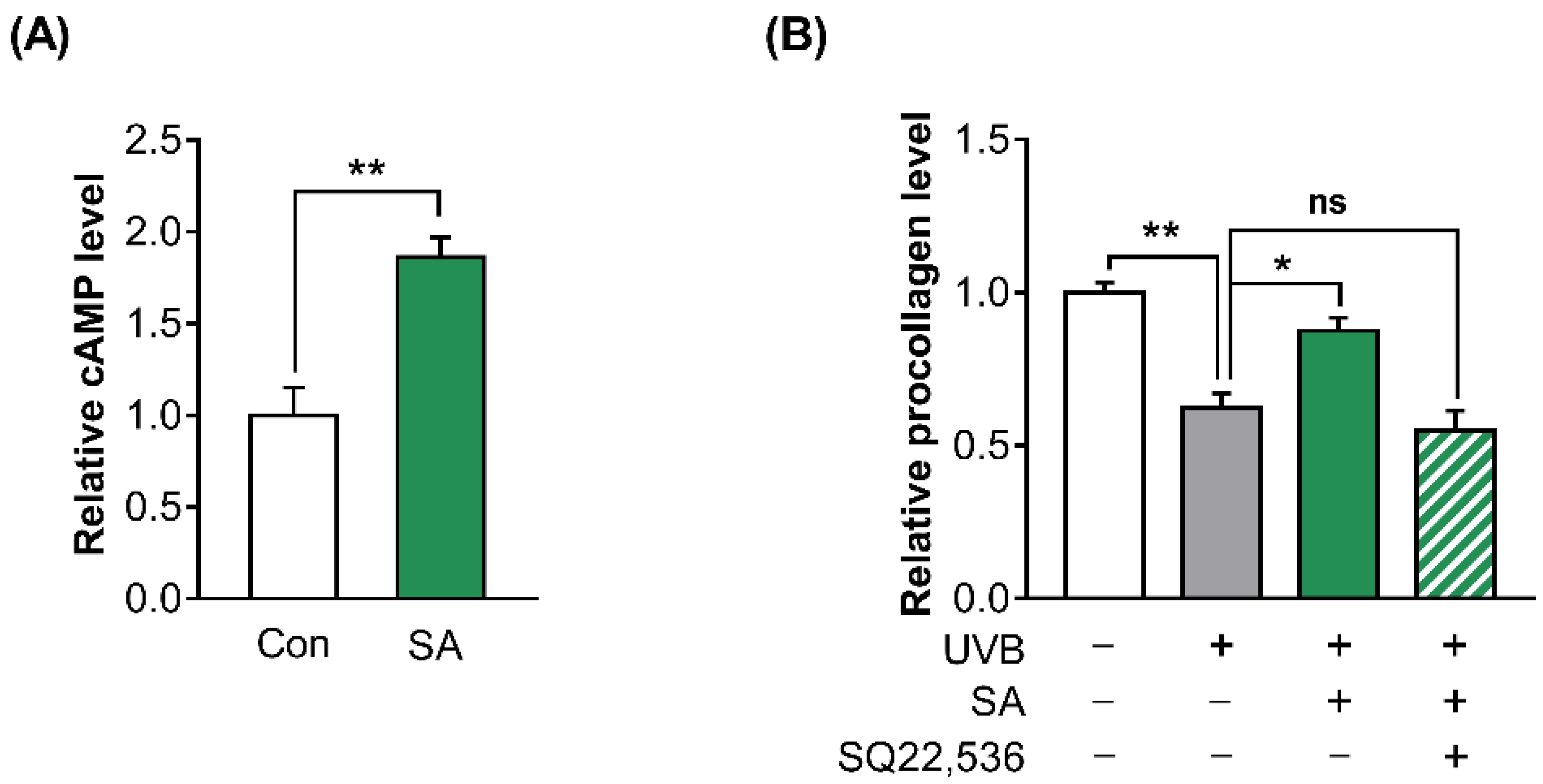
3.5. The Activation of OR10A3 by Suberic Acid Increased the Production of Collagen via Downstream cAMP in UVB-Exposed Hs68 Cells
3.6. Suberic Acid Increased Collagen Production in UVB-Exposed Hs68 Cells through the Akt-Dependent Signaling Pathway
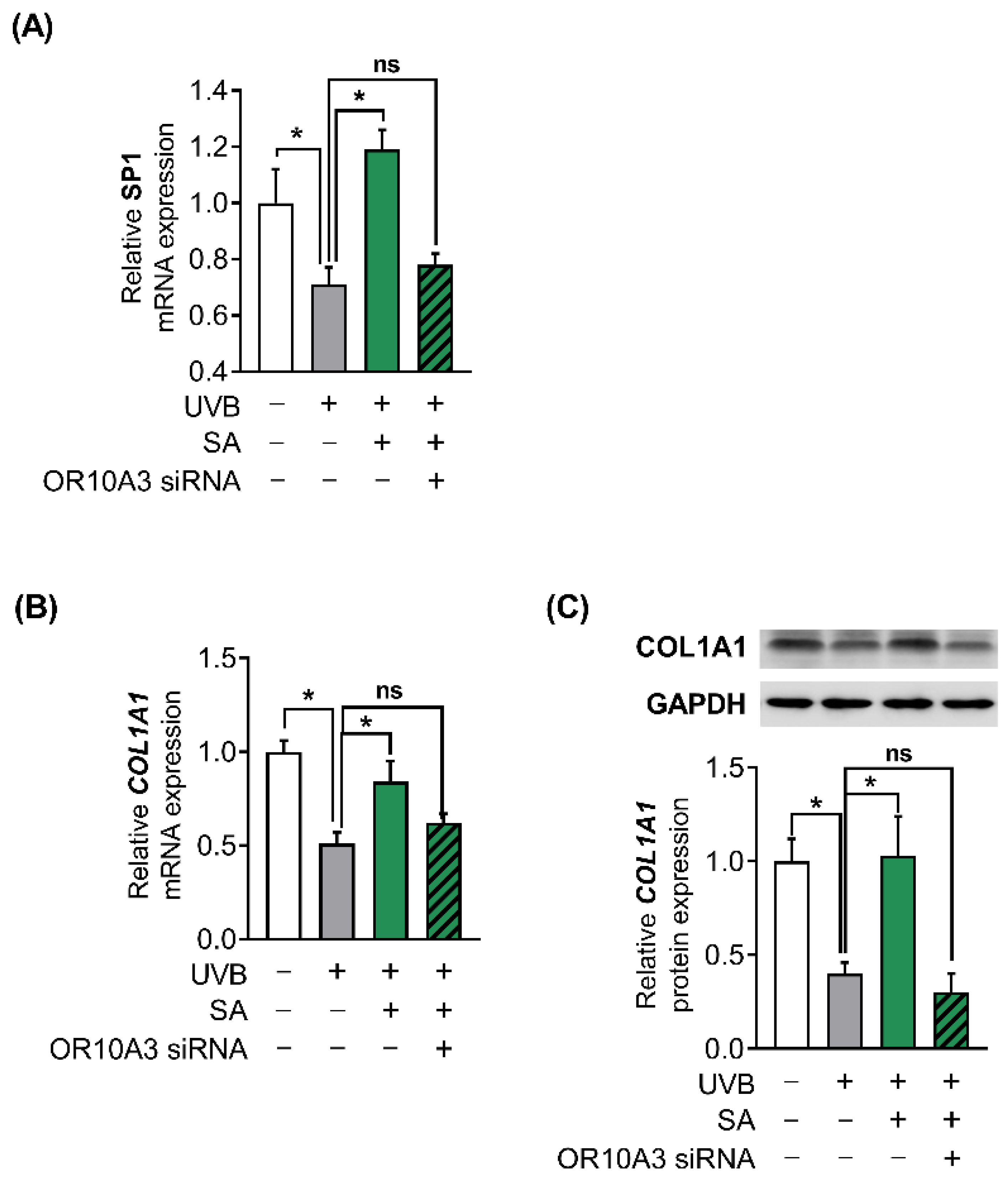
3.7. The Activation of OR10A3 by Suberic Acid Enhanced Gene Expression of Akt Downstream Transcription Factor SP1 and Its Target Gene COL1A1 in UVB-Exposed Hs68 Cells
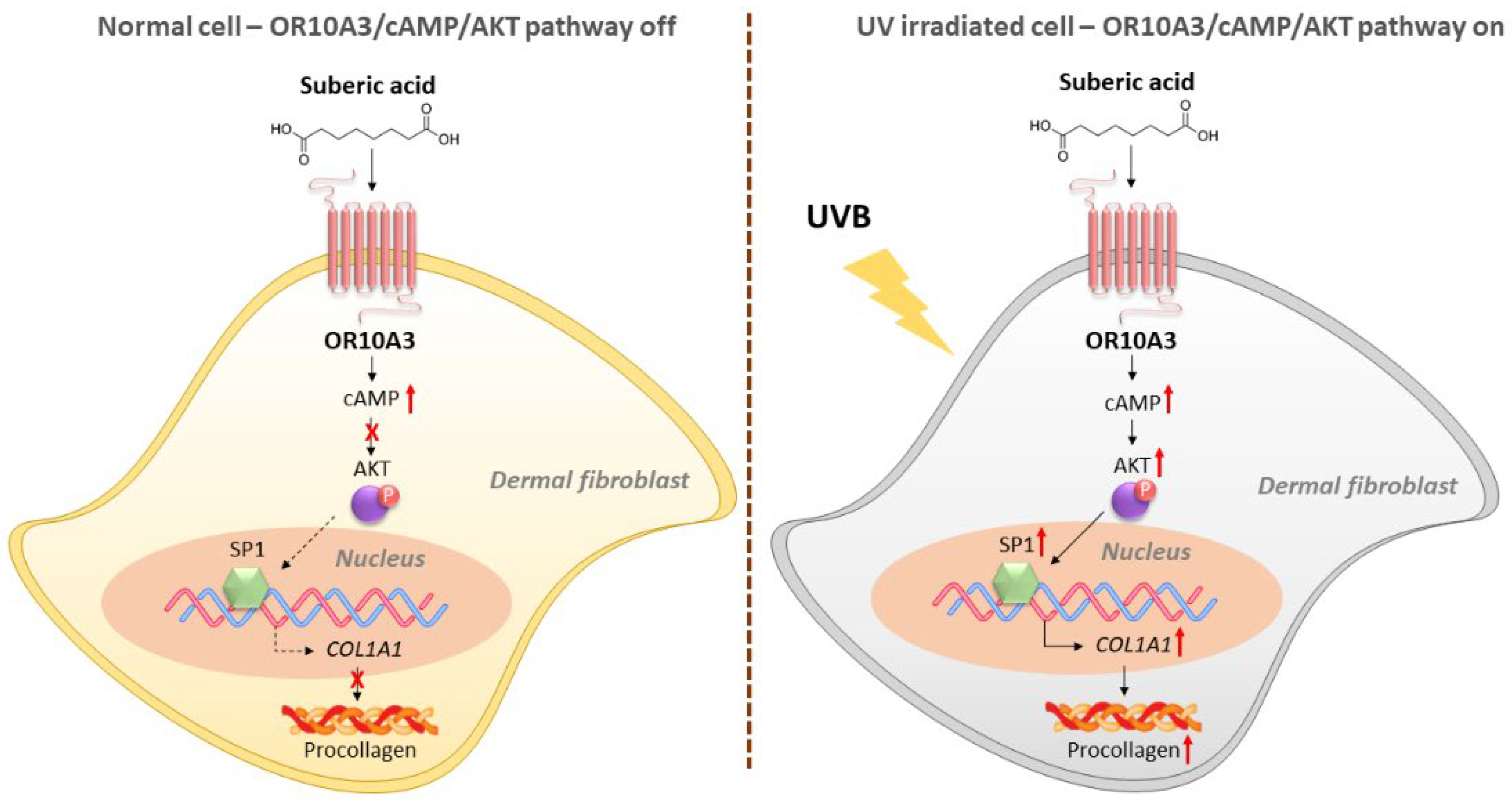
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.B.; Bhatnagar, R.S.; Li, S.; Oreffo, R.O. Biomimetic collagen scaffolds for human bone cell growth and differentiation. Tissue Eng. 2004, 10, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Andrades, J.A.; Han, B.; Becerra, J.; Sorgente, N.; Hall, F.L.; Nimni, M.E. A recombinant human TGF-β1 fusion protein with collagen-binding domain promotes migration, growth, and differentiation of bone marrow mesenchymal cells. Exp. Cell Res. 1999, 250, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Canty, E.G.; Kadler, K.E. Procollagen trafficking, processing and fibrillogenesis. J. Cell Sci. 2005, 118, 1341–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleischmajer, R.; Perlish, J.; Burgeson, R.; Shaikh-Bahai, F.; Timpl, R. Type I and type III collagen interactions during fibrillogenesis. Ann. N. Y. Acad. Sci. 1990, 580, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, H.; Fu, N.; Chen, L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell. Physiol. 2018, 233, 2104–2115. [Google Scholar] [CrossRef]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-J.; Depoortere, I.; Hatt, H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2019, 18, 116–138. [Google Scholar] [CrossRef]
- Tham, E.H.; Dyjack, N.; Kim, B.E.; Rios, C.; Seibold, M.A.; Leung, D.Y.; Goleva, E. Expression and function of the ectopic olfactory receptor OR10G7 in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 1838–1848.e1834. [Google Scholar] [CrossRef]
- Chéret, J.; Bertolini, M.; Ponce, L.; Lehmann, J.; Tsai, T.; Alam, M.; Hatt, H.; Paus, R. Olfactory receptor OR2AT4 regulates human hair growth. Nat. Commun. 2018, 9, 3624. [Google Scholar] [CrossRef]
- Gelis, L.; Jovancevic, N.; Veitinger, S.; Mandal, B.; Arndt, H.-D.; Neuhaus, E.M.; Hatt, H. Functional characterization of the odorant receptor 51E2 in human melanocytes. J. Biol. Chem. 2016, 291, 17772–17786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayorinde, F.O.; Osman, G.; Shepard, R.L.; Powers, F.T. Synthesis of azelaic acid and suberic acid fromvernonia galamensis oil. J. Am. Oil Chem. Soc. 1988, 65, 1774–1777. [Google Scholar] [CrossRef]
- Johnson, R.W.; Pollock, C.M.; Cantrell, R.R. Dicarboxylic acids. In Kirk-Othmer Encyclopedia of Chemical Technology; Wiley: Hoboken, NJ, USA, 2000; pp. 1–18. [Google Scholar] [CrossRef]
- de Moraes, M.S.; Guerreiro, G.; Sitta, A.; de Moura Coelho, D.; Manfredini, V.; Wajner, M.; Vargas, C.R. Oxidative damage in mitochondrial fatty acids oxidation disorders patients and the in vitro effect of l-carnitine on DNA damage induced by the accumulated metabolites. Arch. Biochem. Biophys. 2020, 679, 108206. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Shimojo, N.; Nakanishi, T.; Naka, K.; Okuda, K. Measurements of urinary adipic acid and suberic acid using high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 1994, 655, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Inouye, M.; Mio, T.; Sumino, K. Dicarboxylic acids as markers of fatty acid peroxidation in diabetes. Atherosclerosis 2000, 148, 197–202. [Google Scholar] [CrossRef]
- Kang, W.; Choi, D.; Park, T. Dietary suberic acid protects against UVB-induced skin photoaging in hairless mice. Nutrients 2019, 11, 2948. [Google Scholar] [CrossRef] [Green Version]
- Abaffy, T.; Malhotra, A.; Luetje, C.W. The molecular basis for ligand specificity in a mouse olfactory receptor: A network of functionally important residues. J. Biol. Chem. 2007, 282, 1216–1224. [Google Scholar] [CrossRef] [Green Version]
- Abaffy, T.; Matsunami, H.; Luetje, C.W. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 2006, 97, 1506–1518. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.-T.; Hou, M.-C.; Xiao, Z.-X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Fujita, Y.; Takahashi, T.; Suzuki, A.; Kawashima, K.; Nara, F.; Koishi, R. Deorphanization of Dresden G protein-coupled receptor for an odorant receptor. J. Recept. Signal Transduct. 2007, 27, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.A. New paradigms in GPCR drug discovery. Biochem. Pharmacol. 2015, 98, 541–555. [Google Scholar] [CrossRef] [Green Version]
- Hauser, A.S.; Chavali, S.; Masuho, I.; Jahn, L.J.; Martemyanov, K.A.; Gloriam, D.E.; Babu, M.M. Pharmacogenomics of GPCR drug targets. Cell 2018, 172, 41–54.e19. [Google Scholar] [CrossRef] [Green Version]
- Liccardo, F.; Luini, A.; Di Martino, R. Endomembrane-based signaling by GPCRs and G-proteins. Cells 2022, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, Z.; Firestein, S.; Rogers, M.E. The state of the art of odorant receptor deorphanization: A report from the orphanage. J. Gen. Physiol. 2014, 143, 527–542. [Google Scholar] [CrossRef] [Green Version]
- Maßberg, D.; Hatt, H. Human Olfactory Receptors: Novel Cellular Functions Outside of the Nose. Physiol. Rev. 2018, 98, 1739–1763. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Lefkimmiatis, K.; Zaccolo, M. cAMP signaling in subcellular compartments. Pharmacol. Ther. 2014, 143, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.A.; Akamine, P.; Radzio-Andzelm, E.; Madhusudan, A.; Taylor, S.S. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 2001, 101, 2243–2270. [Google Scholar] [CrossRef]
- Shabb, J.B. Physiological substrates of cAMP-dependent protein kinase. Chem. Rev. 2001, 101, 2381–2412. [Google Scholar] [CrossRef]
- Taylor, S.S.; Buechler, J.A.; Yonemoto, W. cAMP-dependent protein kinase: Framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 1990, 59, 971–1005. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Ladilov, Y. Emerging Role of cAMP/AMPK Signaling. Cells 2022, 11, 308. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef]
- Kim, S.-H.; Yoon, Y.C.; Lee, A.S.; Kang, N.; Koo, J.; Rhyu, M.-R.; Park, J.-H. Expression of human olfactory receptor 10J5 in heart aorta, coronary artery, and endothelial cells and its functional role in angiogenesis. Biochem. Biophys. Res. Commun. 2015, 460, 404–408. [Google Scholar] [CrossRef]
- Mathew-Steiner, S.S.; Roy, S.; Sen, C.K. Collagen in wound healing. Bioengineering 2021, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Mian, M.; Beghe, F.; Mian, E. Collagen as a Pharmacological Approach in Wound-Healing. Int. J. Tissue React. 1992, 14, 1–9. [Google Scholar] [PubMed]
- Dalesio, N.M.; Barreto Ortiz, S.F.; Pluznick, J.L.; Berkowitz, D.E. Olfactory, taste, and photo sensory receptors in non-sensory organs: It just makes sense. Front. Physiol. 2018, 9, 1673. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene Description | Sequences (5′→3) |
|---|---|
| Collagen type I alpha 1 chain (COL1A1) | F: ACATGTTCAGCTTTGTGGACC |
| R: TGTACGCAGGTGATTGGTGG | |
| Olfactory receptor family 1 subfamily L member 8 (OR1L8) | F: GCCCTGTGCTGAAATTGTCC |
| R: GGCTTTGCGTTTCCCAGAAG | |
| OR2H2 | F: CCATCTCACTGTGGTCACCCTCTTC |
| R: GAATGCCCTGGTTACCTCCTTGTTC | |
| OR10A3 | F: ATCTGGCTACTCACCCGAAAC |
| R: AGATGAGCGGATTGAGCAGAG | |
| OR10A4 | F: CACCTCTTGGTTGTCTCTCTCTTC |
| R: CCTTCAGTTTCCATCTAAGCCAATC | |
| OR10A6 | F: GTCAACAGAGAAAGGTTCGGG |
| R: TGGTGTCTTGGATAGATCACTG | |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | F: GACAGTCAGCCGCATCTTCT |
| R: CGCCCAATACGACCAAATC |
| Gene Name | siRNA Sequence (Sense [S], Antisense [A]) | |
|---|---|---|
| OR1L8 | #1 | S: CGUCUCACCUUCUGUGACU A: AGUCACAGAAGGUGAGACG |
| #2 | S: CCUACGCUGUCAAGGACCA A: UGGUCCUUGACAGCGUAGG | |
| OR2H2 | #1 | S: CUGUUUCACCACGAGUUGU A: ACAACUCGUGGUGAAACAG |
| #2 | S: GUCAUUGGGCUAGUGGAGU A: ACUCCACUAGCCCAAUGAC | |
| OR10A3 | #1 | S: CUCUGAACUACCCAGUGAU A: AUCACUGGGUAGUUCAGAG |
| #2 | S: CUCAUCUAUAGCUUACGAA A: UUCGUAAGCUAUAGAUGAG | |
| OR10A4 | #1 | S: CUGAUCAUUCAAGACACAA A: UUGUGUCUUGAAUGAUCAG |
| #2 | S: CUCUCUCUUCUAUAGCACU A: AGUGCUAUAGAAGAGAGAG | |
| OR10A6 | #1 | S: CUGAAUCUGCUUAUCUACA A: UGUAGAUAAGCAGAUUCAG |
| #2 | S: CAUCCUCUCAACUACCAAA A: UUUGGUAGUUGAGAGGAUG | |
| Non-targeting siRNA (NT) | S: GAACUGAUGACAGGGAGGC A: GCCUCCCUGUCAUCAGUUC | |
| Olfactory Receptor (OR) Name | FPKM | Vina Score |
|---|---|---|
| OR1L8 | 0.035 | −5.5 |
| OR2A1/42 | 0.192 | −4.8 |
| OR2A4/7 | 0.413 | −4.9 |
| OR2AE1 | 0.254 | −4.4 |
| OR2B2 | 0.070 | −4.7 |
| OR2H2 | 0.029 | −5.7 |
| OR6A2 | 0.041 | −5.2 |
| OR10A3 | 0.035 | −5.7 |
| OR10A4 | 0.032 | −5.8 |
| OR10A5 | 0.017 | −5.1 |
| OR10A6 | 0.034 | −5.6 |
| OR51B5 | 0.063 | −4.9 |
| OR51I1 | 0.581 | −4.5 |
| OR52DI | 0.274 | −4.6 |
| OR56B4 | 0.026 | −5.2 |
| Name | Description | Score |
|---|---|---|
| BRD-75430629 | PI3K Inhibitor | −97.92 |
| Etilefrine | Adrenergic recetptor agonist | 94.47 |
| 2-(4-methoxybenzylthio)-6-methylpyrimidin-4-ol | Matrix metalloproteinase inhibitor | 93.53 |
| Iloprost | Prostanoid receptor agonist | 91.53 |
| UNC-0321 | Histone lysine methyltransferase inhibitor | −90.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, W.; Choi, D.; Son, B.; Park, S.; Park, T. Activation of OR10A3 by Suberic Acid Promotes Collagen Synthesis in UVB-Irradiated Dermal Fibroblasts via the cAMP-Akt Pathway. Cells 2022, 11, 3961. https://doi.org/10.3390/cells11243961
Kang W, Choi D, Son B, Park S, Park T. Activation of OR10A3 by Suberic Acid Promotes Collagen Synthesis in UVB-Irradiated Dermal Fibroblasts via the cAMP-Akt Pathway. Cells. 2022; 11(24):3961. https://doi.org/10.3390/cells11243961
Chicago/Turabian StyleKang, Wesuk, Dabin Choi, Bomin Son, Soyoon Park, and Taesun Park. 2022. "Activation of OR10A3 by Suberic Acid Promotes Collagen Synthesis in UVB-Irradiated Dermal Fibroblasts via the cAMP-Akt Pathway" Cells 11, no. 24: 3961. https://doi.org/10.3390/cells11243961
APA StyleKang, W., Choi, D., Son, B., Park, S., & Park, T. (2022). Activation of OR10A3 by Suberic Acid Promotes Collagen Synthesis in UVB-Irradiated Dermal Fibroblasts via the cAMP-Akt Pathway. Cells, 11(24), 3961. https://doi.org/10.3390/cells11243961










