Abstract
The VosA-VelB hetero-dimeric complex plays a pivotal role in regulating development and secondary metabolism in Aspergillus nidulans. In this work, we characterize a new VosA/VelB-activated gene called vadH, which is predicted to encode a 457-amino acid length protein containing four adjacent C2H2 zinc-finger domains. Mutational inactivation of vosA or velB led to reduced mRNA levels of vadH throughout the lifecycle, suggesting that VosA and VelB have a positive regulatory effect on the expression of vadH. The deletion of vadH resulted in decreased asexual development (conidiation) but elevated production of sexual fruiting bodies (cleistothecia), indicating that VadH balances asexual and sexual development in A. nidulans. Moreover, the vadH deletion mutant exhibited elevated susceptibility to hyperosmotic stress compared to wild type and showed elevated production of the mycotoxin sterigmatocystin (ST). Genome-wide expression analyses employing RNA-Seq have revealed that VadH is likely involved in regulating more genes and biological pathways in the developmental stages than those in the vegetative growth stage. The brlA, abaA, and wetA genes of the central regulatory pathway for conidiation are downregulated significantly in the vadH null mutant during asexual development. VadH also participates in regulating the genes, mat2, ppgA and lsdA, etc., related to sexual development, and some of the genes in the ST biosynthetic gene cluster. In summary, VadH is a putative transcription factor with four C2H2 finger domains and is involved in regulating asexual/sexual development, osmotic stress response, and ST production in A. nidulans.
1. Introduction
Aspergillus spp. are widely distributed in nature and closely related to human life. public health, the fermentation and food processing industry including A. oryzae and A. niger, as well as some plant and human pathogens such as A. flavus and A. fumigatus [1]. The production of conidia is the primary mode of reproduction in the genus Aspergillus. Conidial formation is an elaborate process, which involves cell differentiation, gene expression, and signal transduction [2]. Conidia are an important carrier for the transmission and infection of Aspergillus, and its developmental process is also related to mycotoxin biosynthesis [3]. Therefore, elucidating the regulatory network of conidial development and exploring the functions of critical regulatory proteins are crucial for understanding the law of growth and development of Aspergillus, rational utilization of beneficial industrial strains and effective control of pathogens.
Aspergillus nidulans is an important model organism for studying fungal development, secondary metabolism, and its regulatory pathway. The mechanism of conidiation has been well studied in A. nidulans [4,5]. In A. nidulans, BrlA, AbaA, and WetA constitute the central regulatory pathway of conidial formation, which can sequentially activate conidial development and control the expression of specific genes involved in asexual development [2]. The activation of BrlA is a fundamental step to initiate conidiation [4,6]. Then, BrlA can activate AbaA, which controls the differentiation and function of phialide [7]. In the late stage, WetA is activated by AbaA, which regulates the expression of several spore-specific genes and conidial wall synthesis [8]. In addition, it is reported that velvet family proteins also play an essential role in the asexual development of A. nidulans [9,10].
Velvet family proteins are fungal-specific proteins with the velvet domain, including VosA, VeA, VelB and VelC, which are highly conserved in filamentous fungi [10,11]. In Aspergillus, velvet proteins participate in regulating growth and development, pigmentation, primary and secondary metabolism [9,12,13]. These proteins often form various complexes, such as VosA-VelB, VelB-VeA-LaeA, VosA-VelC and VelB-VelB, which play different roles in coordinating the growth, differentiation, and secondary metabolism of Aspergillus [12,13]. Among these complexes, the VosA-VelB complex plays a crucial role in regulating conidia maturation and germination, trehalose biosynthesis and conidial wall synthesis [13,14,15].
The VosA-VelB protein complex has a DNA-binding domain similar to that of mammalian NF-κB transcription factor, which can recognize the cis-acting motif specifically in the promoter region of its target genes [15]. Chromatin immunoprecipitation (ChIP) showed that VosA-VelB could bind to promoter sequences of more than 150 genes, including genes activated by VosA-VelB, such as tpsA and treA genes related to trehalose synthesis [15]. Another VosA/VelB-activated gene, vadA, is a novel bifunctional regulatory factor in A. nidulans, which is involved in regulating conidial germination, trehalose synthesis, β-glucan synthesis, oxidative stress, and sterigmatocystin synthesis [16]. In addition, VosA-VelB also has negative regulatory effects on many genes, including brlA and wetA in the central regulatory pathway of conidia, and it can also inhibit β-glucan synthesis in conidia and ascospores by directly binding to the promoter region of fksA gene encoding β-1, 3-glucan synthase [17].
While several target genes of VosA-VelB have been functionally characterized, many remain to be investigated. In this study, we have identified another VosA-VelB target gene vadH (VosA/VelB-Activated Developmental gene H; AN6503 AspGD). The promoter region of vadH is bound by both VosA and VelB in conidia. The vadH gene is predicted to encode a highly conserved transcription factor (VadH) harboring four C2H2 zinc finger domains. The homologous proteins of VadH are universal in the fungal kingdom, and some have been well characterized. Saccharomyces cerevisiae Azf1, a homolog of VadH, participates in activating the genes related to carbon and energy metabolism when glucose exists, and switches to maintaining cell wall integrity when glucose is depleted [18]. In Magnaporthe oryzae, Cos1 is involved in regulating the development of conidiophores and melanin biosynthesis [19]. The VadH homolog CgAzf1 coordinates melanin production, conidium development, appressorium formation and virulence in Colletotrichum gloeosporioides [20]. In this work, the biological functions of vadH have been characterized by gene knockout, overexpression, and transcriptome analyses.
2. Materials and Methods
2.1. Strains and Culture Conditions
All fungal strains used in this study are listed in Table S1 [13,21,22], and media are prepared as previously described [9]. Briefly, minimal media with glucose (MMG) and MMG with 0.5% yeast extract (MMYE) were used for general purposes, and sexual medium (SM) was used for enhancing sexual development. To determine the number of conidia and cleistothecia, the wild type (WT: A. nidulans FGSC4), mutants, and complemented strains were inoculated and cultured on solid MMG, MMYE or SM for seven days at 37 °C. Micrographs were taken by a Zeiss M2Bio microscope. For the overexpression of vadH under the niiA promoter, strains were inoculated on a non-inducing medium (MMG containing 0.2% ammonium tartrate as a nitrogen source) or an inducing medium (MMG containing 0.6% sodium nitrate as a nitrogen source) and incubated at 37 °C for three days.
2.2. Nucleic Acid Isolation and Manipulation
Total RNA isolation was performed as previously described [23]. For asexual and sexual development, cultures from mutants and WT were collected and transferred on solid MMG and SM, respectively. Then, the plates were exposed to air for asexual developmental induction or tightly sealed and blocked from light for sexual developmental induction. For asexual development, samples were collected at 18 and 36 h for RNA isolation. Samples were harvested at 36 and 72 h after transfer for sexual development. For vegetative growth, one milliliter of conidial suspension (106 conidia/mL) was inoculated in 100 mL liquid MMG and incubated at 37 °C. The mycelium was collected at 18 and 36 h post-inoculation (hpi) for RNA isolation. Quantitative reverse transcription-PCR was used to analyze the expression levels of vadH. The primers were listed in Table S2. The quantitative reverse transcription PCR (qRT-PCR) was carried out by the Fast SYBR Green Master Mix. Gene expression levels were normalized using the endogenous control gene actin. The average normalized expression level was calculated using the 2−ΔΔCt method [24]. All the experiments were repeated three times.
2.3. Target Deletion of VadH
Genomic DNA extraction was performed as previously described [25,26]. The primers used in this section are listed in Table S2. The vadH-deletion mutant strain (ΔvadH) was generated by double-joint PCR (DJ-PCR) as previously described [23]. The up- and down-stream sequences of the vadH gene were amplified from A. nidulans FGSC4 genomic DNA the by PCR using the primer pairs OXL-1/OXL-2 and OXL-3/OXL-4. The A. fumigatus pyrG marker was amplified using the primer pair OHS-694/OHS-695 from A. fumigatus AF293 genomic DNA. The vadH deletion cassette was amplified with primer pair OXL-5/OXL-6 and introduced into A. nidulans RJMP1.59 [21]. Protoplasts were generated from A. nidulans RJMP1.59 by the Vinoflow FCE lysing enzyme (Novozymes) [25]. For the complementation of ΔvadH, the vadH gene sequence, including its predicted promoter, was amplified with the primer pair OXL-15/OXL-16 and attached to a pHS13 vector [13]. To generate the overexpressing strain, the vadH ORF derived from the genomic DNA was amplified using the primer pair OXL-31/OXL-32. The PCR product was then attached into pHS11 and introduced into A. nidulans RJMP1.59. The vadH-overexpressing strains were screened by qRT-PCR.
2.4. Spore Viability Determination
To test spore viability, conidia obtained from two-day-old cultured WT, mutant and complementary strains were spread on solid MMG and cultured at 37 °C. Then, conidia were collected after culturing for seven days. About 100 conidia were coated onto solid MMG and incubated at 37 °C for 48 h in triplicate. Survival rates were determined as the ratio of the number of viable colonies to the number of conidia inoculated.
2.5. Osmotic Stress Assays
For stress tests, the strains were inoculated on the solid MMG medium including sorbitol (1.0 M), glycerol (1.0 M) and NaCl (1.0 M) and grown at 37 °C for seven days. Moreover, ten microliters of serially diluted conidia suspensions (10 to 105 conidia/mL) were spotted on the solid MMG medium including sorbitol (1.0 M), glycerol (1.0 M) and NaCl (1.0 M), and incubated at 37 °C for 48 h. The plates without the stress factors served as controls. The inhibition rates were calculated as follows: Inhibition rate (%) = (Dck − Dt)/Dck × 100%. Where Dck is the colony diameter of the strain in the control, and Dt is the colony diameter of the strain in the treatment group.
2.6. Sterigmatocystin (ST) Determination
ST extraction and determination were performed as previously described [16]. Briefly, 106 conidia from the strains were inoculated in 2 mL liquid MMG and cultured at 37 °C for seven days. ST was extracted by adding 2 mL of CHCl3. The organic phase (CHCl3) was separated through centrifugation and transferred to a new glass bottle. The extracting solution was evaporated in a fume hood and dissolved in 1 mL acetonitrile:methanol (50:50, v/v). After filtering through a millipore filter (0.45 μm), the samples were analyzed by high-performance liquid chromatography with diode-array detection (HPLC-DAD, Agilent Technologies, Waldbronn, Germany).
2.7. RNA Sequencing (RNA-Seq)
The isolation of RNA samples was performed as previously described [23]. RNA-Seq analyses of VadH include three aspects: vegetative growth, asexual and sexual development. The preparation of samples in three stages was conducted as previously described [9]. Samples for vegetative growth were collected at 18 hpi, and samples for asexual and sexual development were obtained at 18 and 36 hpi, respectively. A MGISEQ-2000 platform (BGI, Shenzhen, China) was used to analyze the samples. The genome of A. nidulans FGSC A4 from AspGD (http://www.aspergillusgenome.org/, accessed on 1 Septemper 2020) was used as a reference. Data processing and analyses were performed as described previously [27]. The results of RNA-Seq were verified by qRT-PCR according to the published method [27].
2.8. Statistical Analysis
Statistical differences between WT and mutant strains were evaluated by Student’s unpaired t-test. Data are reported as mean ± SD, and statistical significance was defined as p < 0.05.
3. Results
3.1. Characterization of VadH
The gene vadH (AN6503) is predicted to encode a 457-amino acids (aa) protein, which contains four adjacent C2H2 zinc-finger domains from the position 239 to 352 (Figure 1A). The homologous proteins of VadH are ubiquitous in fungi, and all of them harbor four C2H2-type domains (Figure 1A,B). VadH is homologous with CgAzf1 in Colletotrichum gloeosporioides [20], Cos1 in Magnaporthe oryzae [19], and Azf1 in Saccharomyces cerevisiae [18].
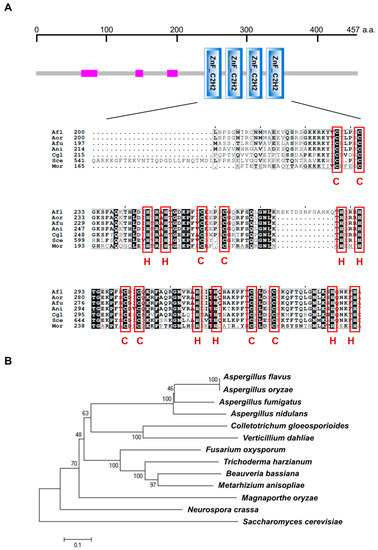
Figure 1.
Overview of the VadH protein. (A) Protein domain analysis of VadH. The pink rectangle is a region of low compositional complexity. The below figure presents the alignment of C2H2 zinc-finger domains from A. nidulans VadH (Ani: AN6503) and its homologs from other fungi. (B) Phylogenetic analysis of VadH. The phylogenetic tree was constructed using Clustal_W and MEGA 6.0 with homologous sequences of VadH from A. flavus (Afl: AFL2T_05677), A. oryzae (Aor: AO090701000019), A. fumigatus (Afu: Afu6g05160), Colletotrichum gloeosporioides (Cgl: AUS82351.1), Verticillium dahlia (XP_009658472.1), Fusarium oxysporum (EWZ45422.1), Trichoderma harzianum (KKP06506.1), Beauveria bassiana (XP_008598185.1), Metarhizium anisopliae (KFG78752.1), Magnaporthe oryzae (Mor: XP_003719876.1), Neurospora crassa (XP_961139.2) and Saccharomyces cerevisiae (Sce: NP_014756.3).
To evaluate the effects of VosA and VelB on the expression of vadH, mRNA levels of vadH were determined in the ΔvosA and ΔvelB strains. As shown in Figure 2A, the expression levels of vadH in ΔvosA and ΔvelB are significantly lower than those of WT in different stages, suggesting that VosA and VelB can positively regulate the expression of vadH. In the wild type, the expression levels of vadH during asexual and sexual development are apparently higher than those in the stage of vegetative growth (Figure 2B).
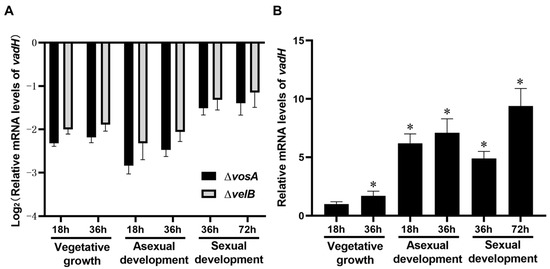
Figure 2.
Relative mRNA levels of vadH. (A) Relative expression levels of vadH in ΔvosA and ΔvelB. (B) Relative expression levels of vadH in WT during three stages. The expression level of vadH at 18 h of the vegetative growth stage was set to 1. The asterisks represent significant level (* p < 0.05).
3.2. VadH Balances Asexual and Sexual Development
To study the function of vadH, we generated the vadH-deletion mutant (ΔvadH) and the complemented strain (C’vadH), and the results related to the verification of the mutant were shown in Figure S1. For colony growth, no significant difference was found between ΔvadH and the wild type on MMG and MMYE, whereas the colony color of ΔvadH was much lighter than that of strain WT (Figure 3A,B). ΔvadH produced fewer conidia than WT on both MMG and MMYE (Figure 3C), and the spore viability of ΔvadH was also slightly lower than that of the WT (Figure 3D). Compared with WT, the mutant ΔvadH exhibited significantly decreased conidial germination (Figure S2). For sexual development, WT, ΔvadH and C’vadH were inoculated on SM, and the number of cleistothecia was determined (Figure 3E). As shown in Figure 3F, the mutant ΔvadH produced significantly more cleistothecia than WT and ΔvadH on MMG, and MMYE, suggesting that VadH is involved in regulating sexual development in A. nidulans. However, there was no significant difference in cleistothecia yields on SM between WT and ΔvadH.
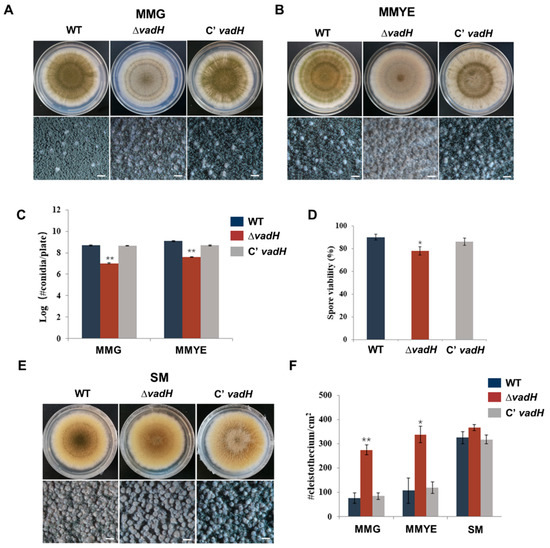
Figure 3.
Developmental phenotypes of the ΔvadH mutant. (A,B) Colony morphology of WT, ΔvadH, and the complemented strain on MMG (A) or MMYE (B) and grown for 7 days. The bottom panel shows close-up views of the colony in the top panel (Bar = 0.5 mm). (C) Statistical analyses of conidial yields on MMG and MMYE. (D) Statistical analyses of spore viabilities. (E) Colony morphology of WT, ΔvadH, and the complemented strain on SM and grown for 7 days. The bottom panel shows close-up views of the colony in the top panel (Bar = 0.5 mm). (F) Statistical analyses of cleistothecia on MMG, MMYE and SM. The asterisks represent significant level (* p < 0.05, ** p < 0.001).
3.3. Disruption of VadH Led to Elevated Sensitivity to Osmotic Stress
For osmotic stress assays, the strains were inoculated on the solid MMG including sorbitol (1.0 M), glycerol (1.0 M) and NaCl (1.0 M), and cultured at 37 °C for 7 days. As shown in Figure 4A,B, ΔvadH was more sensitive to hyper-osmotic stress than WT, and it had an obvious difference in colony color on the stress plates. ΔvadH hardly formed conidia on the MMG with 1.0 M NaCl and produced noticeably fewer conidia than the wild type on the MMG with 1.0 M glycerol and 1.0 M sorbitol. Regarding conidial suspension assays, 102 conidia of ΔvadH could not form a colony on the MMG plates with glycerol (1.0 M), sorbitol (1.0 M) and NaCl (1.0 M) (Figure 4C). These results propose that the vadH gene is involved in regulating osmotic stress response of A. nidulans.
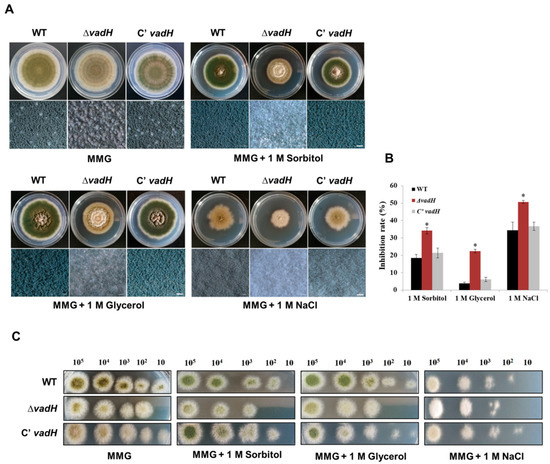
Figure 4.
Effects of the hyperosmotic medium on the growth of WT, ΔvadH and the complemented strain. (A) Comparison of the growth of strains on the MMG added with 1.0 M Sorbitol, 1.0 M Glycerol and 1.0 M NaCl and cultured at 37 °C for 7 days. The bottom panel shows close-up views of the colony in the top panel (Bar = 0.5 mm). (B) The inhibition rate of stress factors against WT, ΔvadH and the complemented strain. The asterisks represent significant level (* p < 0.05). (C) The growth of serially diluted conidia on the MMG added with 1.0 M Sorbitol, 1.0 M Glycerol and 1.0 M NaCl, culturing at 37 °C for 2 days.
3.4. Deleting VadH Leads to an Increase in ST Production
To test the effect of vadH on ST production, ST was extracted from WT, ΔvadH and C’vadH and analyzed using HPLC. As shown in Figure 5, the ΔvadH mutant produced more ST than WT and C’vadH, suggesting that VadH may negatively regulate ST production in A. nidulans.
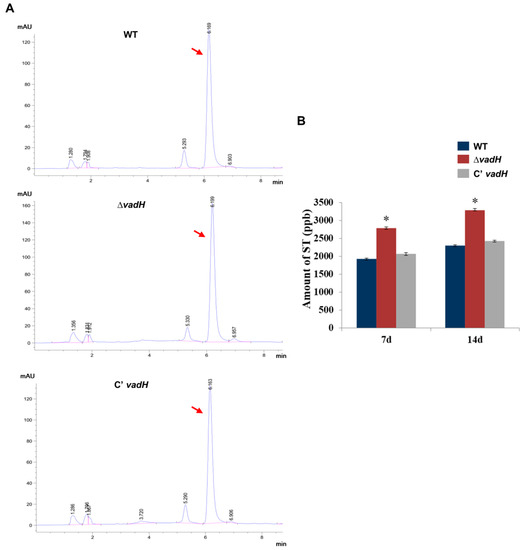
Figure 5.
HPLC analysis of ST yield. (A) Determination of ST production of WT, ΔvadH and the complemented strain using HPLC. Arrows indicate the peak of ST. (B) Statistical analyses of ST yields. The asterisk represents significant level (* p < 0.05).
3.5. Overexpression of VadH Suppresses Sexual Development
As mentioned above, the deletion of vadH leads to enhanced sexual development on MMG and MMYE. To further validate the function of VadH in fungal development, we generated the vadH-overexpression strain (OEvadH) and analyzed its phenotypes (Figure 6A). On the non-inducing medium, there was no significant differences in the number of conidia and cleistothecia between OEvadH and WT (Figure 6B,C). However, when induced, overexpression of vadH led to significantly reduced production of cleistothecia (Figure 6C). The expression level of vadH in OEvadH on the inducing medium was verified by qRT-PCR (Figure S3). Collectively, these results suggest that VadH is essential for sexual development, and it may act as a suppressor of sexual development.
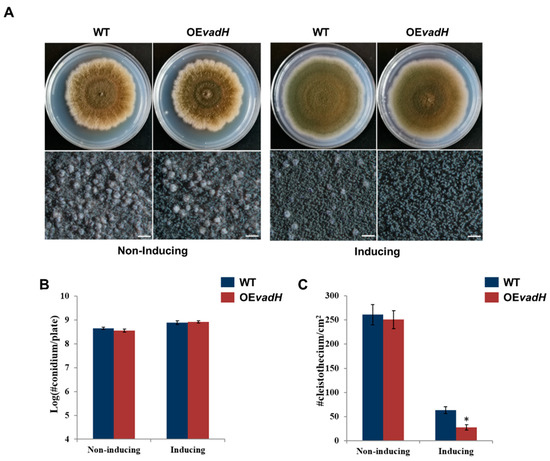
Figure 6.
Effects of overexpression of vadH on the development of A. nidulans. (A) WT and OEvadH were inoculated onto solid MMG (non-inducing) and MMT (100 mM threonine, inducing) and cultured at 37 °C for 7 days. The bottom panel shows close-up views of the colony in the top panel (Bar = 0.5 mm). (B) Statistical analyses of conidial yields under non-induced and induced conditions. (C) Statistical analyses of cleistothecia under non-induced and induced conditions. The asterisk represents significant level (* p < 0.05).
3.6. Transcriptomic Analyses of VadH
Genome-wide expression analyses of WT and ΔvadH under three stages were performed by employing RNA-Seq. The related data have been submitted to GenBank (PRJNA905844). For the vegetative growth stage, 895 DEGs were obtained, in which 328 DEGs were upregulated and 567 DEGs were downregulated. Nevertheless, for asexual and sexual development stages, more than 1200 DEGs were identified, suggesting that VadH is involved in regulating more genes in the developmental stages than those in vegetative growth (Figure 7A). The Venn diagram indicates that 126 DEGs, including 63 up-regulated and 63 down-regulated DEGs, participate in the vegetative growth, asexual and sexual development stage simultaneously (Figure 7B). The RNA-Seq results in the three stages were verified by qRT-PCR, and the expression levels of selected DEGs from three stages showed the same trend as those in RNA-Seq with all the correlation coefficients being more than 95% (Figure S4).
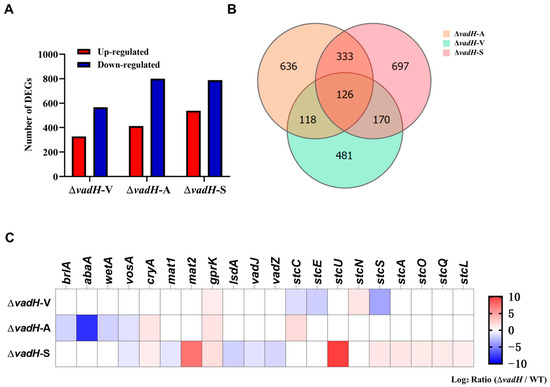
Figure 7.
Results of RNA-Seq analyses. (A) DEGs statistics of ΔvadH at in three stages. ΔvadH-V: vegetative growth, ΔvadH-A: asexual development, ΔvadH-S: sexual development. (B) A Venn diagram of DEGs in the three stages. (C) A heatmap of partial DEGs involved in asexual/sexual development and ST production.
Based on the RNA-Seq results, we further analyzed the expression of DEGs related to sporulation and ST biosynthesis. As shown in Figure 7C, brlA, abaA and wetA in the central regulatory pathway are downregulated significantly in the asexual development stage of ΔvadH, especially abaA. In the process of sexual development, the genes mat2 and ppgA related to sexual sporulation are upregulated, and lsdA, vadJ and vadZ are downregulated in ΔvadH; some of the genes (stuA, stuL, stuO, stuQ, stuS, stuU) in the gene cluster of ST biosynthesis are also upregulated to varying degrees.
Then, we performed the KEGG pathway analysis for DEGs in three stages (Figure 8). In the vegetative growth stage, six significant enrichment pathways were obtained, which were mainly related to amino acid metabolism and aflatoxin biosynthesis (Figure 8A). Regarding asexual and sexual development, more biological pathways were affected by VadH, including some pathways related to the metabolism of fatty acid, nitrogen and carbohydrate (Figure 8B,C). The numbers of significant enrichment pathways in the asexual and sexual development are more than twice as much as those in the vegetative growth stage.
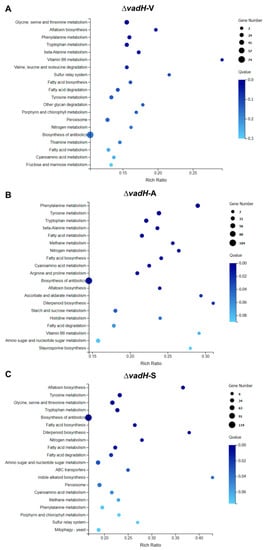
Figure 8.
KEGG pathway enrichment of DEGs during the vegetative growth (A), asexual developmental (B), and sexual developmental (C) phases.
4. Discussion
The velvet proteins are fungal-specific transcript factors that coordinate both development and secondary metabolism [11,15]. Previous studies indicated that the target genes regulated by VosA/VelB complex could be divided into VosA/VelB-activated developmental genes (VADs) and VosA/VelB-inhibited developmental genes (VIDs), and many VADs and VIDs are regulatory factors involved in the development process of A. nidulans [15,17]. Here, we characterized a new VAD gene vadH in A. nidulans, which encodes a zinc finger protein with four adjacent C2H2-type domains. The expression levels of vadH were noticeably decreased in the mutants ΔvosA or ΔvelB, suggesting that both VosA and VelB have a positive regulatory effect on vadH. It is observed that the homologous proteins of VadH can play various roles in different fungi [18,19,20]. In A. nidulans, VadH is involved in asexual and sexual development, osmotic stress response and ST production.
Our study indicated that VadH could balance the asexual and sexual development of A. nidulans. VadH could inhibit conidial production and stimulate sexual development on MMY and MMYE, and overexpression of vadH obviously suppresses the formation of cleistothecia. VadH can negatively regulate the expression of two genes related to sexual development, mat2 and ppgA, which may affect cleistothecium formation to a certain extent. It has been reported that Mat2 and PpgA can act as activators in the sexual development of A. nidulans [28]. In addition, VadH can also positively regulate the expression of lsdA, vadJ and vadZ, which have proved to function as repressors in sexual sporulation [29,30,31]. In A. nidulans, the central regulatory pathway activates conidial development and regulates the expression of specific genes in conidiation. In the mutant ΔvadH, the genes brlA, abaA, and wetA are all downregulated in the asexual development stage, which will inhibit the conidial production of ΔvadH. Especially abaA related to the differentiation of phialide is downregulated dramatically, which may affect the formation of conidia to a great extent. Therefore, VadH may function as a positive regulator in asexual sporulation and a negative one in sexual development. It is reported that VadA (AN5709), a member of VADs, also participates in the balance between asexual and sexual development [16]. The ΔvadA mutant exhibits increased production of cleistothecia, and overexpression of vadA leads to increased conidial production. Similar to vadH, the other two VADs, VadJ (AN3214, a histidine kinase) and VadZ (AN8774, a C6 transcription factor) also act as activators of asexual development and repressors of sexual development [30,31]. Interestingly, one member of VIDs, VidA (AN2498) proved to be essential for proper asexual and sexual development in A. nidulans. Deletion of vidA can lower the production of conidia, and slightly increases the yield of cleistothecia [32]. These reported genes regulated directly by the VosA-VelB complex all have a function of balancing asexual and sexual development, suggesting that the complex does play a crucial role in the development process of A. nidulans.
VadH participates in the osmotic pressure response of A. nidulans. Deletion of vadH leads to elevated sensitivity to hyperosmotic stress, and the asexual sporulation of ΔvadH is also suppressed significantly on the hyperosmotic media. The KEGG analysis shows that the expression of more than 60 genes in the mitogen-activated protein kinase (MAPK) signaling pathway is influenced by VadH, and many of them are involved in cell wall integrity and high osmolarity pathways. We speculate that deleting vadH may change cell wall integrity, subsequently affecting the osmotic pressure response of A. nidulans. In S. cerevisiae, Azf1 also have the function of maintaining cell wall integrity when glucose is depleted [18]. However, the VadH orthologues, Cos1 and CgAzf1 are not related to osmotic pressure response in M. oryzae and C. gloeosporioides, respectively [19,20]. Further study will be needed to elucidate the precise mechanism of VadH coordinating the hyperosmotic stress. Regarding secondary metabolism, VadH proves to be involved in regulating ST production. Disruption of vadH leads to elevated ST production, and the RNA-Seq results also indicate that VadH can negatively regulate the expression of several genes related to the biosynthesis of ST. It has been found that Cos1 and CgAzf1 are also relevant to secondary metabolism, and both of them can regulate melanin production [19,20]. Similar to vadH, the VADs, vadA, vadJ and vadZ, also exhibit the function of suppressing ST production [16,30,31], whereas the VID gene vidA is not involved in the biosynthesis of ST [32].
5. Conclusions
Taken together, we propose a working model for VadH regulating asexual/sexual development and secondary metabolism (Figure 9). The VosA-VelB complex can positively regulate the expression of vadH. Then, VadH functions as a positive regulator for asexual sporulation through the central regulatory pathway, and it also acts as a negative regulator for the production of cleistothecia and sterigmatocystin. In summary, a newly identified VAD gene vadH is predicted to encode a C2H2-type transcription factor, which is involved in balancing asexual/sexual development and regulating osmotic stress and sterigmatocystin production. These findings provide further evidence for the crucial roles of VADs in the development and secondary metabolism of A. nidulans. Understanding the mechanism of VadH will contribute to revealing the precise regulatory networks of the VosA-VelB complex.
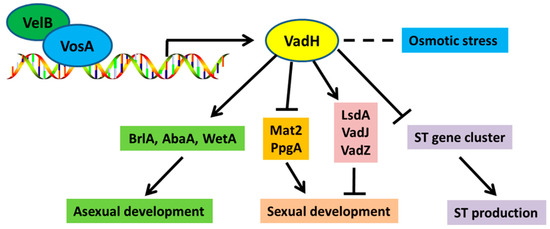
Figure 9.
A genetic model of VadH regulating asexual/sexual development and secondary metabolism of A. nidulans.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11243998/s1. Figure S1: Verification of the gene-knockout mutant and complementary strain; Figure S2: Conidial germination of WT, ΔvadH and the complemented strain; Figure S3: Verification of the mRNA level of vadH in OEvadH; Figure S4: The qRT-PCR verification of RNA-Seq data in three stages; Table S1: Aspergillus strains used in this study; Table S2: Primers used in this study.
Author Contributions
Conceptualization, X.L., Z.L. and J.-H.Y.; investigation, X.L., Y.Z. and J.L.; formal analysis, X.L., H.M., H.-S.P. and Z.L.; supervision, Z.L. and J.-H.Y.; funding acquisition, X.L., Z.L. and J.-H.Y.; writing—original draft preparation, X.L. and Z.L.; writing—review and editing, H.-S.P. and J.-H.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the Natural Science Foundation of Hainan Province (2019RC074) and the National Natural Science Foundation of China (32160041 and 32160371). This work was also supported by the fund of China Scholarship Council. The work at UW-Madison was supported by the National Institute of Food and Agriculture, United States Department of Agriculture, Hatch project 7000326 to Jae-Hyuk Yu.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Bennett, J.W. An overview of the genus Aspergillus. Aspergillus Mol. Biol. Genom. 2010, 1–17. [Google Scholar]
- Park, H.S.; Yu, J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Ebbole, D.J. The Conidium. In Cellular and Molecular Biology of Filamentous Fungi; ASM Press: Washington, DC, USA, 2010; pp. 577–590. [Google Scholar]
- Adams, T.H.; Wieser, J.K.; Yu, J.H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H. Regulation of development in Aspergillus nidulans and Aspergillus fumigates. Mycobiology 2010, 38, 229–237. [Google Scholar] [CrossRef]
- Han, S.; Adams, T. Complex control of the developmental regulatory locus brlA in Aspergillus nidulans. Mol. Genet. Genom. 2001, 266, 260–270. [Google Scholar] [CrossRef]
- Andrianopoulos, A.; Timberlake, W.E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 1994, 14, 2503–2515. [Google Scholar] [PubMed]
- Marshall, M.A.; Timberlake, W.E. Aspergillus nidulans wetA activates spore-specific gene expression. Mol. Cell. Biol. 1991, 11, 55–62. [Google Scholar]
- Park, H.S.; Nam, T.Y.; Han, K.H.; Kim, S.C.; Yu, J.H. VelC positively controls sexual development in Aspergillus nidulans. PLoS ONE 2014, 9, e89883. [Google Scholar] [CrossRef]
- Park, H.S.; Yu, J.H. Velvet Regulators in Aspergillus spp. Microbiol. Biotechnol. Lett. 2016, 44, 409–419. [Google Scholar] [CrossRef]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Ni, M.; Jeong, K.C.; Kim, Y.H.; Yu, J.H. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS ONE 2012, 7, e45935. [Google Scholar] [CrossRef] [PubMed]
- Sarikaya Bayram, O.; Bayram, O.; Valerius, O.; Park, H.S.; Irniger, S.; Gerke, J.; Ni, M.; Han, K.H.; Yu, J.H.; Braus, G.H. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 2010, 6, e1001226. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Y.L.; Gerke, J.; Park, H.S.; Bayram, O.; Neumann, P.; Ni, M.; Dickmanns, A.; Kim, S.C.; Yu, J.H.; Braus, G.H.; et al. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-kappaB. PLoS Biol. 2013, 11, e1001750. [Google Scholar] [CrossRef]
- Park, H.S.; Lee, M.K.; Kim, S.C.; Yu, J.H. The role of VosA/VelB-activated developmental gene vadA in Aspergillus nidulans. PLoS ONE 2017, 12, e0177099. [Google Scholar] [CrossRef]
- Park, H.S.; Yu, Y.M.; Lee, M.K.; Maeng, P.J.; Kim, S.C.; Yu, J.H. Velvet-mediated repression of beta-glucan synthesis in Aspergillus nidulans spores. Sci. Rep. 2015, 5, 10199. [Google Scholar] [CrossRef]
- Slattery, M.G.; Liko, D.; Heideman, W. The function and properties of the Azf1 transcriptional regulator change with growth conditions in Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 313–320. [Google Scholar] [CrossRef]
- Zhou, Z.Z.; Li, G.H.; Lin, C.H.; He, C.Z. Conidiophore Stalk less1 encodes a putative zinc-finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae. Mol. Plant Microbe Interact. 2009, 22, 402–410. [Google Scholar] [CrossRef]
- Li, X.; Ke, Z.; Yu, X.; Liu, Z.; Zhang, C. Transcription factor CgAzf1 regulates melanin production, conidial development and infection in Colletotrichum gloeosporioides. Antonie Van Leeuwenhoek 2019, 112, 1095–1104. [Google Scholar] [CrossRef]
- Shaaban, M.I.; Bok, J.W.; Lauer, C.; Keller, N.P. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell 2010, 9, 1816–1824. [Google Scholar] [CrossRef]
- Kwon, N.J.; Shin, K.S.; Yu, J.H. Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Dominguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Yu, J.H. Multi-copy genetic screen in Aspergillus nidulans. Methods Mol. Biol. 2012, 944, 183–190. [Google Scholar] [PubMed]
- Lee, M.K.; Park, H.S.; Han, K.H.; Hong, S.B.; Yu, J.H. High molecular weight genomic DNA mini-prep for filamentous fungi. Fungal Genet. Biol. 2017, 104, 1–5. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Liu, Z.; Zhang, C. The function and transcriptome analysis of a bZIP transcription factor CgAP1 in Colletotrichum gloeosporioides. Microbiol. Res. 2017, 197, 39–48. [Google Scholar] [CrossRef]
- Paoletti, M.; Seymour, F.A.; Alcocer, M.J.C.; Kaur, N.; Calvo, A.M.; Archer, D.B.; Dyer, P.S. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 2007, 17, 1384–1389. [Google Scholar] [CrossRef]
- Lee, D.W.; Kim, S.; Kim, S.J.; Han, D.M.; Jahng, K.Y.; Chae, K.S. The lsdA gene is necessary for sexual development inhibition by a salt in Aspergillus nidulans. Curr. Genet. 2001, 39, 237–243. [Google Scholar]
- Zhao, Y.; Lee, M.K.; Lim, J.; Moon, H.; Park, H.S.; Zheng, W.; Yu, J.H. The putative sensor histidine kinase VadJ coordinates development and sterigmatocystin production in Aspergillus nidulans. J. Microbiol. 2021, 59, 746–752. [Google Scholar] [CrossRef]
- Zhao, Y.; Lee, M.K.; Lim, J.; Moon, H.; Park, H.S.; Zheng, W.; Yu, J.H. The velvet-activated putative C6 transcription factor VadZ regulates development and sterigmatocystin production in Aspergillus nidulans. Fungal Biol. 2022, 126, 421–428. [Google Scholar] [CrossRef]
- Kim, M.J.; Jung, W.H.; Son, Y.E.; Yu, J.H.; Lee, M.K.; Park, H.S. The velvet repressed vidA gene plays a key role in governing development in Aspergillus nidulans. J. Microbiol. 2019, 57, 893–899. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).