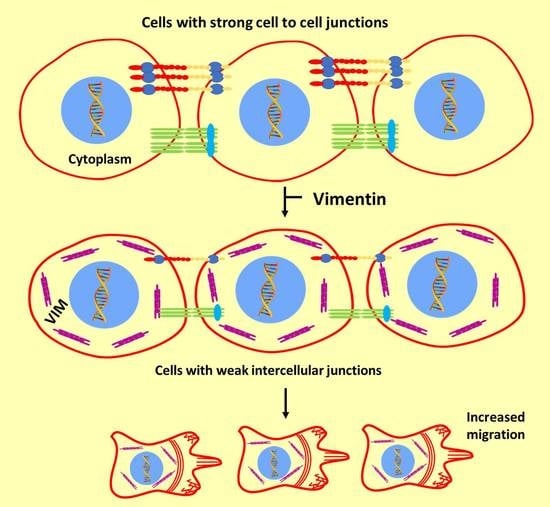
Transcriptome Analysis Reveals Vimentin-Induced Disruption of Cell–Cell Associations Augments Breast Cancer Cell Migration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cell Lines
2.2. Antibodies
2.3. RNA Extraction and cDNA Synthesis
2.4. Quantitative PCR
2.5. Protein Extraction and Western Blotting
2.6. Plasmid Constructs, cDNA Cloning, Retrovirus Production and Spinfection
2.7. Analysis of Cell Parameters
2.8. Colony Assay
2.9. CyQUANTTM Cell Proliferation and Cell Adhesion Assays
2.10. MTT Assay
2.11. Wound Healing Assay
2.12. Chemotactic Migration
2.13. RNA-Seq Analysis and Bioinformatics
3. Statistical Analysis
4. Results
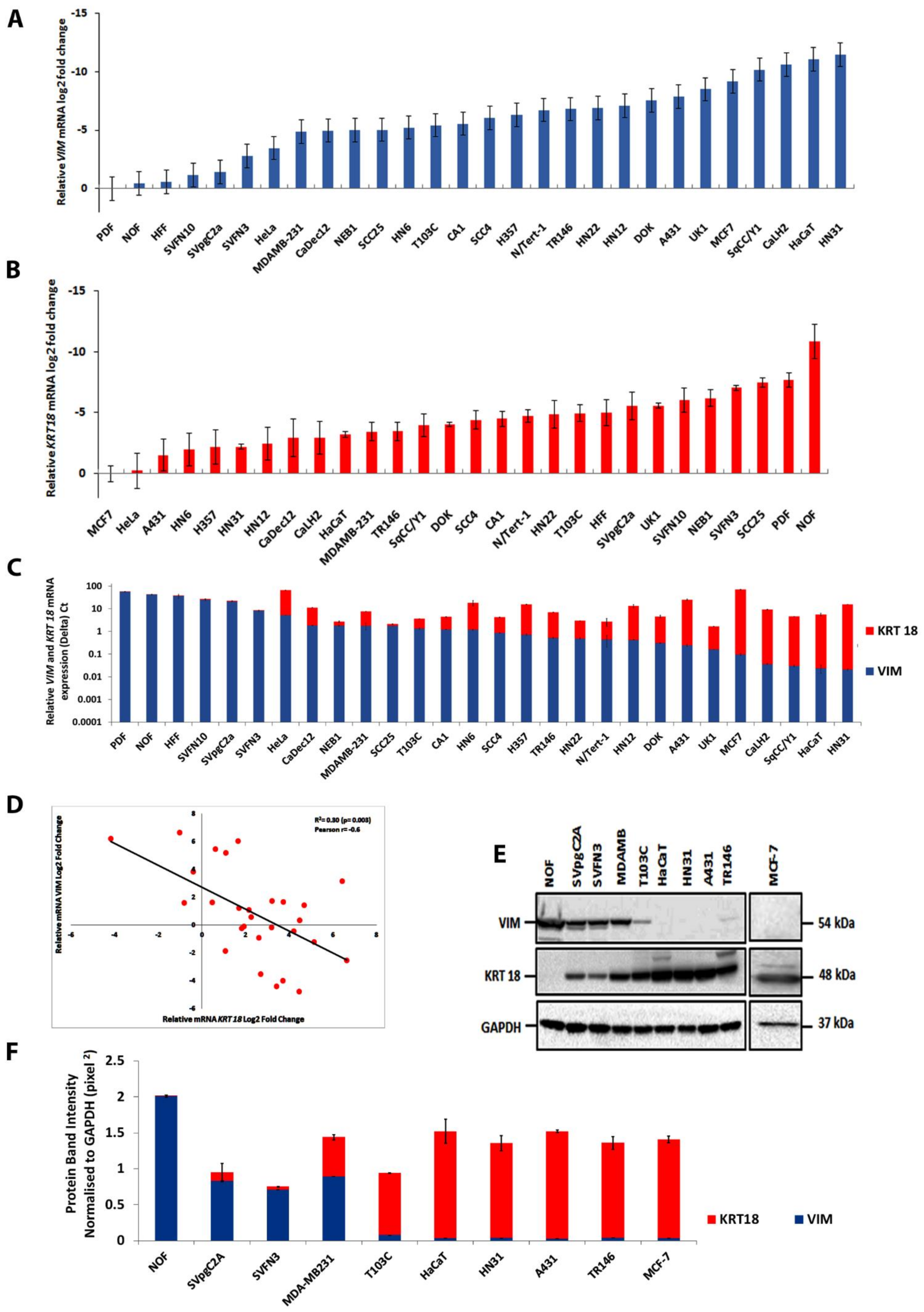
4.1. Screening Human Cell Lines for Vimentin and Keratin K18 Expression
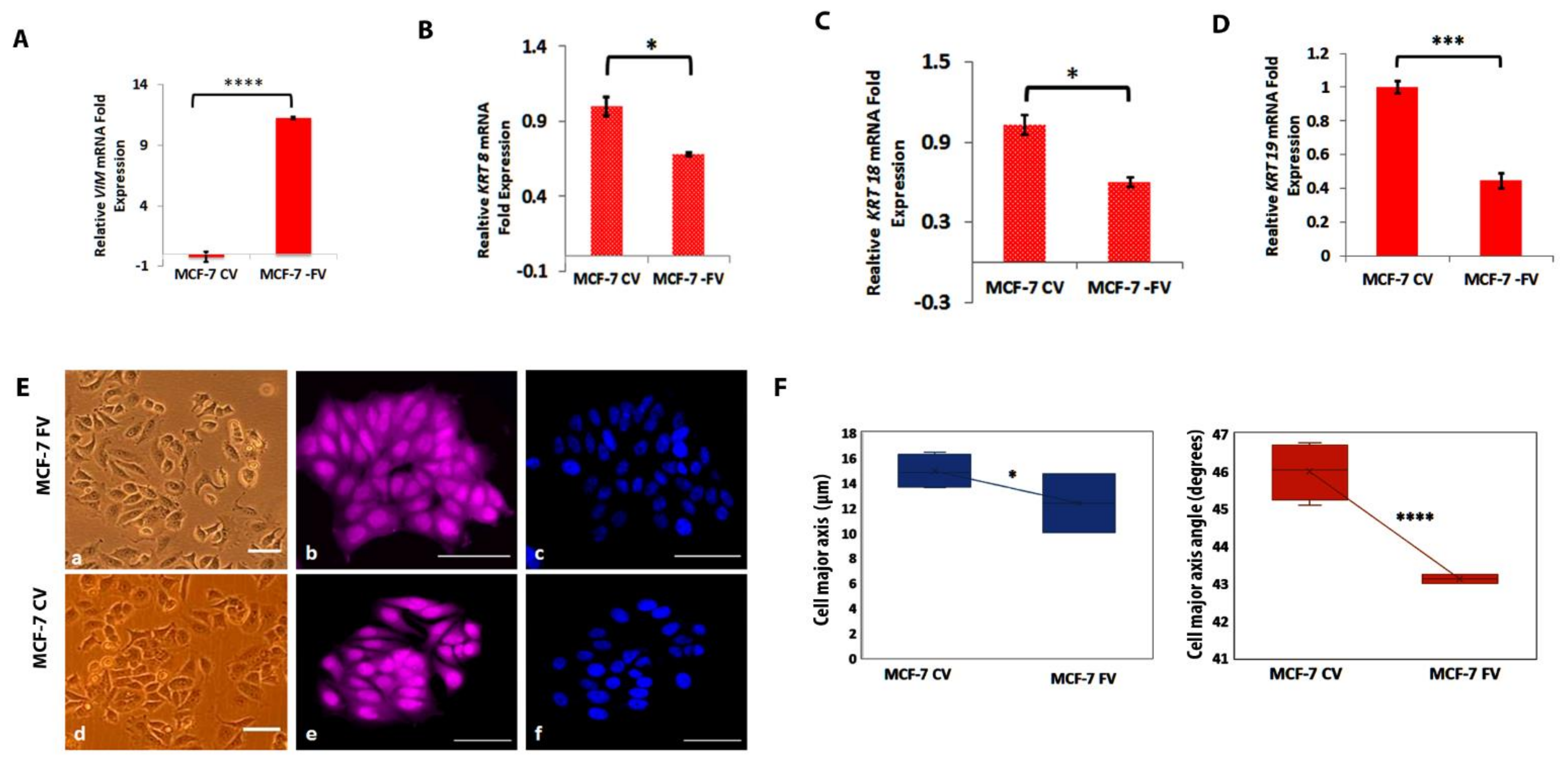
4.2. Ectopic Expression of Vimentin in MCF-7 Cells Downregulated KRT8, KRT18 and KRT19
4.3. Ectopically Expressed Vimentin Alters MCF-7 Morphology
4.4. Vimentin Does Not Change Cell Proliferation or Adhesion but Induces Cell Migration
4.5. RNA-Seq and Bioinformatics Analyses
4.6. Functional Enrichment Analysis of DEGs
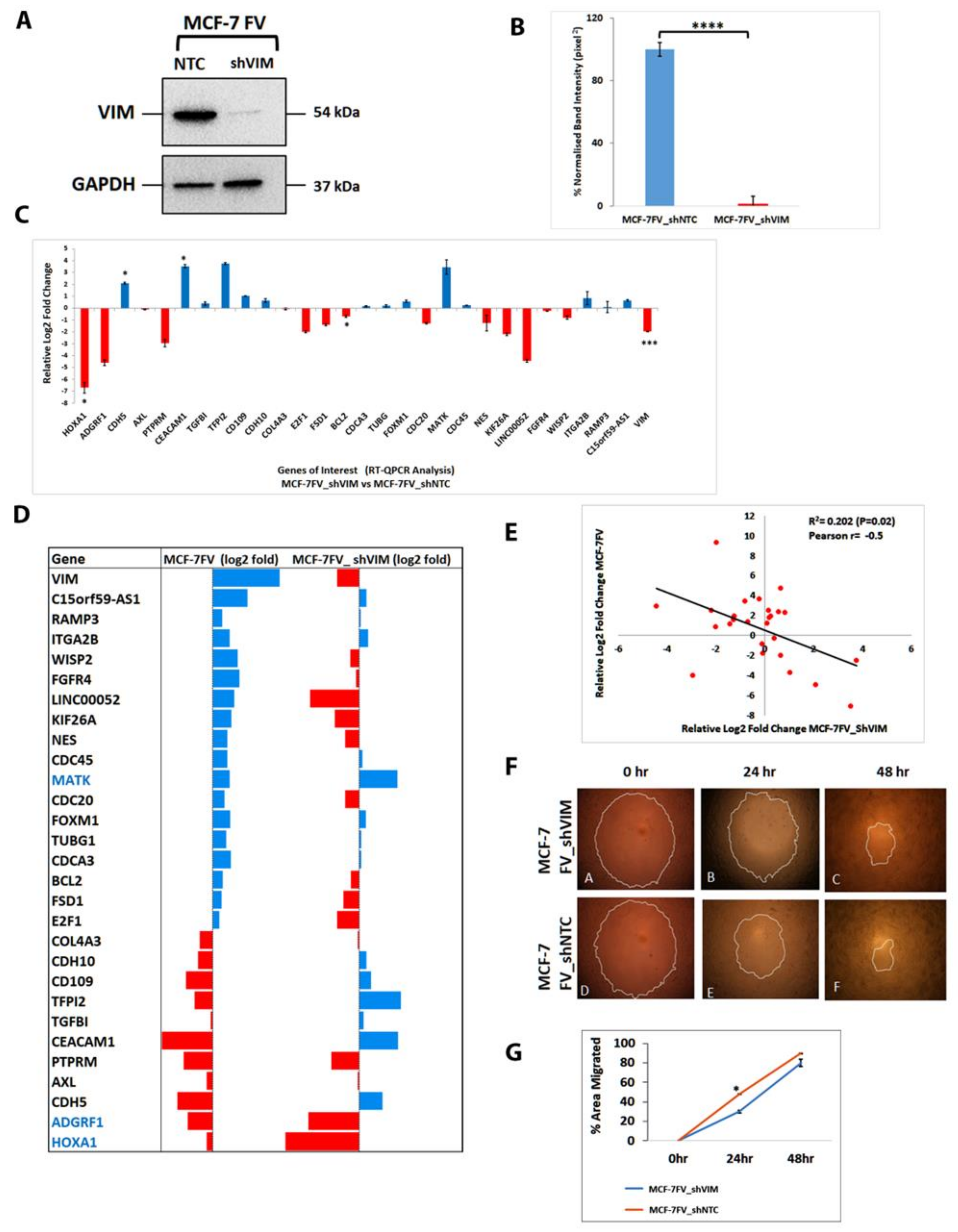
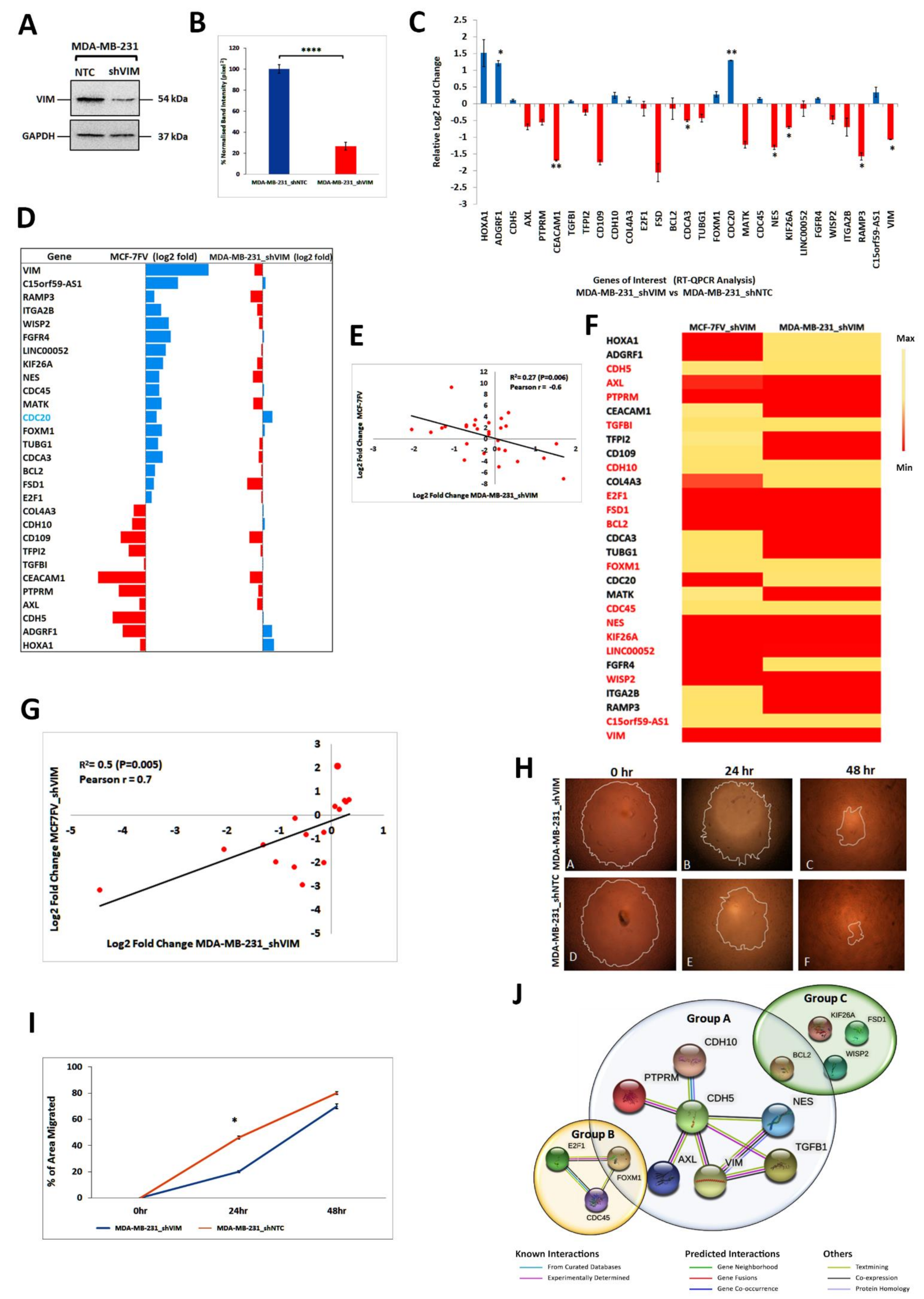
4.7. shRNA Mediated Downregulation of Vimentin in MCF-7FV Suppressed Its Migration and Reversed the Profile of DEGs of Interest
4.8. Vimentin Downregulation in MDA-MB-231 Cells Induced Changes Similar to Those in MCF-7FV_shVIM
4.9. Vimentin Responsive Genes Common in MCF-7FV_shVIM and MDA-MB-231_shVIM
4.10. Downregulation of CDH5 in MCF-7 Increases Cell Migration
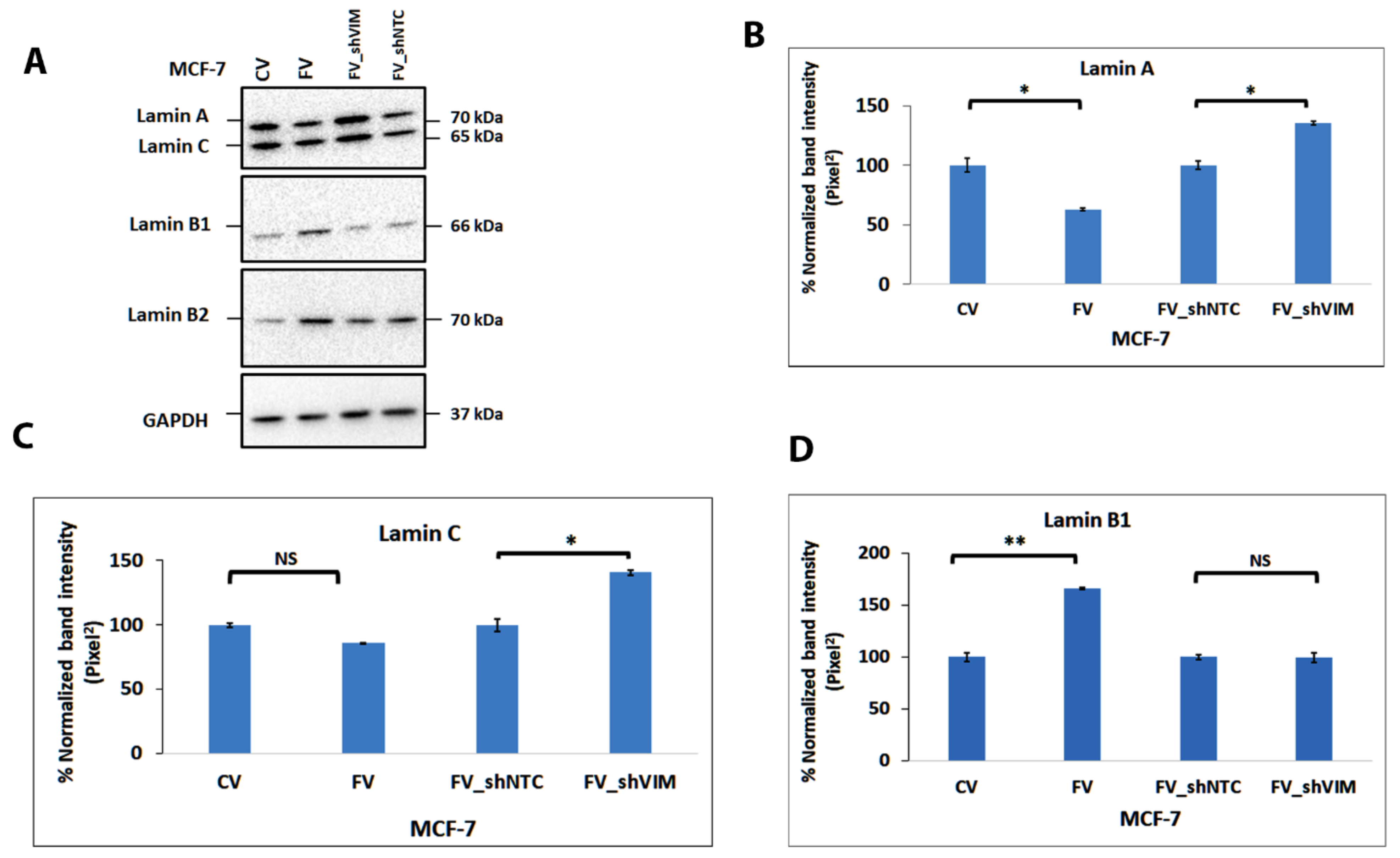
4.11. Vimentin Expression in MCF-7 Alters the Lamin Expression
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saez, A.; Gonzalez-Granado, J.M. Recent Advances in Intermediate Filaments. Int. J. Mol. Sci. 2022, 23, 5308. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.T.; Coulombe, P.A.; Kwan, R.; Omary, M.B. Types I and II Keratin Intermediate Filaments. Cold Spring Harb. Perspect. Biol. 2018, 10, a018275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usman, S.; Waseem, N.H.; Nguyen, T.K.N.; Mohsin, S.; Jamal, A.; Teh, M.T.; Waseem, A. Vimentin Is at the Heart of Epithelial Mesenchymal Transition (EMT) Mediated Metastasis. Cancers 2021, 13, 4985. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Lin, H.H.; Tang, M.J.; Wang, Y.K. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget 2015, 6, 15966–15983. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, N.; Tokunaga, E.; Kitao, H.; Hisamatsu, Y.; Taketani, K.; Akiyoshi, S.; Okada, S.; Aishima, S.; Morita, M.; Maehara, Y. Vimentin as a poor prognostic factor for triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2013, 139, 739–746. [Google Scholar] [CrossRef]
- Shi, R.; Liu, L.; Wang, F.; He, Y.; Niu, Y.; Wang, C.; Zhang, X.; Zhang, X.; Zhang, H.; Chen, M.; et al. Downregulation of cytokeratin 18 induces cellular partial EMT and stemness through increasing EpCAM expression in breast cancer. Cell. Signal 2020, 76, 109810. [Google Scholar] [CrossRef]
- Velez-delValle, C.; Marsch-Moreno, M.; Castro-Munozledo, F.; Galvan-Mendoza, I.J.; Kuri-Harcuch, W. Epithelial cell migration requires the interaction between the vimentin and keratin intermediate filaments. Sci. Rep. 2016, 6, 24389. [Google Scholar] [CrossRef] [Green Version]
- Steinert, P.M.; Marekov, L.N.; Parry, D.A. Diversity of intermediate filament structure. Evidence that the alignment of coiled-coil molecules in vimentin is different from that in keratin intermediate filaments. J. Biol. Chem. 1993, 268, 24916–24925. [Google Scholar] [CrossRef]
- Biskou, O.; Casanova, V.; Hooper, K.M.; Kemp, S.; Wright, G.P.; Satsangi, J.; Barlow, P.G.; Stevens, C. The type III intermediate filament vimentin regulates organelle distribution and modulates autophagy. PLoS ONE 2019, 14, e0209665. [Google Scholar] [CrossRef]
- Keeling, M.C.; Flores, L.R.; Dodhy, A.H.; Murray, E.R.; Gavara, N. Actomyosin and vimentin cytoskeletal networks regulate nuclear shape, mechanics and chromatin organization. Sci. Rep. 2017, 7, 5219. [Google Scholar] [CrossRef]
- Gao, Y.; Sztul, E. A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. J. Cell Biol. 2001, 152, 877–894. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Barlan, K.; Chou, Y.H.; Grin, B.; Lakonishok, M.; Serpinskaya, A.S.; Shumaker, D.K.; Herrmann, H.; Gelfand, V.I.; Goldman, R.D. The dynamic properties of intermediate filaments during organelle transport. J. Cell Sci. 2009, 122, 2914–2923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matveeva, E.A.; Chernoivanenko, I.S.; Minin, A.A. Vimentin intermediate filaments protect mitochondria from oxidative stress. Biochem. Suppl. Ser. A Membr. Cell Biol. 2010, 4, 321–331. [Google Scholar] [CrossRef]
- Lian, N.; Wang, W.; Li, L.; Elefteriou, F.; Yang, X. Vimentin inhibits ATF4-mediated osteocalcin transcription and osteoblast differentiation. J. Biol. Chem. 2009, 284, 30518–30525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Sala, D.; Oeste, C.L.; Martinez, A.E.; Carrasco, M.J.; Garzon, B.; Canada, F.J. Vimentin filament organization and stress sensing depend on its single cysteine residue and zinc binding. Nat. Commun. 2015, 6, 7287. [Google Scholar] [CrossRef] [Green Version]
- Pattabiraman, S.; Azad, G.K.; Amen, T.; Brielle, S.; Park, J.E.; Sze, S.K.; Meshorer, E.; Kaganovich, D. Vimentin protects differentiating stem cells from stress. Sci. Rep. 2020, 10, 19525. [Google Scholar] [CrossRef]
- Challa, A.A.; Stefanovic, B. A novel role of vimentin filaments: Binding and stabilization of collagen mRNAs. Mol. Cell. Biol. 2011, 31, 3773–3789. [Google Scholar] [CrossRef] [Green Version]
- Ostrowska-Podhorodecka, Z.; Ding, I.; Norouzi, M.; McCulloch, C.A. Impact of Vimentin on Regulation of Cell Signaling and Matrix Remodeling. Front. Cell Dev. Biol. 2022, 10, 869069. [Google Scholar] [CrossRef]
- Terriac, E.; Coceano, G.; Mavajian, Z.; Hageman, T.A.; Christ, A.F.; Testa, I.; Lautenschlager, F.; Gad, A.K. Vimentin Levels and Serine 71 Phosphorylation in the Control of Cell-Matrix Adhesions, Migration Speed, and Shape of Transformed Human Fibroblasts. Cells 2017, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Burgstaller, G.; Gregor, M.; Winter, L.; Wiche, G. Keeping the vimentin network under control: Cell-matrix adhesion-associated plectin 1f affects cell shape and polarity of fibroblasts. Mol. Biol. Cell 2010, 21, 3362–3375. [Google Scholar] [CrossRef]
- Kim, H.; Nakamura, F.; Lee, W.; Shifrin, Y.; Arora, P.; McCulloch, C.A. Filamin A is required for vimentin-mediated cell adhesion and spreading. Am. J. Physiol. Cell Physiol. 2010, 298, C221–C236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, I.; Chu, D.; Chou, Y.H.; Goldman, R.D.; Matsudaira, P. Integrating the actin and vimentin cytoskeletons. adhesion-dependent formation of fimbrin-vimentin complexes in macrophages. J. Cell Biol. 1999, 146, 831–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patteson, A.E.; Carroll, R.J.; Iwamoto, D.V.; Janmey, P.A. The vimentin cytoskeleton: When polymer physics meets cell biology. Phys. Biol. 2020, 18, 011001. [Google Scholar] [CrossRef] [PubMed]
- Burke, B.; Stewart, C.L. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 2013, 14, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Georgatos, S.D.; Blobel, G. Lamin B constitutes an intermediate filament attachment site at the nuclear envelope. J. Cell Biol. 1987, 105, 117–125. [Google Scholar] [CrossRef] [Green Version]
- Vahabikashi, A.; Sivagurunathan, S.; Nicdao, F.A.S.; Han, Y.L.; Park, C.Y.; Kittisopikul, M.; Wong, X.; Tran, J.R.; Gundersen, G.G.; Reddy, K.L.; et al. Nuclear lamin isoforms differentially contribute to LINC complex-dependent nucleocytoskeletal coupling and whole-cell mechanics. Proc. Natl. Acad. Sci. USA 2022, 119, e2121816119. [Google Scholar] [CrossRef]
- Messica, Y.; Laser-Azogui, A.; Volberg, T.; Elisha, Y.; Lysakovskaia, K.; Eils, R.; Gladilin, E.; Geiger, B.; Beck, R. The role of Vimentin in Regulating Cell Invasive Migration in Dense Cultures of Breast Carcinoma Cells. Nano Lett. 2017, 17, 6941–6948. [Google Scholar] [CrossRef]
- Mendez, M.G.; Kojima, S.; Goldman, R.D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010, 24, 1838–1851. [Google Scholar] [CrossRef] [Green Version]
- Vuoriluoto, K.; Haugen, H.; Kiviluoto, S.; Mpindi, J.P.; Nevo, J.; Gjerdrum, C.; Tiron, C.; Lorens, J.B.; Ivaska, J. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 2011, 30, 1436–1448. [Google Scholar] [CrossRef] [Green Version]
- Paccione, R.J.; Miyazaki, H.; Patel, V.; Waseem, A.; Gutkind, J.S.; Zehner, Z.E.; Yeudall, W.A. Keratin down-regulation in vimentin-positive cancer cells is reversible by vimentin RNA interference, which inhibits growth and motility. Mol. Cancer Ther. 2008, 7, 2894–2903. [Google Scholar] [CrossRef]
- Hendrix, M.J.; Seftor, E.A.; Seftor, R.E.; Trevor, K.T. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am. J. Pathol. 1997, 150, 483–495. [Google Scholar] [PubMed]
- Mackenzie, I.C. Growth of malignant oral epithelial stem cells after seeding into organotypical cultures of normal mucosa. J. Oral. Pathol. Med. 2004, 33, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.J.; Piper, K.; Common, J.; Fortune, F.; Mackenzie, I.C. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J. Oral. Pathol. Med. 2007, 36, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Locke, M.; Heywood, M.; Fawell, S.; Mackenzie, I.C. Retention of intrinsic stem cell hierarchies in carcinoma-derived cell lines. Cancer Res. 2005, 65, 8944–8950. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, P.S.; Sundqvist, K.; Betsholtz, C.; Hoglund, P.; Wiman, K.G.; Zhivotovsky, B.; Bertolero, F.; Liu, Y.; Grafstrom, R.C. Characterization of human buccal epithelial cells transfected with the simian virus 40 T-antigen gene. Carcinogenesis 1995, 16, 2515–2521. [Google Scholar] [CrossRef]
- Rheinwald, J.G.; Beckett, M.A. Defective terminal differentiation in culture as a consistent and selectable character of malignant human keratinocytes. Cell 1980, 22, 629–632. [Google Scholar] [CrossRef]
- Rupniak, H.T.; Rowlatt, C.; Lane, E.B.; Steele, J.G.; Trejdosiewicz, L.K.; Laskiewicz, B.; Povey, S.; Hill, B.T. Characteristics of four new human cell lines derived from squamous cell carcinomas of the head and neck. J. Natl. Cancer Inst. 1985, 75, 621–635. [Google Scholar]
- Chang, S.E.; Foster, S.; Betts, D.; Marnock, W.E. DOK, a cell line established from human dysplastic oral mucosa, shows a partially transformed non-malignant phenotype. Int. J. Cancer 1992, 52, 896–902. [Google Scholar] [CrossRef]
- Easty, D.M.; Easty, G.C.; Carter, R.L.; Monaghan, P.; Butler, L.J. Ten human carcinoma cell lines derived from squamous carcinomas of the head and neck. Br. J. Cancer 1981, 43, 772–785. [Google Scholar] [CrossRef] [Green Version]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors, Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Lucey, B.P.; Nelson-Rees, W.A.; Hutchins, G.M. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch. Pathol. Lab. Med. 2009, 133, 1463–1467. [Google Scholar] [CrossRef] [PubMed]
- Cailleau, R.; Young, R.; Olive, M.; Reeves, W.J., Jr. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 1974, 53, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Soule, H.D.; Vazguez, J.; Long, A.; Albert, S.; Brennan, M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Qadir, F.; Aziz, M.A.; Sari, C.P.; Ma, H.; Dai, H.; Wang, X.; Raithatha, D.; Da Silva, L.G.L.; Hussain, M.; Poorkasreiy, S.P.; et al. Transcriptome reprogramming by cancer exosomes: Identification of novel molecular targets in matrix and immune modulation. Mol. Cancer 2018, 17, 97. [Google Scholar] [CrossRef] [Green Version]
- Gemenetzidis, E.; Bose, A.; Riaz, A.M.; Chaplin, T.; Young, B.D.; Ali, M.; Sugden, D.; Thurlow, J.K.; Cheong, S.C.; Teo, S.H.; et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS ONE 2009, 4, e4849. [Google Scholar] [CrossRef]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef] [Green Version]
- Eiselleova, L.; Peterkova, I.; Neradil, J.; Slaninova, I.; Hampl, A.; Dvorak, P. Comparative study of mouse and human feeder cells for human embryonic stem cells. Int. J. Dev. Biol. 2008, 52, 353–363. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, Z.; Rajthala, S.; Sapkota, D.; Dongre, H.; Parajuli, H.; Suliman, S.; Das, R.; Li, L.; Bindoff, L.A.; et al. Metabolic reprogramming of normal oral fibroblasts correlated with increased glycolytic metabolism of oral squamous cell carcinoma and precedes their activation into carcinoma associated fibroblasts. Cell Mol. Life Sci. 2020, 77, 1115–1133. [Google Scholar] [CrossRef]
- Brem, H.; Golinko, M.S.; Stojadinovic, O.; Kodra, A.; Diegelmann, R.F.; Vukelic, S.; Entero, H.; Coppock, D.L.; Tomic-Canic, M. Primary cultured fibroblasts derived from patients with chronic wounds: A methodology to produce human cell lines and test putative growth factor therapy such as GMCSF. J. Transl. Med. 2008, 6, 75. [Google Scholar] [CrossRef] [Green Version]
- Bryan, D.; Sexton, C.J.; Williams, D.; Leigh, I.M.; McKay, I.A. Oral keratinocytes immortalized with the early region of human papillomavirus type 16 show elevated expression of interleukin 6, which acts as an autocrine growth factor for the derived T103C cell line. Cell Growth Differ. 1995, 6, 1245–1250. [Google Scholar]
- Dickson, M.A.; Hahn, W.C.; Ino, Y.; Ronfard, V.; Wu, J.Y.; Weinberg, R.A.; Louis, D.N.; Li, F.P.; Rheinwald, J.G. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell Biol. 2000, 20, 1436–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morley, S.M.; D’Alessandro, M.; Sexton, C.; Rugg, E.L.; Navsaria, H.; Shemanko, C.S.; Huber, M.; Hohl, D.; Heagerty, A.I.; Leigh, I.M.; et al. Generation and characterization of epidermolysis bullosa simplex cell lines: Scratch assays show faster migration with disruptive keratin mutations. Br. J. Dermatol. 2003, 149, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Yeudall, W.A.; Crawford, R.Y.; Ensley, J.F.; Robbins, K.C. MTS1/CDK4I is altered in cell lines derived from primary and metastatic oral squamous cell carcinoma. Carcinogenesis 1994, 15, 2683–2686. [Google Scholar] [CrossRef] [PubMed]
- Aldehlawi, H.; Usman, S.; Lalli, A.; Ahmad, F.; Williams, G.; Teh, M.T.; Waseem, A. Serum lipids, retinoic acid and phenol red differentially regulate expression of keratins K1, K10 and K2 in cultured keratinocytes. Sci. Rep. 2020, 10, 4829. [Google Scholar] [CrossRef] [Green Version]
- Usman, S.; Aldehlawi, H.; Nguyen, T.K.N.; Teh, M.T.; Waseem, A. Impact of N-Terminal Tags on De Novo Vimentin Intermediate Filament Assembly. Int. J. Mol. Sci. 2022, 23, 6349. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [Green Version]
- Edelheit, O.; Hanukoglu, A.; Hanukoglu, I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Swift, S.; Lorens, J.; Achacoso, P.; Nolan, G.P. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr. Protoc. Immunol. 1999, 31, 10–17. [Google Scholar] [CrossRef]
- Aldehlawi, H.; Niemiec, K.A.; Avisetti, D.R.; Lalli, A.; Teh, M.T.; Waseem, A. The monoclonal antibody EPR1614Y against the stem cell biomarker keratin K15 lacks specificity and reacts with other keratins. Sci. Rep. 2019, 9, 1943. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- LCC_Omen. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM5400803 (accessed on 1 December 2022).
- Chavez, K.J.; Garimella, S.V.; Lipkowitz, S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010, 32, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Polioudaki, H.; Agelaki, S.; Chiotaki, R.; Politaki, E.; Mavroudis, D.; Matikas, A.; Georgoulias, V.; Theodoropoulos, P.A. Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC Cancer 2015, 15, 399. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.A.; Kirschmann, D.A.; Cerhan, J.R.; Folberg, R.; Seftor, E.A.; Sellers, T.A.; Hendrix, M.J. Association between keratin and vimentin expression, malignant phenotype, and survival in postmenopausal breast cancer patients. Clin. Cancer Res. 1999, 5, 2698–2703. [Google Scholar] [PubMed]
- Langa, F.; Kress, C.; Colucci-Guyon, E.; Khun, H.; Vandormael-Pournin, S.; Huerre, M.; Babinet, C. Teratocarcinomas induced by embryonic stem (ES) cells lacking vimentin: An approach to study the role of vimentin in tumorigenesis. J. Cell. Sci. 2000, 113 Pt 19, 3463–3472. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.M.; Wen, Y.J.; Yang, H.B.; Qin, G.; Tian, L.; Deng, H.X.; Wen, B. Enhanced expression of human vimentin intermediate filaments in hepatocellular carcinoma cells decreases their proliferative and invasive abilities in vitro. Zhonghua Zhong Liu Za Zhi 2008, 30, 408–412. [Google Scholar] [PubMed]
- Kim, J.; Jang, J.; Yang, C.; Kim, E.J.; Jung, H.; Kim, C. Vimentin filament controls integrin alpha5beta1-mediated cell adhesion by binding to integrin through its Ser38 residue. FEBS Lett. 2016, 590, 3517–3525. [Google Scholar] [CrossRef] [PubMed]
- Havel, L.S.; Kline, E.R.; Salgueiro, A.M.; Marcus, A.I. Vimentin regulates lung cancer cell adhesion through a VAV2-Rac1 pathway to control focal adhesion kinase activity. Oncogene 2015, 34, 1979–1990. [Google Scholar] [CrossRef] [Green Version]
- McInroy, L.; Maatta, A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem. Biophys. Res. Commun. 2007, 360, 109–114. [Google Scholar] [CrossRef]
- Chu, Y.W.; Seftor, E.A.; Romer, L.H.; Hendrix, M.J. Experimental coexpression of vimentin and keratin intermediate filaments in human melanoma cells augments motility. Am. J. Pathol. 1996, 148, 63–69. [Google Scholar] [PubMed]
- Zhang, Y.; Stefanovic, B. LARP6 Meets Collagen mRNA: Specific Regulation of Type I Collagen Expression. Int. J. Mol. Sci. 2016, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Tsuruta, D.; Jones, J.C. The vimentin cytoskeleton regulates focal contact size and adhesion of endothelial cells subjected to shear stress. J. Cell Sci. 2003, 116, 4977–4984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrowska-Podhorodecka, Z.; Ding, I.; Lee, W.; Tanic, J.; Abbasi, S.; Arora, P.D.; Liu, R.S.; Patteson, A.E.; Janmey, P.A.; McCulloch, C.A. Vimentin tunes cell migration on collagen by controlling beta1 integrin activation and clustering. J. Cell Sci. 2021, 134, jcs254359. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Lu, M.; Jolly, M.K.; Tsarfaty, I.; Onuchic, J.; Ben-Jacob, E. The three-way switch operation of Rac1/RhoA GTPase-based circuit controlling amoeboid-hybrid-mesenchymal transition. Sci. Rep. 2014, 4, 6449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Engeland, N.C.A.; Suarez Rodriguez, F.; Rivero-Muller, A.; Ristori, T.; Duran, C.L.; Stassen, O.; Antfolk, D.; Driessen, R.C.H.; Ruohonen, S.; Ruohonen, S.T.; et al. Vimentin regulates Notch signaling strength and arterial remodeling in response to hemodynamic stress. Sci. Rep. 2019, 9, 12415. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.W.; Wu, T.H.; Lin, T.Y.; Chen, M.H.; Yeh, C.T.; Pan, T.L. Characterization of the Roles of Vimentin in Regulating the Proliferation and Migration of HSCs during Hepatic Fibrogenesis. Cells 2019, 8, 1184. [Google Scholar] [CrossRef] [Green Version]
- Xiong, D.; Sheng, Y.; Ding, S.; Chen, J.; Tan, X.; Zeng, T.; Qin, D.; Zhu, L.; Huang, A.; Tang, H. LINC00052 regulates the expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC cells invasion and migration. Oncotarget 2016, 7, 47593–47608. [Google Scholar] [CrossRef] [Green Version]
- Mirza, M.K.; Sun, Y.; Zhao, Y.D.; Potula, H.H.; Frey, R.S.; Vogel, S.M.; Malik, A.B.; Zhao, Y.Y. FoxM1 regulates re-annealing of endothelial adherens junctions through transcriptional control of beta-catenin expression. J. Exp. Med. 2010, 207, 1675–1685. [Google Scholar] [CrossRef]
- Li, L.; Backer, J.; Wong, A.S.; Schwanke, E.L.; Stewart, B.G.; Pasdar, M. Bcl-2 expression decreases cadherin-mediated cell-cell adhesion. J. Cell Sci. 2003, 116, 3687–3700. [Google Scholar] [CrossRef] [Green Version]
- Sivagurunathan, S.; Vahabikashi, A.; Yang, H.; Zhang, J.; Vazquez, K.; Rajasundaram, D.; Politanska, Y.; Abdala-Valencia, H.; Notbohm, J.; Ming Guo, M.; et al. Expression of vimentin alters cell mechanics, cell-cell adhesion, and gene expression profiles suggesting the induction of a hybrid EMT in human mammary epithelial cells. Front. Cell Dev. Biol. 2022, 10, 929495. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, T.; Kim, E.; Riehl, B.D.; Esfahani, A.M.; Rosenbohm, J.; Yang, R.; Duan, B.; Lim, J.Y. The LINC complex, mechanotransduction, and mesenchymal stem cell function and fate. J. Biol. Eng. 2019, 13, 68. [Google Scholar] [CrossRef]
- Shin, J.Y.; Worman, H.J. Molecular Pathology of Laminopathies. Annu. Rev. Pathol. 2022, 17, 159–180. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.A.; Martin, C.A.; Greene, P.; Marsh, J.A.; Genomics England Research Consortium; Blyth, M.; Cox, H.; Donnelly, D.; Greenhalgh, L.; Greville-Heygate, S.; et al. Heterozygous lamin B1 and lamin B2 variants cause primary microcephaly and define a novel laminopathy. Genet. Med. 2021, 23, 408–414. [Google Scholar] [CrossRef]
- Dubik, N.; Mai, S. Lamin A/C: Function in Normal and Tumor Cells. Cancers 2020, 12, 3688. [Google Scholar] [CrossRef] [PubMed]
- Patteson, A.E.; Vahabikashi, A.; Pogoda, K.; Adam, S.A.; Mandal, K.; Kittisopikul, M.; Sivagurunathan, S.; Goldman, A.; Goldman, R.D.; Janmey, P.A. Vimentin protects cells against nuclear rupture and DNA damage during migration. J. Cell Biol. 2019, 218, 4079–4092. [Google Scholar] [CrossRef] [PubMed]
- Turgay, Y.; Eibauer, M.; Goldman, A.E.; Shimi, T.; Khayat, M.; Ben-Harush, K.; Dubrovsky-Gaupp, A.; Sapra, K.T.; Goldman, R.D.; Medalia, O. The molecular architecture of lamins in somatic cells. Nature 2017, 543, 261–264. [Google Scholar] [CrossRef] [PubMed]





| Cell Line | Insert | Drug Selection | Retrovirus Used |
|---|---|---|---|
| MCF-7CV | Control vector | Hygromycin | pLPChygro |
| MCF-7FV | Full-length vimentin cDNA | Hygromycin | pLPChygro-VIM |
| MCF-7FV_shNTC | Non-target control | Puromycin | shRNA-NTC |
| MCF-7FV_shVIM | Vimentin specific short hairpin | Puromycin | shRNA-VIM |
| MDA-MB-231_shNTC | Non-target control | Puromycin | shRNA-NTC |
| MDA-MB-231_shVIM | Vimentin specific short hairpin | Puromycin | shRNA-VIM |
| MCF-7_shNTC | Non-target control | Puromycin | shRNA-NTC for CDH5 knockdown |
| MCF-7_shCDH5-1 | CDH5 specific short hairpin 1 | Puromycin | shRNA-CDH5-1 |
| MCF-7_shCDH5-2 | CDH5 specific short hairpin 2 | Puromycin | shRNA-CDH5-2 |
| MCF-7_shCDH5-3 | CDH5 specific short hairpin 3 | Puromycin | shRNA-CDH5-3 |
| MCF-7_shCDH5-4 | CDH5 specific short hairpin 4 | Puromycin | shRNA-CDH5-4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usman, S.; Jamal, A.; Bushaala, A.; Waseem, N.H.; Al-Dehlawi, H.; Yeudall, W.A.; Teh, M.-T.; Tummala, H.; Waseem, A. Transcriptome Analysis Reveals Vimentin-Induced Disruption of Cell–Cell Associations Augments Breast Cancer Cell Migration. Cells 2022, 11, 4035. https://doi.org/10.3390/cells11244035
Usman S, Jamal A, Bushaala A, Waseem NH, Al-Dehlawi H, Yeudall WA, Teh M-T, Tummala H, Waseem A. Transcriptome Analysis Reveals Vimentin-Induced Disruption of Cell–Cell Associations Augments Breast Cancer Cell Migration. Cells. 2022; 11(24):4035. https://doi.org/10.3390/cells11244035
Chicago/Turabian StyleUsman, Saima, Ahmad Jamal, Antesar Bushaala, Naushin H. Waseem, Hebah Al-Dehlawi, William Andrew Yeudall, Muy-Teck Teh, Hemanth Tummala, and Ahmad Waseem. 2022. "Transcriptome Analysis Reveals Vimentin-Induced Disruption of Cell–Cell Associations Augments Breast Cancer Cell Migration" Cells 11, no. 24: 4035. https://doi.org/10.3390/cells11244035