Human PSEN1 Mutant Glia Improve Spatial Learning and Memory in Aged Mice
Abstract
1. Introduction
2. Materials and Methods
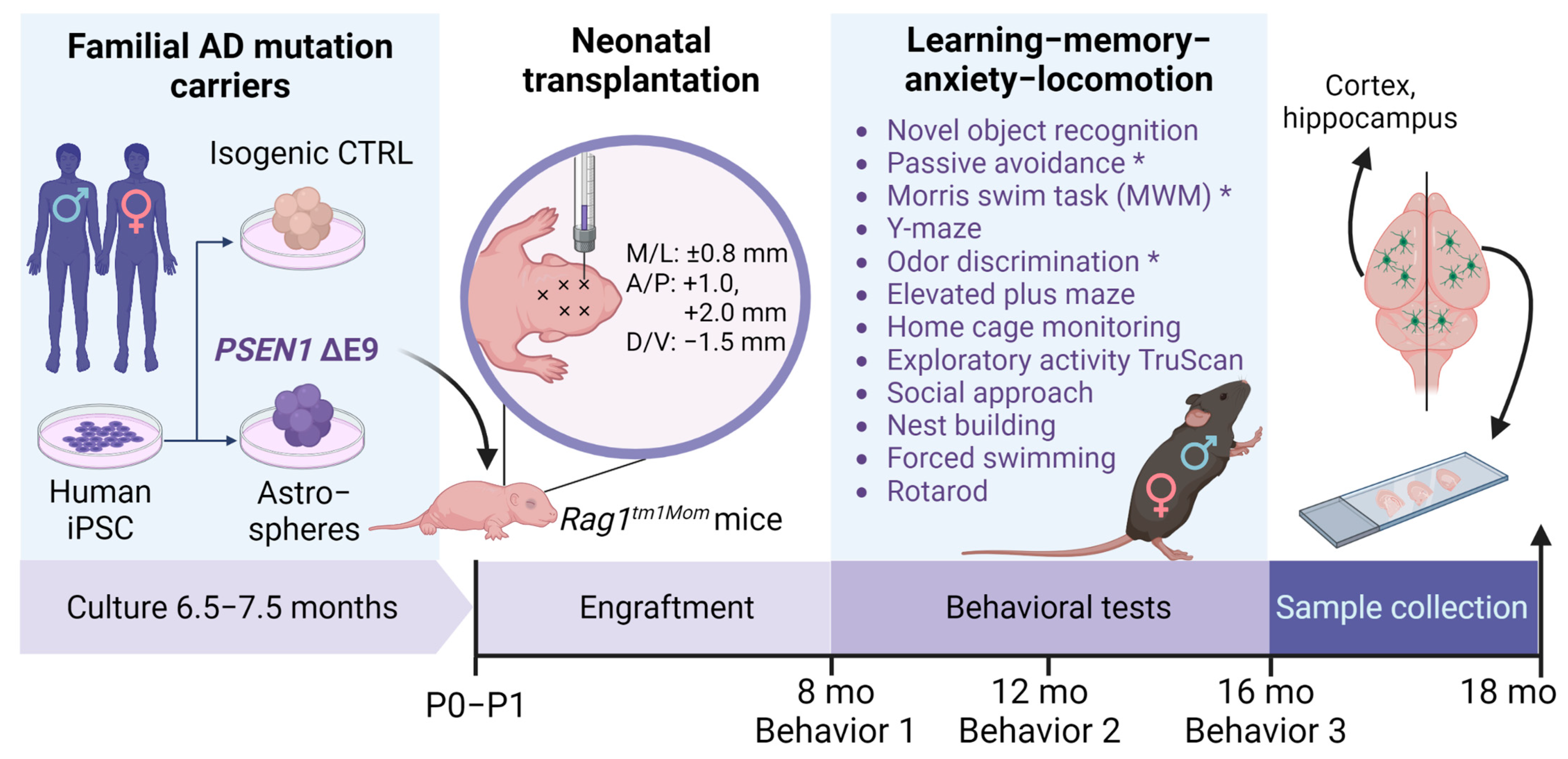
2.1. Human iPS Cells and Mice
2.2. Glial Differentiation
2.3. Transplantation
2.4. Sample Collection
2.5. Behavioral Tests
2.6. Immunohistochemistry
2.7. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR) and RNA Sequencing
2.8. Aβ Measurement by ELISA
2.9. Western Blotting
2.10. Statistics
2.11. Data Availability
3. Results
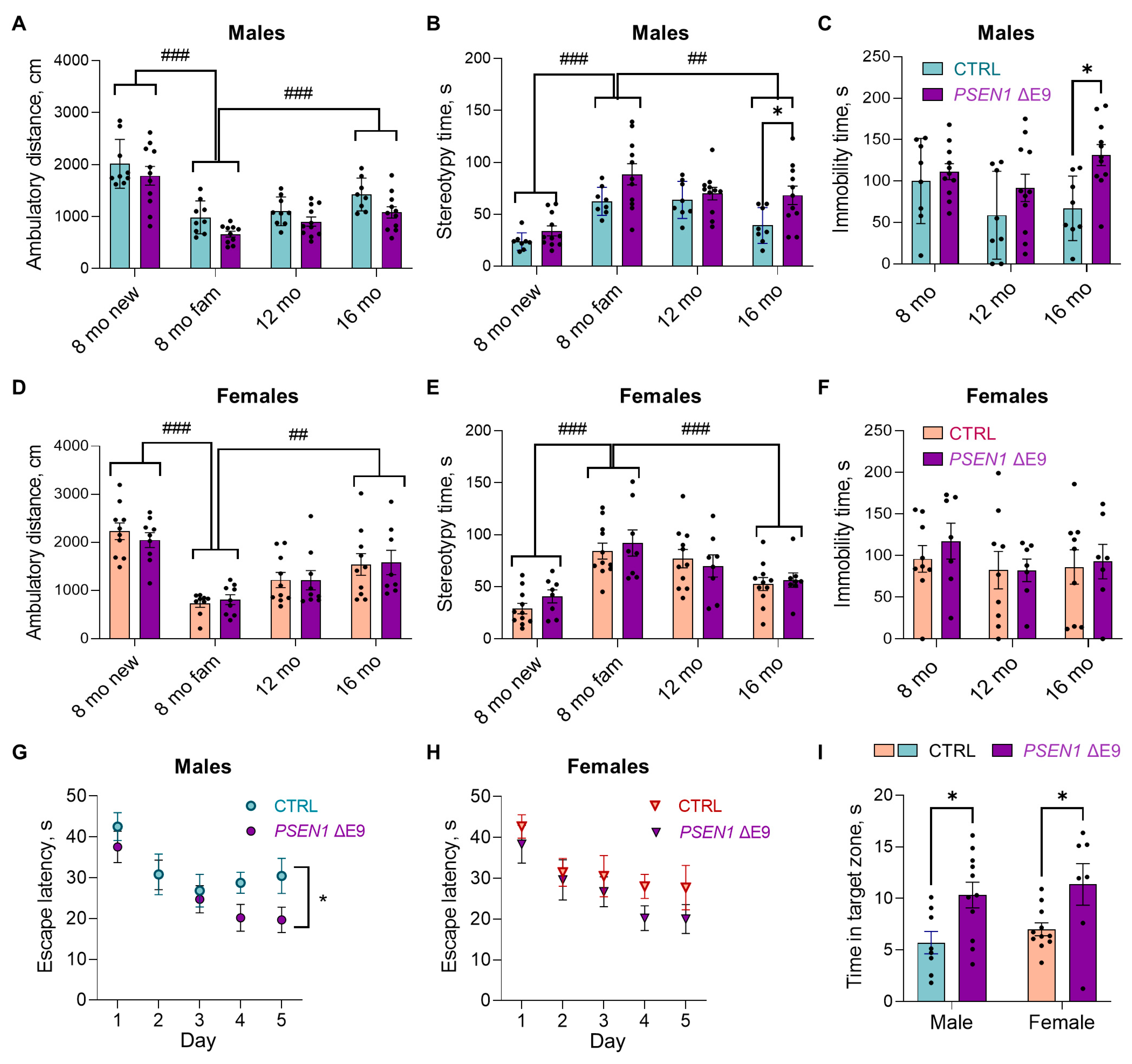
3.1. PSEN1 ΔE9 Mutation in Human Glial Progenitors Improves Spatial Learning and Memory at 16 Months after the Transplantation
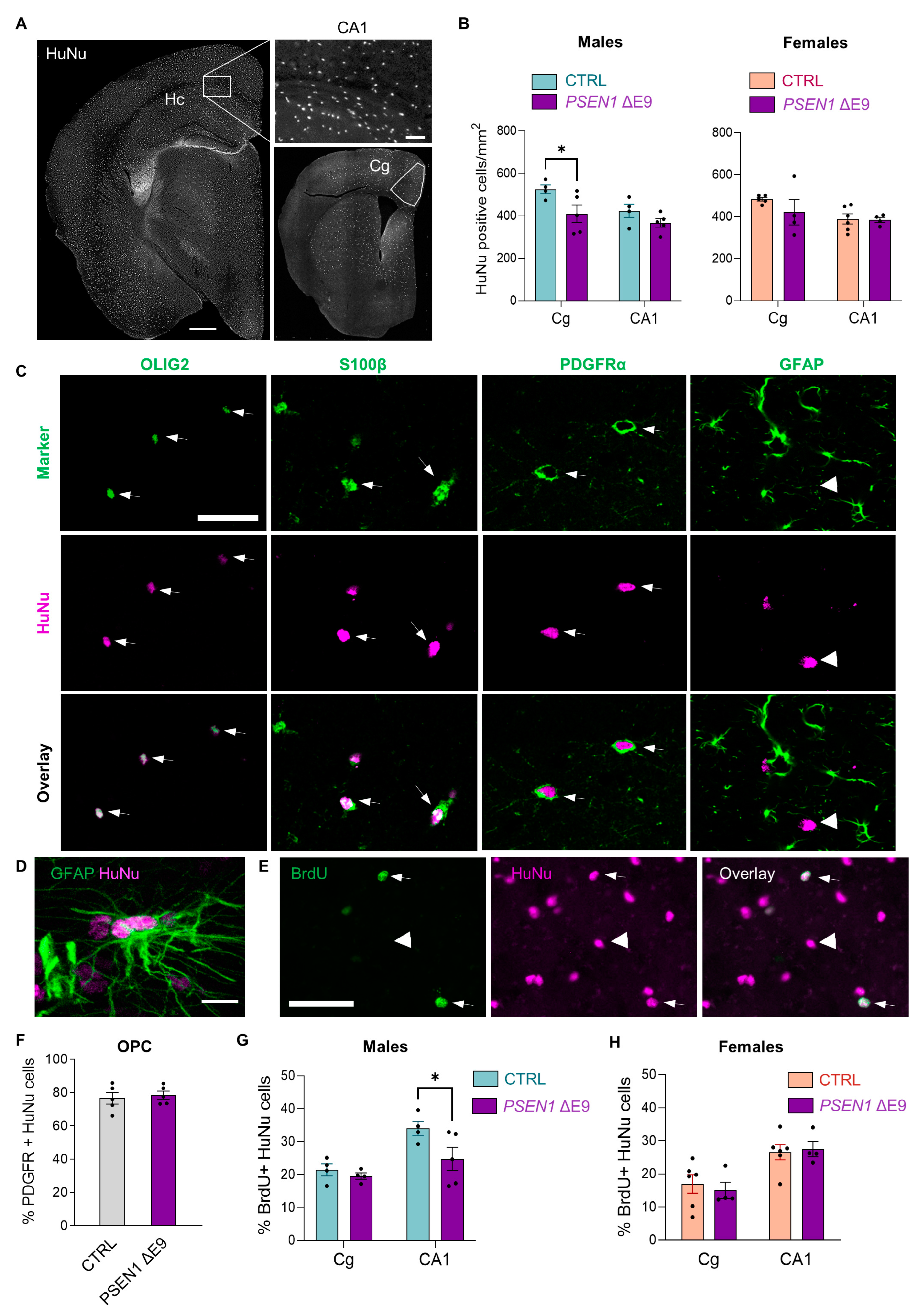
3.2. Transplanted Human Glial Cells Were Distributed throughout the Mouse Brain and Expressed Predominantly OPC Markers
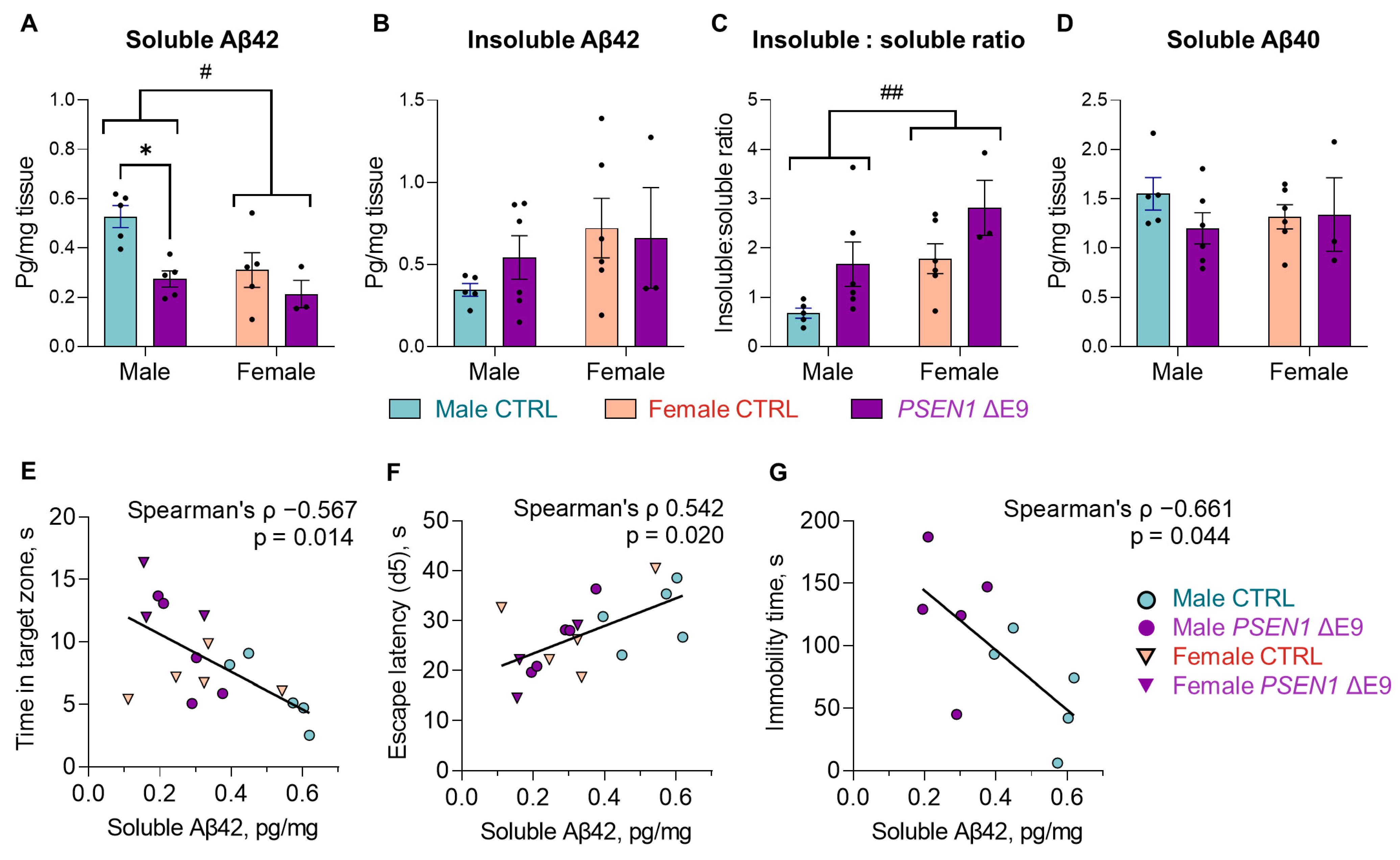
3.3. PSEN1 ΔE9 Mutant Human Cells Converted a Higher Proportion of Aβ42 into Insoluble Form Than the Control Cells
3.4. PSEN1 ΔE9 Mutant Human Glial Cells Exhibit an Altered Transcriptome
3.5. Human Glial Transplantation Decreased Immobility Time in the Forced Swim Test as Compared to the Sham-Transplanted Controls
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hebert, L.E.; Weuve, J.; Scherr, P.A.; Evans, D.A. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 2013, 80, 1778–1783. [Google Scholar] [CrossRef] [PubMed]
- Crook, R.; Verkkoniemi, A.; Perez-Tur, J.; Mehta, N.; Baker, M.; Houlden, H.; Farrer, M.; Hutton, M.; Lincoln, S.; Hardy, J.; et al. A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin 1. Nat. Med. 1998, 4, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Hiltunen, M.; Helisalmi, S.; Mannermaa, A.; Alafuzoff, I.; Koivisto, A.M.; Lehtovirta, M.; Pirskanen, M.; Sulkava, R.; Verkkoniemi, A.; Soininen, H. Identification of a novel 4.6-kb genomic deletion in presenilin-1 gene which results in exclusion of exon 9 in a Finnish early onset Alzheimer’s disease family: An Alu core sequence-stimulated recombination? Eur. J. Hum. Genet. 2000, 8, 259–266. [Google Scholar] [CrossRef][Green Version]
- Dumanchin, C.; Tournier, I.; Martin, C.; Didic, M.; Belliard, S.; Carlander, B.; Rouhart, F.; Duyckaerts, C.; Pellissier, J.F.; Latouche, J.B.; et al. Biological effects of four PSEN1 gene mutations causing Alzheimer disease with spastic paraparesis and cotton wool plaques. Hum. Mutat. 2006, 27, 1063. [Google Scholar] [CrossRef] [PubMed]
- Kumar-Singh, S.; Theuns, J.; Van Broeck, B.; Pirici, D.; Vennekens, K.; Corsmit, E.; Cruts, M.; Dermaut, B.; Wang, R.; Van Broeckhoven, C. Mean age-of-onset of familial alzheimer disease caused by presenilin mutations correlates with both increased Abeta42 and decreased Abeta40. Hum. Mutat. 2006, 27, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Svedruzic, Z.M.; Popovic, K.; Smoljan, I.; Sendula-Jengic, V. Modulation of gamma-secretase activity by multiple enzyme-substrate interactions: Implications in pathogenesis of Alzheimer’s disease. PLoS ONE 2012, 7, e32293. [Google Scholar] [CrossRef]
- Woodruff, G.; Young, J.E.; Martinez, F.J.; Buen, F.; Gore, A.; Kinaga, J.; Li, Z.; Yuan, S.H.; Zhang, K.; Goldstein, L.S. The presenilin-1 DeltaE9 mutation results in reduced gamma-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep. 2013, 5, 974–985. [Google Scholar] [CrossRef]
- Jankowsky, J.L.; Slunt, H.H.; Gonzales, V.; Jenkins, N.A.; Copeland, N.G.; Borchelt, D.R. APP processing and amyloid deposition in mice haplo-insufficient for presenilin 1. Neurobiol. Aging 2004, 25, 885–892. [Google Scholar] [CrossRef]
- Oksanen, M.; Petersen, A.J.; Naumenko, N.; Puttonen, K.; Lehtonen, S.; Gubert Olive, M.; Shakirzyanova, A.; Leskela, S.; Sarajarvi, T.; Viitanen, M.; et al. PSEN1 Mutant iPSC-Derived Model Reveals Severe Astrocyte Pathology in Alzheimer’s Disease. Stem Cell Rep. 2017, 9, 1885–1897. [Google Scholar] [CrossRef]
- Oksanen, M.; Hyotylainen, I.; Trontti, K.; Rolova, T.; Wojciechowski, S.; Koskuvi, M.; Viitanen, M.; Levonen, A.L.; Hovatta, I.; Roybon, L.; et al. NF-E2-related factor 2 activation boosts antioxidant defenses and ameliorates inflammatory and amyloid properties in human Presenilin-1 mutated Alzheimer’s disease astrocytes. Glia 2020, 68, 589–599. [Google Scholar] [CrossRef]
- Windrem, M.S.; Schanz, S.J.; Morrow, C.; Munir, J.; Chandler-Militello, D.; Wang, S.; Goldman, S.A. A competitive advantage by neonatally engrafted human glial progenitors yields mice whose brains are chimeric for human glia. J. Neurosci. 2014, 34, 16153–16161. [Google Scholar] [CrossRef] [PubMed]
- Windrem, M.S.; Osipovitch, M.; Liu, Z.; Bates, J.; Chandler-Militello, D.; Zou, L.; Munir, J.; Schanz, S.; McCoy, K.; Miller, R.H.; et al. Human iPSC Glial Mouse Chimeras Reveal Glial Contributions to Schizophrenia. Cell Stem Cell 2017, 21, 195–208.e196. [Google Scholar] [CrossRef] [PubMed]
- Jantti, H.; Sitnikova, V.; Ishchenko, Y.; Shakirzyanova, A.; Giudice, L.; Ugidos, I.F.; Gomez-Budia, M.; Korvenlaita, N.; Ohtonen, S.; Belaya, I.; et al. Microglial amyloid beta clearance is driven by PIEZO1 channels. J. Neuroinflamm. 2022, 19, 147. [Google Scholar] [CrossRef] [PubMed]
- Koskuvi, M.; Lehtonen, S.; Trontti, K.; Keuters, M.; Wu, Y.C.; Koivisto, H.; Ludwig, A.; Plotnikova, L.; Virtanen, P.L.J.; Rasanen, N.; et al. Contribution of astrocytes to familial risk and clinical manifestation of schizophrenia. Glia 2022, 70, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Deacon, R.M. Assessing nest building in mice. Nat. Protoc. 2006, 1, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Kaidanovich-Beilin, O.; Lipina, T.; Vukobradovic, I.; Roder, J.; Woodgett, J.R. Assessment of social interaction behaviors. J. Vis. Exp. 2011, 48, e2473. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.Y.; Janovjak, H.; Miserez, A.R.; Dobbie, Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 2002, 32, 1372–1374, 1376, 1378–1379. [Google Scholar] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lan, Y.; Xu, J.; Quan, F.; Zhao, E.; Deng, C.; Luo, T.; Xu, L.; Liao, G.; Yan, M.; et al. CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019, 47, D721–D728. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Bryan, K.J.; Lee, H.; Perry, G.; Smith, M.A.; Casadesus, G. Transgenic Mouse Models of Alzheimer’s Disease: Behavioral Testing and Considerations. In Methods of Behavior Analysis in Neuroscience, 2nd ed.; Buccafusco, J.J., Ed.; Frontiers in Neuroscience: Boca Raton, FL, USA, 2009. [Google Scholar]
- Yee, D.M.; Crawford, J.L.; Lamichhane, B.; Braver, T.S. Dorsal Anterior Cingulate Cortex Encodes the Integrated Incentive Motivational Value of Cognitive Task Performance. J. Neurosci. 2021, 41, 3707–3720. [Google Scholar] [CrossRef]
- Totah, N.K.; Kim, Y.B.; Homayoun, H.; Moghaddam, B. Anterior cingulate neurons represent errors and preparatory attention within the same behavioral sequence. J. Neurosci. 2009, 29, 6418–6426. [Google Scholar] [CrossRef]
- Espinosa-Jeffrey, A.; Kumar, S.; Zhao, P.M.; Awosika, O.; Agbo, C.; Huang, A.; Chang, R.; De Vellis, J. Transferrin regulates transcription of the MBP gene and its action synergizes with IGF-1 to enhance myelinogenesis in the md rat. Dev. Neurosci. 2002, 24, 227–241. [Google Scholar] [CrossRef]
- Zhang, Y.; Sloan, S.A.; Clarke, L.E.; Caneda, C.; Plaza, C.A.; Blumenthal, P.D.; Vogel, H.; Steinberg, G.K.; Edwards, M.S.; Li, G.; et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef]
- Sasuclark, A.R.; Khadka, V.S.; Pitts, M.W. Cell-Type Specific Analysis of Selenium-Related Genes in Brain. Antioxidants 2019, 8, 120. [Google Scholar] [CrossRef]
- Chanda, K.; Das, S.; Chakraborty, J.; Bucha, S.; Maitra, A.; Chatterjee, R.; Mukhopadhyay, D.; Bhattacharyya, N.P. Altered Levels of Long NcRNAs Meg3 and Neat1 in Cell And Animal Models Of Huntington’s Disease. RNA Biol. 2018, 15, 1348–1363. [Google Scholar] [CrossRef] [PubMed]
- Feyeux, M.; Bourgois-Rocha, F.; Redfern, A.; Giles, P.; Lefort, N.; Aubert, S.; Bonnefond, C.; Bugi, A.; Ruiz, M.; Deglon, N.; et al. Early transcriptional changes linked to naturally occurring Huntington’s disease mutations in neural derivatives of human embryonic stem cells. Hum. Mol. Genet. 2012, 21, 3883–3895. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Seredenina, T.; Coppola, G.; Kuhn, A.; Geschwind, D.H.; Luthi-Carter, R.; Thomas, E.A. Gene expression profiling of R6/2 transgenic mice with different CAG repeat lengths reveals genes associated with disease onset and progression in Huntington’s disease. Neurobiol. Dis. 2011, 42, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Saroja, S.R.; Sase, A.; Kircher, S.G.; Wan, J.; Berger, J.; Hoger, H.; Pollak, A.; Lubec, G. Hippocampal proteoglycans brevican and versican are linked to spatial memory of Sprague-Dawley rats in the morris water maze. J. Neurochem. 2014, 130, 797–804. [Google Scholar] [CrossRef]
- Evanko, S.P.; Angello, J.C.; Wight, T.N. Formation of hyaluronan- and versican-rich pericellular matrix is required for proliferation and migration of vascular smooth muscle cells. Arter. Thromb. Vasc. Biol. 1999, 19, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Wight, T.N.; Lara, S.; Riessen, R.; Le Baron, R.; Isner, J. Selective deposits of versican in the extracellular matrix of restenotic lesions from human peripheral arteries. Am. J. Pathol. 1997, 151, 963–973. [Google Scholar] [PubMed]
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J.Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019, 570, 332–337. [Google Scholar] [CrossRef]
- Fell, C.W.; Hagelkruys, A.; Cicvaric, A.; Horrer, M.; Liu, L.; Li, J.S.S.; Stadlmann, J.; Polyansky, A.A.; Mereiter, S.; Tejada, M.A.; et al. FIBCD1 is an endocytic GAG receptor associated with a novel neurodevelopmental disorder. EMBO Mol. Med. 2022, 14, e15829. [Google Scholar] [CrossRef] [PubMed]
- Molendijk, M.L.; de Kloet, E.R. Forced swim stressor: Trends in usage and mechanistic consideration. Eur. J. Neurosci. 2022, 55, 2813–2831. [Google Scholar] [CrossRef]
- Yang, J.T.; Wang, Z.J.; Cai, H.Y.; Yuan, L.; Hu, M.M.; Wu, M.N.; Qi, J.S. Sex Differences in Neuropathology and Cognitive Behavior in APP/PS1/tau Triple-Transgenic Mouse Model of Alzheimer’s Disease. Neurosci. Bull. 2018, 34, 736–746. [Google Scholar] [CrossRef]
- Wang, J.; Tanila, H.; Puolivali, J.; Kadish, I.; van Groen, T. Gender differences in the amount and deposition of amyloidbeta in APPswe and PS1 double transgenic mice. Neurobiol. Dis. 2003, 14, 318–327. [Google Scholar] [CrossRef]
- Tomiyama, T.; Nagata, T.; Shimada, H.; Teraoka, R.; Fukushima, A.; Kanemitsu, H.; Takuma, H.; Kuwano, R.; Imagawa, M.; Ataka, S.; et al. A new amyloid beta variant favoring oligomerization in Alzheimer’s-type dementia. Ann. Neurol. 2008, 63, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Muller-Schiffmann, A.; Herring, A.; Abdel-Hafiz, L.; Chepkova, A.N.; Schable, S.; Wedel, D.; Horn, A.H.; Sticht, H.; de Souza Silva, M.A.; Gottmann, K.; et al. Amyloid-beta dimers in the absence of plaque pathology impair learning and synaptic plasticity. Brain 2016, 139, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Dahlgren, K.N.; Manelli, A.M.; Stine, W.B., Jr.; Baker, L.K.; Krafft, G.A.; LaDu, M.J. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J. Biol. Chem. 2002, 277, 32046–32053. [Google Scholar] [CrossRef] [PubMed]
- Torres-Lista, V.; Gimenez-Llort, L. Persistence of behaviours in the Forced Swim Test in 3xTg-AD mice at advanced stages of disease. Behav. Process. 2014, 106, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Balusu, S.; Horre, K.; Thrupp, N.; Snellinx, A.; Serneels, L.; Chrysidou, I.; Arranz, A.M.; Sierksma, A.; Simren, J.; Karikari, T.K.; et al. Long noncoding RNA MEG3 activates neuronal necroptosis in Alzheimer’s disease. BioRxiv 2022. [Google Scholar] [CrossRef]
- Ma, Q.H.; Futagawa, T.; Yang, W.L.; Jiang, X.D.; Zeng, L.; Takeda, Y.; Xu, R.X.; Bagnard, D.; Schachner, M.; Furley, A.J.; et al. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat. Cell Biol. 2008, 10, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Raditsis, A.V.; Milojevic, J.; Melacini, G. Abeta association inhibition by transferrin. Biophys. J. 2013, 105, 473–480. [Google Scholar] [CrossRef]
- Yue, C.; Shan, Z.; Tan, Y.; Yao, C.; Liu, Y.; Liu, Q.; Tan, X.; Du, X. His-Rich Domain of Selenoprotein P Ameliorates Neuropathology and Cognitive Deficits by Regulating TrkB Pathway and Zinc Homeostasis in an Alzheimer Model of Mice. ACS Chem. Neurosci. 2020, 11, 4098–4110. [Google Scholar] [CrossRef]
- Lee, J.Y.; Cole, T.B.; Palmiter, R.D.; Suh, S.W.; Koh, J.Y. Contribution by synaptic zinc to the gender-disparate plaque formation in human Swedish mutant APP transgenic mice. Proc. Natl. Acad. Sci. USA 2002, 99, 7705–7710. [Google Scholar] [CrossRef]
- Datki, Z.; Galik-Olah, Z.; Janosi-Mozes, E.; Szegedi, V.; Kalman, J.; Hunya, A.G.; Fulop, L.; Tamano, H.; Takeda, A.; Adlard, P.A.; et al. Alzheimer risk factors age and female sex induce cortical Abeta aggregation by raising extracellular zinc. Mol. Psychiatry 2020, 25, 2728–2741. [Google Scholar] [CrossRef] [PubMed]
- Kiyohara, A.C.P.; Torres, D.J.; Hagiwara, A.; Pak, J.; Rueli, R.; Shuttleworth, C.W.R.; Bellinger, F.P. Selenoprotein P Regulates Synaptic Zinc and Reduces Tau Phosphorylation. Front. Nutr. 2021, 8, 683154. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavalu, K.; Choi, S.H.; Zhang, X.; Sisodia, S.S. Presenilin 1 mutants impair the self-renewal and differentiation of adult murine subventricular zone-neuronal progenitors via cell-autonomous mechanisms involving notch signaling. J. Neurosci. 2010, 30, 6903–6915. [Google Scholar] [CrossRef] [PubMed]
- Killick, R.; Pollard, C.C.; Asuni, A.A.; Mudher, A.K.; Richardson, J.C.; Rupniak, H.T.; Sheppard, P.W.; Varndell, I.M.; Brion, J.P.; Levey, A.I.; et al. Presenilin 1 independently regulates beta-catenin stability and transcriptional activity. J. Biol. Chem. 2001, 276, 48554–48561. [Google Scholar] [CrossRef] [PubMed]
- Soriano, S.; Kang, D.E.; Fu, M.; Pestell, R.; Chevallier, N.; Zheng, H.; Koo, E.H. Presenilin 1 negatively regulates beta-catenin/T cell factor/lymphoid enhancer factor-1 signaling independently of beta-amyloid precursor protein and notch processing. J. Cell Biol. 2001, 152, 785–794. [Google Scholar] [CrossRef]
- Zhang, Y.; Argaw, A.T.; Gurfein, B.T.; Zameer, A.; Snyder, B.J.; Ge, C.; Lu, Q.R.; Rowitch, D.H.; Raine, C.S.; Brosnan, C.F.; et al. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc. Natl. Acad. Sci. USA 2009, 106, 19162–19167. [Google Scholar] [CrossRef]
- Chew, L.J.; Ming, X.; McEllin, B.; Dupree, J.; Hong, E.; Catron, M.; Fauveau, M.; Nait-Oumesmar, B.; Gallo, V. Sox17 Regulates a Program of Oligodendrocyte Progenitor Cell Expansion and Differentiation during Development and Repair. Cell Rep. 2019, 29, 3173–3186.e3177. [Google Scholar] [CrossRef]
- Dai, Z.M.; Sun, S.; Wang, C.; Huang, H.; Hu, X.; Zhang, Z.; Lu, Q.R.; Qiu, M. Stage-specific regulation of oligodendrocyte development by Wnt/beta-catenin signaling. J. Neurosci. 2014, 34, 8467–8473. [Google Scholar] [CrossRef]
- Liu, J.; Li, Q.; Zhang, K.S.; Hu, B.; Niu, X.; Zhou, S.M.; Li, S.G.; Luo, Y.P.; Wang, Y.; Deng, Z.F. Downregulation of the Long Non-Coding RNA Meg3 Promotes Angiogenesis After Ischemic Brain Injury by Activating Notch Signaling. Mol. Neurobiol. 2017, 54, 8179–8190. [Google Scholar] [CrossRef]
- Ferreira, S.; Pitman, K.A.; Wang, S.; Summers, B.S.; Bye, N.; Young, K.M.; Cullen, C.L. Amyloidosis is associated with thicker myelin and increased oligodendrogenesis in the adult mouse brain. J. Neurosci. Res. 2020, 98, 1905–1932. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, Y.; Liu, Z.; Geng, Q.; Chen, Z.; Zhang, Y. Alterations of myelin morphology and oligodendrocyte development in early stage of Alzheimer’s disease mouse model. Neurosci. Lett. 2017, 642, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.T.; Lu, A.; Craessaerts, K.; Pavie, B.; Sala Frigerio, C.; Corthout, N.; Qian, X.; Lalakova, J.; Kuhnemund, M.; Voytyuk, I.; et al. Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer’s Disease. Cell 2020, 182, 976–991.e919. [Google Scholar] [CrossRef] [PubMed]
- Grubman, A.; Chew, G.; Ouyang, J.F.; Sun, G.; Choo, X.Y.; McLean, C.; Simmons, R.K.; Buckberry, S.; Vargas-Landin, D.B.; Poppe, D.; et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019, 22, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ren, S.Y.; Chen, J.F.; Liu, K.; Li, R.X.; Li, Z.F.; Hu, B.; Niu, J.Q.; Xiao, L.; Chan, J.R.; et al. Myelin degeneration and diminished myelin renewal contribute to age-related deficits in memory. Nat. Neurosci. 2020, 23, 481–486. [Google Scholar] [CrossRef]
- Assmann, A.; Richter, A.; Schutze, H.; Soch, J.; Barman, A.; Behnisch, G.; Knopf, L.; Raschick, M.; Schult, A.; Wustenberg, T.; et al. Neurocan genome-wide psychiatric risk variant affects explicit memory performance and hippocampal function in healthy humans. Eur. J. Neurosci. 2021, 53, 3942–3959. [Google Scholar] [CrossRef]
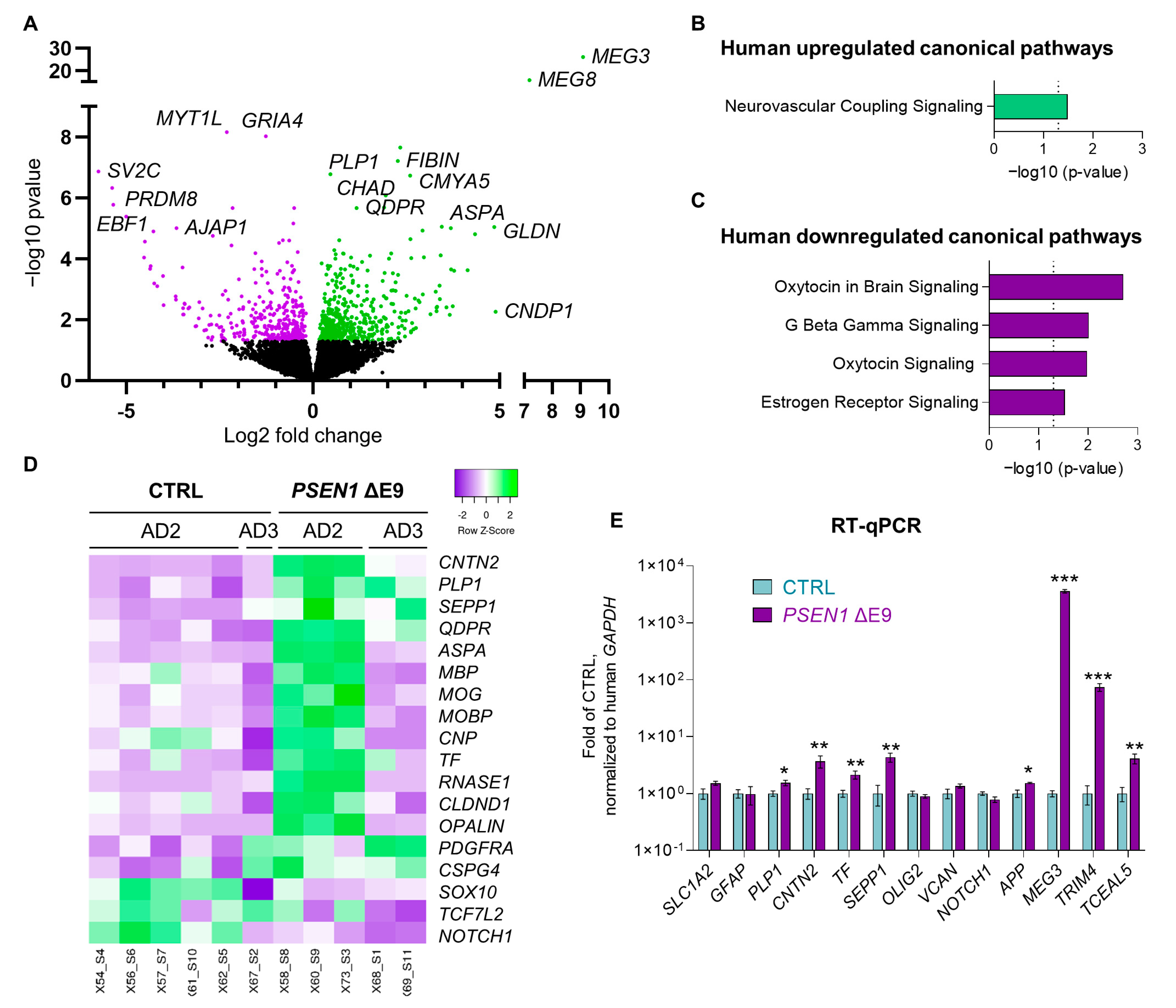
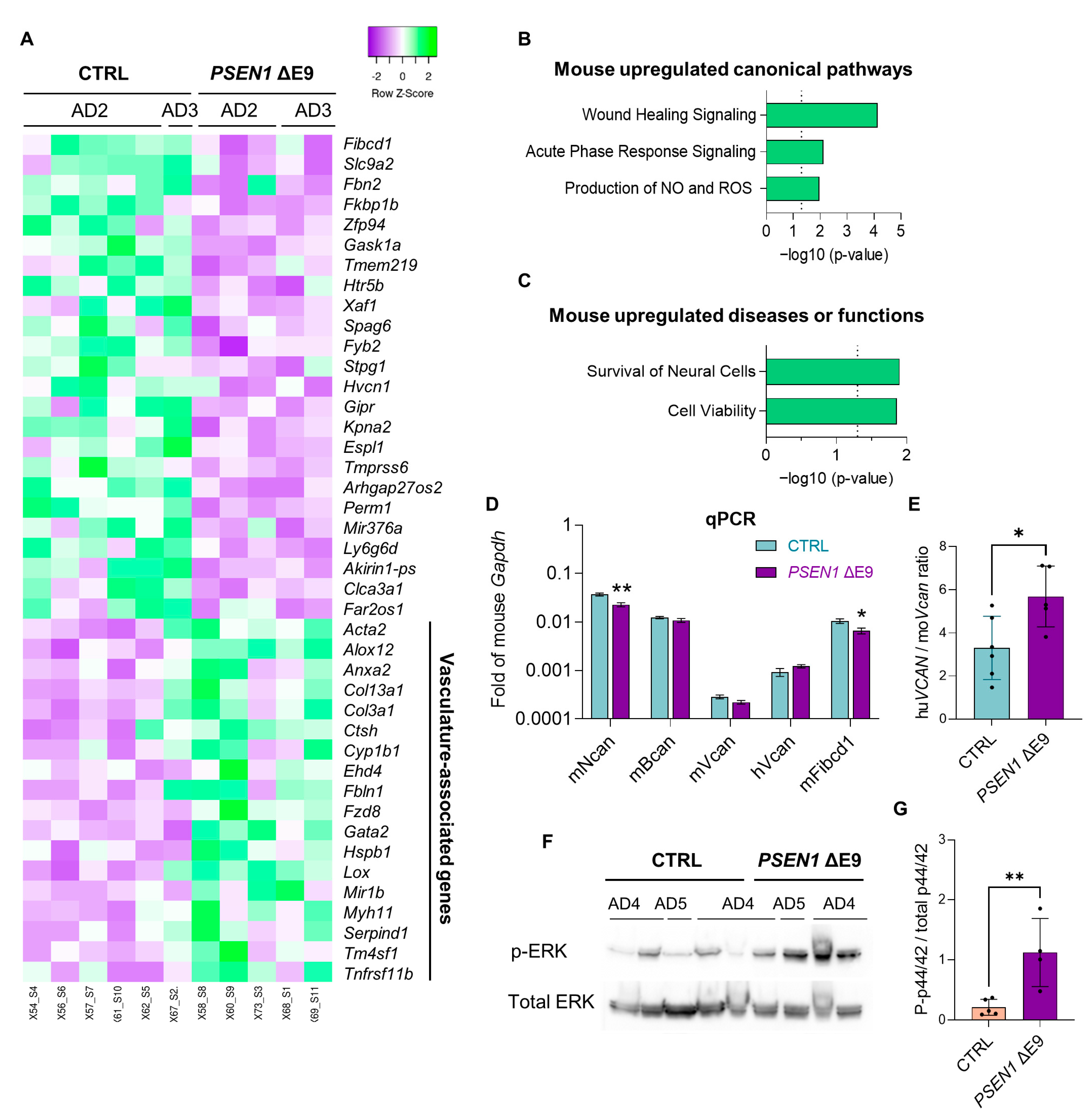

| Line | Sex | Age at Biopsy | PSEN1 Genotype | APOE Genotype | Transplanted Mice Referred to in the Text as |
|---|---|---|---|---|---|
| Batch I (main) | |||||
| AD2 | M | 48 | ΔE9/CTRL | ε3/ε3 | PSEN1 ΔE9 |
| AD2-iso | corrected | ε3/ε3 | CTRL | ||
| AD3 | F | 47 | ΔE9/CTRL | ε3/ε3 | PSEN1 ΔE9 |
| AD3-iso | corrected | ε3/ε3 | CTRL | ||
| Batch II | |||||
| MADGIC 1 | M | 67 | CTRL | ε3/ε3 | Human cells |
| MADGIC 8 | M | 64 | CTRL | ε3/ε3 | Human cells |
| iPSC Genotype | iPSC Line | Transplanted Pups Total | Males | Females |
|---|---|---|---|---|
| Batch I (main) | ||||
| PSEN1 ΔE9 | AD2 | 9 | 4 | 5 |
| AD3 | 13 | 8 | 5 | |
| Isogenic (CTRL) | AD2-iso | 10 | 5 | 5 |
| AD3-iso | 16 | 7 | 9 | |
| Batch II | ||||
| CTRL | MADGIC 1 | 9 | 3 | 6 |
| MADGIC 8 | 7 | 2 | 5 | |
| Sham | 22 | 7 | 15 |
| Behavioral Test | Outcome Measure |
|---|---|
| Home cage monitoring | Normal behavior |
| Exploratory activity TruScan | Locomotion, exploratory activity |
| Nest building | General health |
| Rotarod | Motor coordination |
| Social approach | Social behavior, cognition |
| Elevated plus maze | Anxiety |
| Forced swimming | Despair |
| Spontaneous alternation in a Y-maze | Exploratory behavior, cognitive function |
| Novel object recognition | Memory (perirhinal cortex) |
| Morris swim task (water maze) | Spatial learning |
| Odor discrimination | Memory (piriform cortex) |
| Passive avoidance | Learning, memory |
| Primary Antibodies | Host | Specificities | Supplier | Catalog # | Application | Dilution | RRID # |
| HuNu | mouse | human nuclei | Merck | MAB4383 | IHC | 1:500 | AB_827439 |
| BrdU | rat | species independent | Abcam * | ab6326 | IHC | 1:400 | AB_305426 |
| S100β | rabbit | mouse, rat | Abcam | ab41548 | IHC | 1:5000 | AB_956280 |
| PDGFRα | rabbit | human | Cell Signaling Technology ** | 5241 | IHC | 1:300 | AB_10692773 |
| GFAP | rabbit | human, mouse | Agilent Dako *** | Z0334 | IHC | 1:500 | AB_10013382 |
| Olig2 | rabbit | human, mouse | Merck | AB9610 | IHC | 1:1000 | AB_570666 |
| Aβ peptide | rabbit | human, mouse | Thermo Fisher Scientific | 51–2700 | IHC | 1:1000 | AB_2533902 |
| p-AKT (Ser473) | rabbit | human, mouse | Cell Signaling Technology | 4060 | WB | 1:1000 | AB_2315049 |
| AKT | rabbit | human, mouse | Cell Signaling Technology | 4691 | WB | 1:2000 | AB_915783 |
| p-ERK (Thr202/Tyr204) | rabbit | human, mouse | Cell Signaling Technology | 9101 | WB | 1:1000 | AB_331646 |
| ERK | rabbit | human, mouse | Cell Signaling Technology | 9102 | WB | 1:1000 | AB_330744 |
| β actin | mouse | human, mouse | Merck | A5441 | WB | 1:5000 | AB_476744 |
| Secondary Antibodies | Host | Specificities | Supplier | Catalog # | Application | Dilution | RRID # |
| Alexa Fluor 555 | donkey | mouse | Thermo Fisher Scientific | A31570 | IHC | 1:500 | AB_2536180 |
| Alexa Fluor 488 | donkey | rat | Thermo Fisher Scientific | A21208 | IHC | 1:500 | AB_2535794 |
| Alexa Fluor 488 | donkey | rabbit | Thermo Fisher Scientific | A21206 | IHC | 1:500 | AB_2535792 |
| Alexa Fluor 488 | goat | rabbit | Thermo Fisher Scientific | A11008 | IHC | 1:800 | AB_143165 |
| Alexa Fluor 568 | goat | mouse | Thermo Fisher Scientific | A11004 | IHC | 1:800 | AB_2534072 |
| Biotinylated | goat | mouse | Vector Laboratories | BA-9200 | IHC | 1:500 | AB_2336171 |
| Biotinylated | goat | rabbit | Vector Laboratories | BA-1000 | IHC | 1:500 | AB_2313606 |
| HRP | goat | rabbit | Thermo Fisher Scientific | A16096 | WB | 1:10000 | AB_2534770 |
| HRP | rabbit | mouse | Merck | A9044 | WB | 1:20000 | AB_258431 |
| Target | Specificity | Supplier | Catalog # |
|---|---|---|---|
| SLC1A2 | human | Thermo Fisher Scientific | Hs01102423_m1 |
| GFAP | human | Thermo Fisher Scientific | Hs00909233_m1 |
| PLP1 | human | Thermo Fisher Scientific | Hs00166914_m1 |
| CNTN2 | human | Thermo Fisher Scientific | Hs00543989_m1 |
| TF | human | Thermo Fisher Scientific | Hs00169070_m1 |
| SEPP1 | human | Thermo Fisher Scientific | Hs01032845_m1 |
| OLIG2 | human | Thermo Fisher Scientific | Hs00377820_m1 |
| VCAN | human | Thermo Fisher Scientific | Hs00171642_m1 |
| NOTCH1 | human | Thermo Fisher Scientific | Hs01062014_m1 |
| APP | human | Thermo Fisher Scientific | Hs00169098_m1 |
| MEG3 | human | Thermo Fisher Scientific | Hs00292028_m1 |
| TRIM4 | human | Thermo Fisher Scientific | Hs00982695_m1 |
| TCEAL5 | human | Thermo Fisher Scientific | Hs01383798_m1 |
| GAPDH | human | Thermo Fisher Scientific | Hs99999905_m1 |
| Ncan | mouse | Thermo Fisher Scientific | Mm00484007_m1 |
| Bcan | mouse | Thermo Fisher Scientific | Mm00476090_m1 |
| Vcan | mouse | Thermo Fisher Scientific | Mm01283063_m1 |
| Fibcd1 | mouse | Thermo Fisher Scientific | Mm00619124_m1 |
| Gapdh | mouse | Thermo Fisher Scientific | Mm99999915_g1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jäntti, H.; Oksanen, M.; Kettunen, P.; Manta, S.; Mouledous, L.; Koivisto, H.; Ruuth, J.; Trontti, K.; Dhungana, H.; Keuters, M.; et al. Human PSEN1 Mutant Glia Improve Spatial Learning and Memory in Aged Mice. Cells 2022, 11, 4116. https://doi.org/10.3390/cells11244116
Jäntti H, Oksanen M, Kettunen P, Manta S, Mouledous L, Koivisto H, Ruuth J, Trontti K, Dhungana H, Keuters M, et al. Human PSEN1 Mutant Glia Improve Spatial Learning and Memory in Aged Mice. Cells. 2022; 11(24):4116. https://doi.org/10.3390/cells11244116
Chicago/Turabian StyleJäntti, Henna, Minna Oksanen, Pinja Kettunen, Stella Manta, Lionel Mouledous, Hennariikka Koivisto, Johanna Ruuth, Kalevi Trontti, Hiramani Dhungana, Meike Keuters, and et al. 2022. "Human PSEN1 Mutant Glia Improve Spatial Learning and Memory in Aged Mice" Cells 11, no. 24: 4116. https://doi.org/10.3390/cells11244116
APA StyleJäntti, H., Oksanen, M., Kettunen, P., Manta, S., Mouledous, L., Koivisto, H., Ruuth, J., Trontti, K., Dhungana, H., Keuters, M., Weert, I., Koskuvi, M., Hovatta, I., Linden, A.-M., Rampon, C., Malm, T., Tanila, H., Koistinaho, J., & Rolova, T. (2022). Human PSEN1 Mutant Glia Improve Spatial Learning and Memory in Aged Mice. Cells, 11(24), 4116. https://doi.org/10.3390/cells11244116







