Lactobacillus salivarius SNK-6 Regulates Liver Lipid Metabolism Partly via the miR-130a-5p/MBOAT2 Pathway in a NAFLD Model of Laying Hens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Sequencing of Bacteria
2.2. Animal Models
2.3. Histological Analysis
2.4. Oil-Red O Staining
2.5. Detection of Lipid Levels and Enzymatic Activity
2.6. Supernatants Collection
2.7. Cell Culture and Transfection
2.8. Dual Luciferase Reporter Assay
2.9. Quantitative Real-Time RT-PCR
2.10. Western Blotting
2.11. Untargeted Metabolomic Analysis
2.12. SCFA Quantification
2.13. Bile Acid Composition
2.14. Amino Acid Composition
2.15. Statistical Analysis
3. Results
3.1. Genome Sequencing of L. salivarius SNK-6
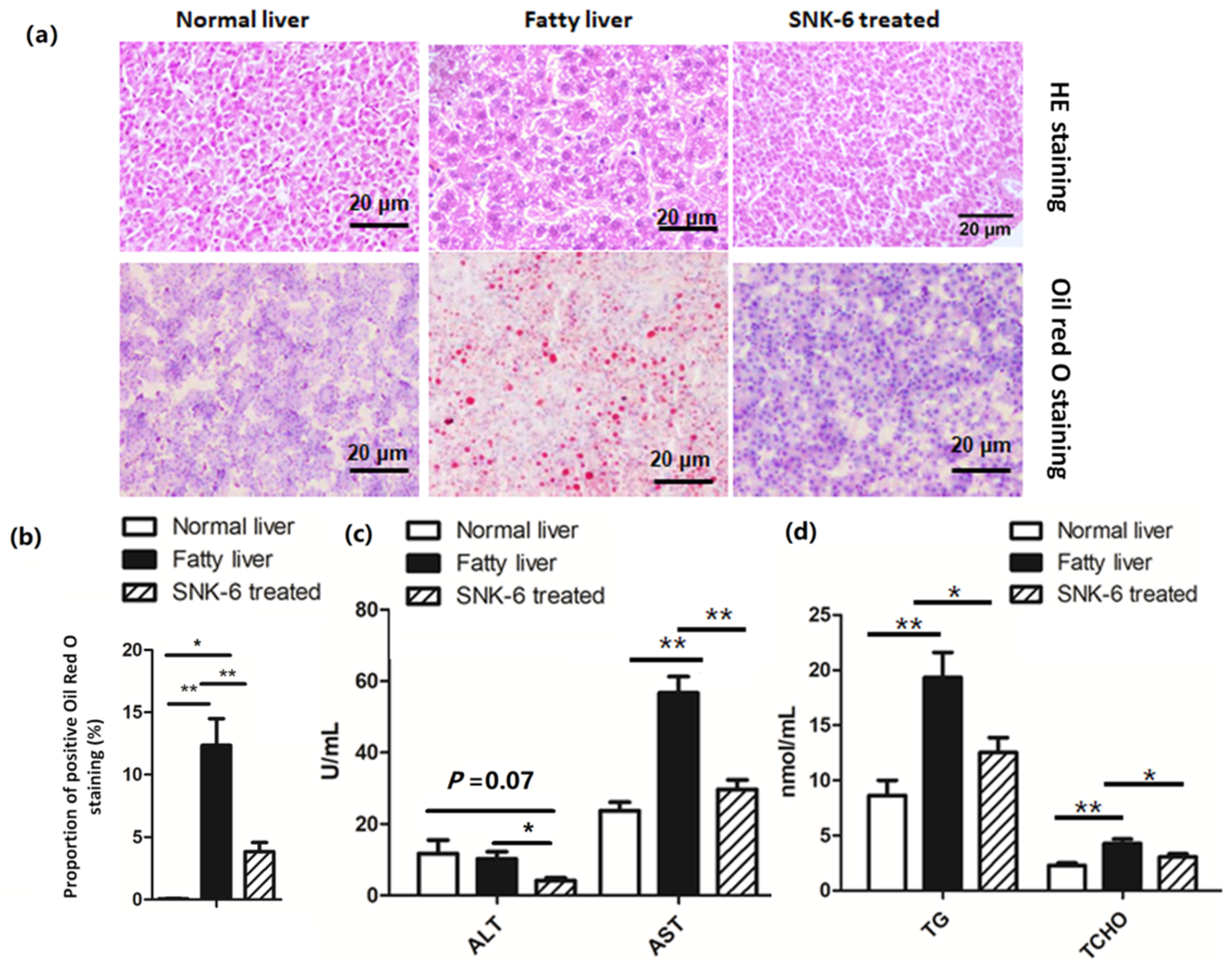
3.2. L. salivarius SNK-6 Alleviates Fat Deposition in the NAFLD Model in Laying Hens
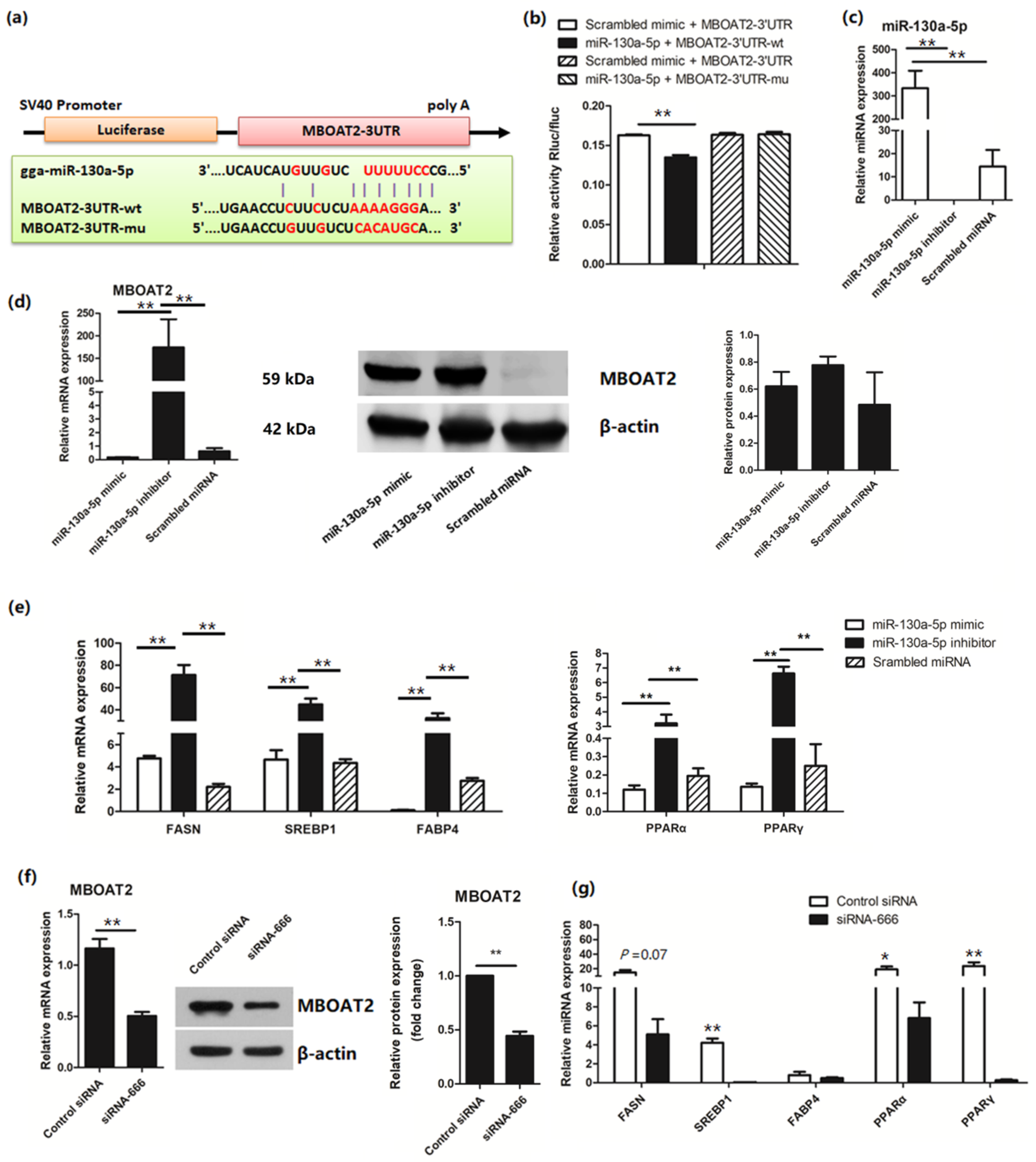
3.3. miR-130a-5p Targets the MBOAT2/ PPAR/SREBP Pathway
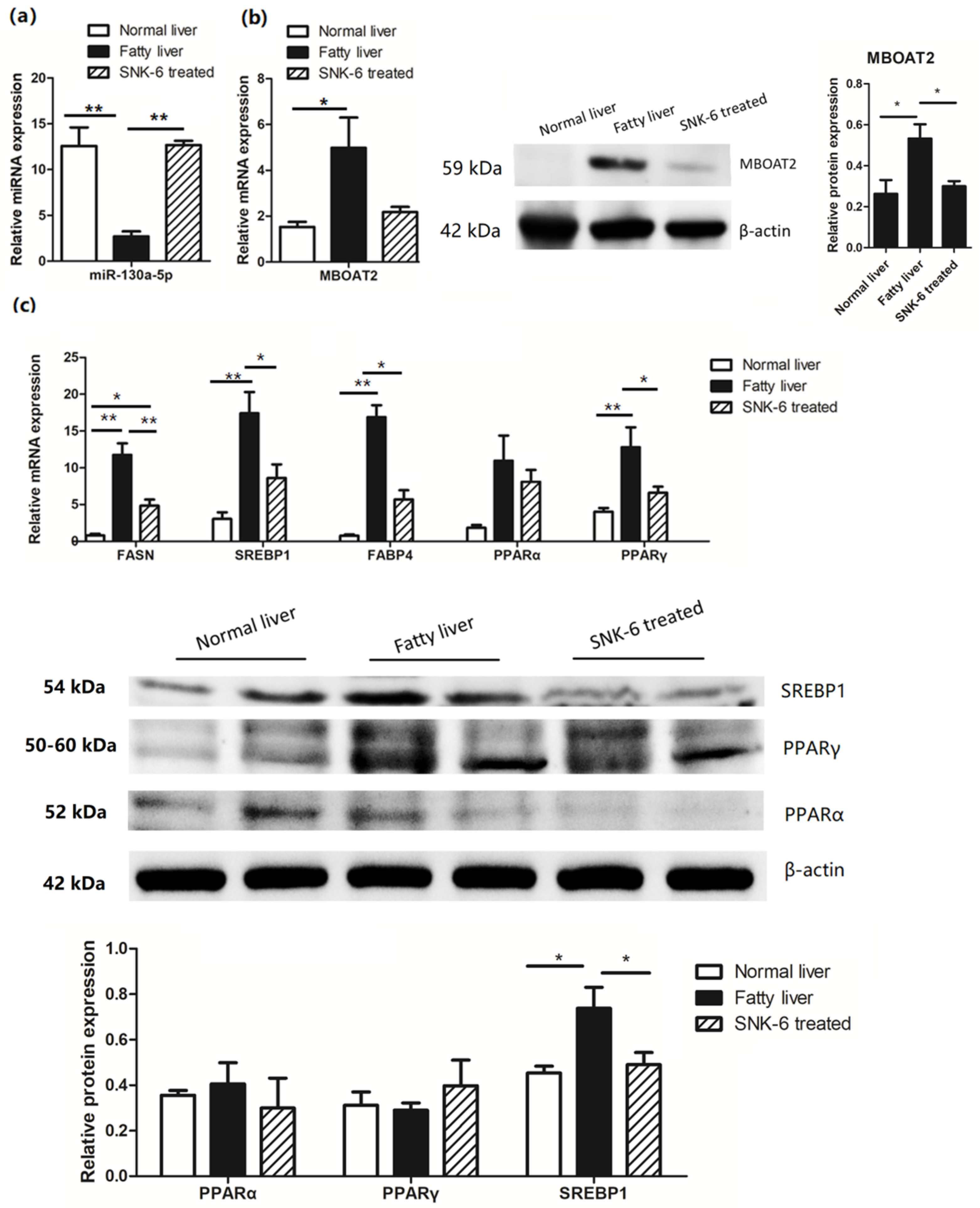
3.4. L. salivarius SNK-6 Regulates the miR-130a-5p/MBOAT2 Pathway in the NAFLD Model
3.5. L. salivarius SNK-6 Culture Medium Inhibits fat Deposition in Hepatocytes
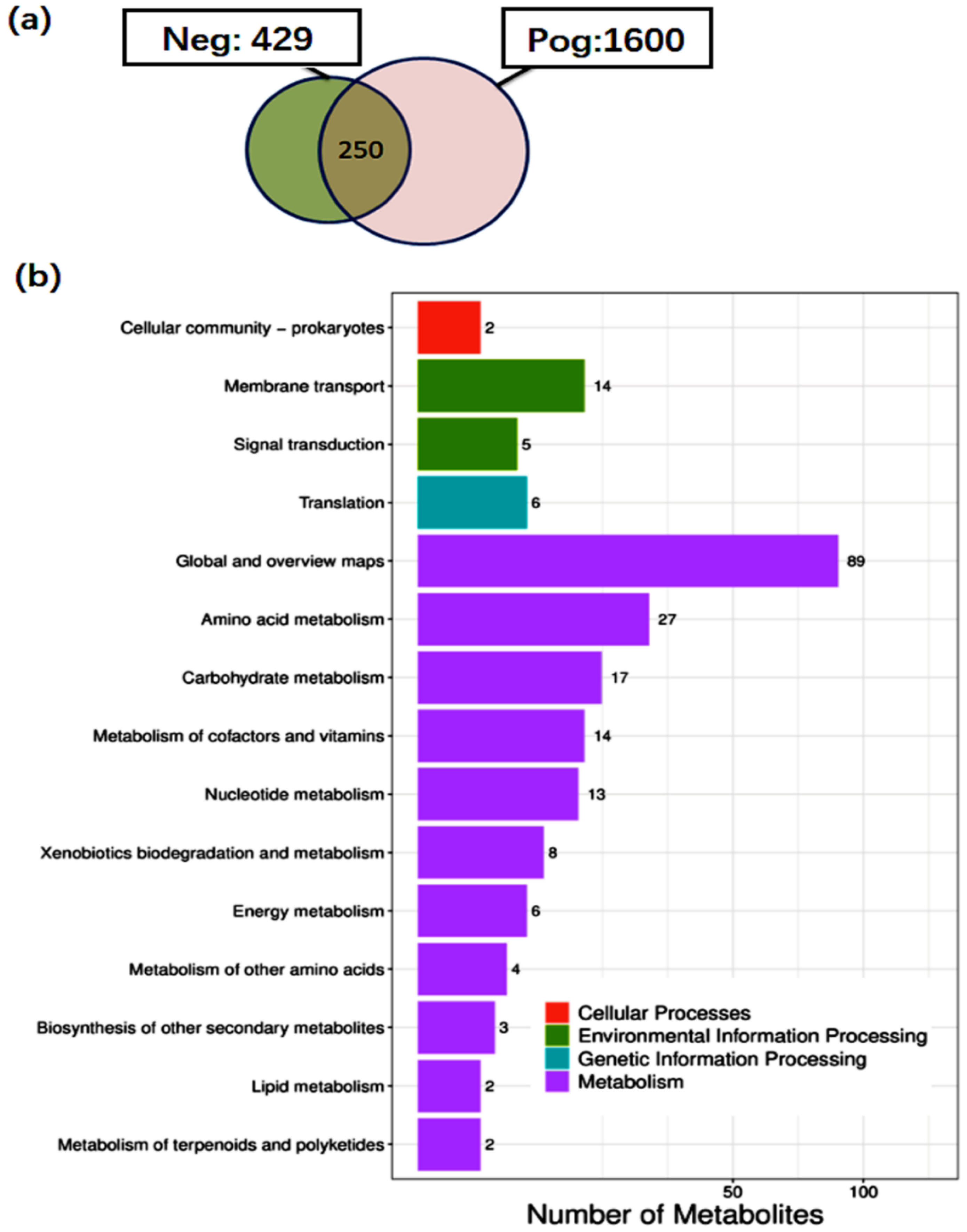
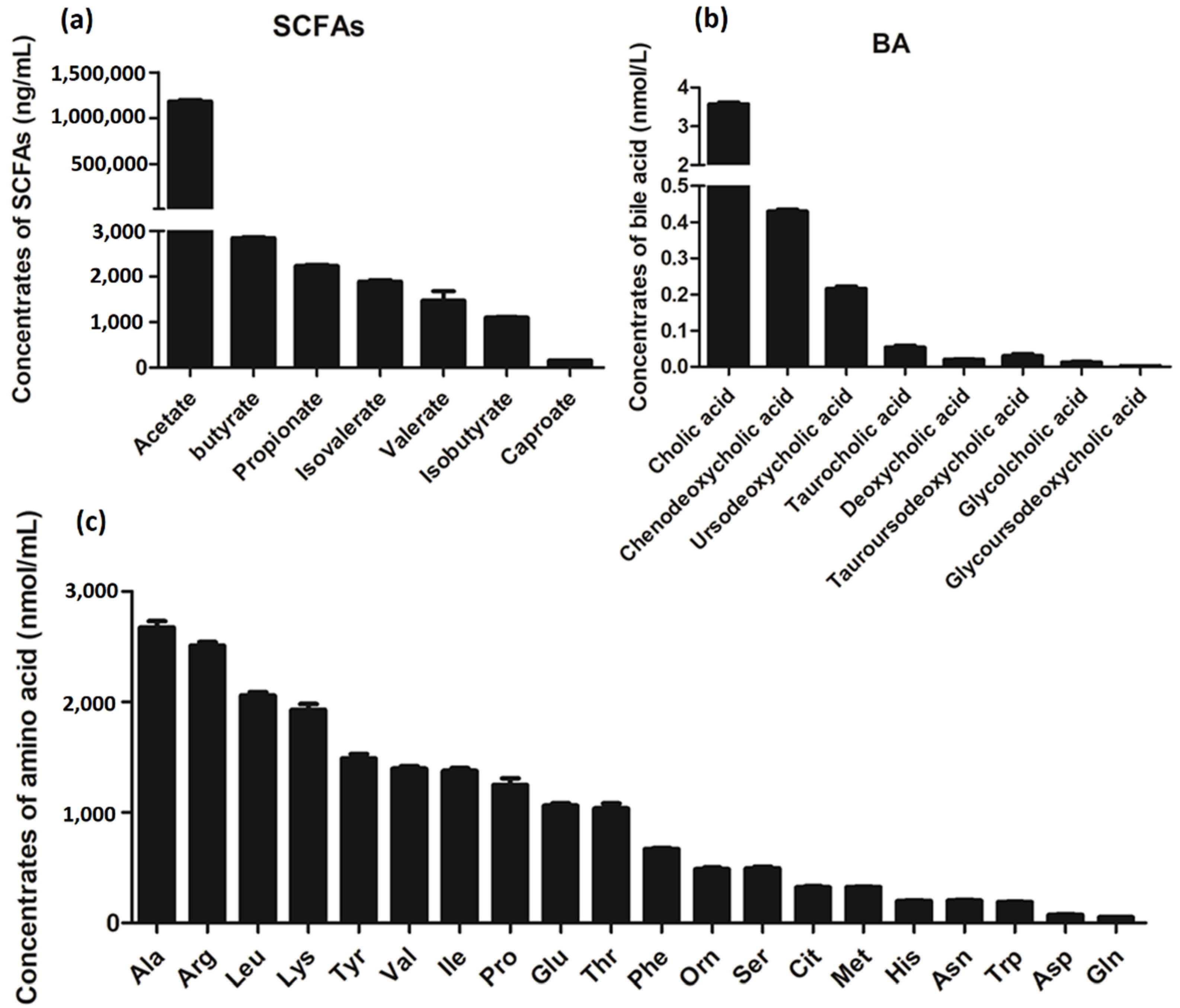
3.6. Metabolite Analysis of L. salivarius SNK-6
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bibbò, S.; Ianiro, G.; Dore, M.P.; Simonelli, C.; Newton, E.E.; Cammarota, G. Gut microbiota as a driver of inflammation in nonalcoholic fatty liver disease. Mediat. Inflamm. 2018, 2018, 9321643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Perumpail, B.J.; Li, A.A.; John, N.; Sallam, S.; Shah, N.D.; Kwong, W.; Cholankeril, G.; Kim, D.; Ahmed, A. The therapeutic implications of the gut microbiome and probiotics in patients with NAFLD. Diseases 2019, 7, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, P.C.; Shen, Y.F.; Zhuge, A.X.; Lv, L.X.; Zhu, X.L.; Yuan, Y.; Yang, L.Y.; Wang, K.C.; Li, B.; Li, L.J. Dose-dependent relationship between protection of thioacetamide-induced acute liver injury and hyperammonemia and concentration of Lactobacillus salivarius Li01 in mice. Microbiol. Spectr. 2021, 9, e0184721. [Google Scholar] [CrossRef]
- Zhuge, A.X.; Li, S.J.; Yuan, Y.; Li, B.; Li, L.; Li, L.J. The synergy of dietary supplements Lactobacillus salivarius LI01 and Bifidobacterium longum TC01 in alleviating liver failure in rats treated with D-galactosamine. Food Funct. 2021, 12, 10239–10252. [Google Scholar] [CrossRef]
- Zhuge, A.X.; Li, B.; Yuan, Y.; Lv, L.X.; Li, Y.; Wu, J.; Yang, L.Y.; Bian, X.; Wang, K.; Wang, Q.; et al. Lactobacillus salivarius LI01 encapsulated in alginate-pectin microgels ameliorates D-galactosamine-induced acute liver injury in rats. Appl. Microbiol. Biotechnol. 2020, 104, 7437–7455. [Google Scholar] [CrossRef]
- Yang, L.Y.; Bian, X.Y.; Wu, W.R.; Lv, L.X.; Li, Y.T.; Ye, J.Z.; Jiang, X.W.; Wang, Q.; Shi, D.; Fang, D.Q.; et al. Protective effect of Lactobacillus salivarius Li01 on thioacetamide-induced acute liver injury and hyperammonaemia. Microb. Biotechnol. 2020, 13, 1860–1876. [Google Scholar] [CrossRef]
- Shini, A.; Shini, S.; Bryden, W.L. Fatty liver haemorrhagic syndrome occurrence in laying hens: Impact of production system. Avian Pathol. 2019, 48, 25–34. [Google Scholar] [CrossRef]
- Leveille, G.A.; Romsos, D.R.; Yeh, Y.Y.; O’Hea, E.K. Lipid biosynthesis in the Chick. A consideration of site of synthesis, influence of diet and possible regulatory mechanisms. Poult. Sci. 1975, 54, 1075–1093. [Google Scholar] [CrossRef]
- Wang, X.; Xing, C.; Yang, F.; Zhou, S.; Li, G.; Zhang, C.; Cao, H.; Hu, G. Abnormal expression of liver autophagy and apoptosis-related mRNA in fatty liver haemorrhagic syndrome and improvement function of resveratrol in laying hens. Avian. Pathol. 2020, 49, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Xing, C.; Cao, H.; Zhang, C.; Luo, J.; Guo, X.; Hu, G. Insulin resistance and metabonomics analysis of fatty liver haemorrhagic syndrome in laying hens induced by a high-energy low-protein diet. Sci. Rep. 2019, 9, 10141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Ko, M.L.; Huang, C.C.; Park, S.Y.; Hong, M.P.; Wu, C.; Ko, G.Y. Chicken embryos as a potential new model for early onset type I diabetes. J. Diabetes Res. 2014, 2014, 354094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, K.; Zhao, Q.; Wang, J.; Qi, G.H.; Wu, S.G.; Zhang, H.J. Effects of pyrroloquinoline quinone on lipid metabolism and anti-oxidative capacity in a high-fat-diet metabolic dysfunction-associated fatty liver disease chick model. Int. J. Mol. Sci. 2021, 22, 1458. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.T.; Chen, Y.J.; Chen, C.Y.; Tsai, M.H.; Han, C.L.; Chen, Y.J.; Mersmann, H.J.; Ding, S.T. Identification of potential plasma biomarkers for nonalcoholic fatty liver disease by integrating transcriptomics and proteomics in laying hens. J. Nutr. 2017, 147, 293–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, R.B.; Liu, B.H.; Xie, Y.L.; Li, Z.Y.; Huang, W.H.; Yuan, J.Y.; He, G.Z.; Chen, Y.X.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [Green Version]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.D.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Zhu, L.H.; Liao, R.R.; Huang, J.W.; Yan, H.X.; Xiao, C.F.; Yang, Y.Z.; Wang, H.Y.; Yang, C.S. The miR-216/miR-217 cluster regulates lipid metabolism in laying hens with fatty liver syndrome via PPAR/SREBP signaling pathway. Front. Vet. Sci. 2022, 9, 913841. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Dunphy, L.J.; Grimes, K.; Wase, N.; Kolling, G.; Papin, J.A. Untargeted metabolomics reveals species-specific metabolite production and shared nutrient consumption by pseudomonas aeruginosa and dtaphylococcus aureus. mSystems 2021, 6, e0048021. [Google Scholar] [CrossRef] [PubMed]
- John, C.; Werner, P.; Worthmann, A.; Wegner, K.; Todter, K.; Scheja, L.; Rohn, S.; Heeren, J.; Fischer, M. A liquid chromatography-tandem mass spectrometry-based method for the simultaneous determination of hydroxy sterols and bile acids. J. Chromatogr. A 2014, 1371, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Agbu, P.; Carthew, R.W. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef]
- Zheng, Y.S.; Xiang, L.G.; Chen, M.L.; Xiang, C. H MicroRNA130a inhibits the proliferation, migration and invasive ability of hepatocellular carcinoma cells by downregulating Rho-kinase 2. Mol. Med. Rep. 2018, 18, 3077–3084. [Google Scholar]
- Gijón, M.A.; Riekhof, W.R.; Zarini, S.; Murphy, R.C.; Voelker, D.R. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J. Biol. Chem. 2008, 283, 30235–30245. [Google Scholar] [CrossRef] [Green Version]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef]
- Boeckmans, J.; Natale, A.; Rombaut, M.; Buyl, K.; Rogiers, V.; De Kock, J.; Vanhaecke, T.; Rodrigues, R.M. Anti-NASH drug development hitches a lift on PPAR agonism. Cells 2019, 9, 37. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.F.; Mullican, S.E.; DiSpirito, J.R.; Peed, L.C.; Lazar, M.A. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proc. Natl. Acad. Sci. USA 2013, 110, 18656–18661. [Google Scholar] [CrossRef] [Green Version]
- Chyau, C.C.; Wang, H.F.; Zhang, W.J.; Chen, C.C.; Huang, S.H.; Chang, C.C.; Peng, R.Y. Antrodan alleviates high-fat and high-fructose diet-induced fatty liver disease in C57BL/6 mice model via AMPK/Sirt1/SREBP-1c/PPARγ pathway. Int. J. Mol. Sci. 2020, 21, 360. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J.; et al. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, S.; Iwasaki, Y.; Matsumoto, S.; Satoh, T.; Ozawa, A.; Yamada, E.; Kakizaki, S.; Trejo, J.A.O.; Uchiyama, Y.; Yamada, M.; et al. Administration of small-molecule guanabenz acetate attenuates fatty liver and hyperglycemia associated with obesity. Sci. Rep. 2020, 10, 13671. [Google Scholar] [CrossRef] [PubMed]
- Coppola, S.; Avagliano, C.; Calignano, A.; Berni Canani, R. The protective role of butyrate against Obesity and Obesity-Related Diseases. Molecules 2021, 26, 682. [Google Scholar] [CrossRef] [PubMed]
- Jiao, A.R.; Yu, B.; He, J.; Yu, J.; Zheng, P.; Luo, Y.H.; Luo, J.Q.; Mao, X.B.; Chen, D.W. Short chain fatty acids could prevent fat deposition in pigs via regulating related hormones and genes. Food Funct. 2020, 11, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Sheng, L.L.; Zhong, J.; Tao, X.; Zhu, W.Z.; Ma, J.L.; Yan, J.; Zhao, A.H.; Zheng, X.J.; Wu, G.S.; et al. Desulfovibrio vulgaris, a potent acetic acid-producing bacterium, attenuates nonalcoholic fatty liver disease in mice. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.J.; Lin, C.L.; Zhang, Y.P.; Deng, Y.J.; Liu, C.; Yang, Q.H. Probiotic mixture of Lactobacillus and Bifidobacterium alleviates systemic adiposity and inflammation in non-alcoholic fatty liver disease rats through Gpr109a and the commensal metabolite butyrate. Inflammopharmacology 2018, 26, 1051–1055. [Google Scholar] [CrossRef]
- Chavez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile acid control of metabolism and inflammation in obesity, Type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology 2017, 152, 1679–1694.e3. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile acid metabolism and signaling in liver disease and therapy. Liver Res. 2017, 1, 3–9. [Google Scholar] [CrossRef]
- Ozcan, U.; Cao, Q.; Yilmaz, E.; Lee, A.H.; Iwakoshi, N.N.; Ozdelen, E.; Tuncman, G.; Gorgun, C.; Glimcher, L.H.; Hotamisligil, G.S. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 2004, 306, 457–461. [Google Scholar] [CrossRef] [Green Version]
- Cummings, B.P.; Bettaieb, A.; Graham, J.L.; Kim, J.; Ma, F.G.; Shibata, N.; Stanhope, K.L.; Giulivi, C.; Hansen, F.; Jelsing, J.; et al. Bile-acid-mediated decrease in endoplasmic reticulum stress: A potential contributor to the metabolic benefits of ileal interposition surgery in UCD-T2DM rats. Dis. Model Mech. 2013, 6, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Thakur, J.; Pal, S.; Mishra, D.; Rani, P.; Kumar, S.; Saini, A.; Singh, A.; Yadav, K.; Srivastava, A.; et al. Cholic-acid-derived amphiphiles can prevent and degrade fungal biofilms. ACS Appl. Bio Mater. 2021, 4, 7332–7341. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, S.; Wenniger, L.M.; Paulusma, C.C.; van Vliet, S.J.; Jefferson, D.M.; Elferink, R.P.; Beuers, U. A biliary HCO3-umbrella constitutes a protective mechanism against bile acid-induced injury in human cholangiocytes. Hepatology 2012, 55, 173–783. [Google Scholar] [CrossRef] [PubMed]
- Beuers, U. Drug insight: Mechanisms and sites of action of ursodeoxycholic acid in cholestasis. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.S.; Yan, L.; Guo, Z.H.; Chen, Y.; Li, M.; Huang, C.S.; Chen, Z.H.; Meng, X.Y. Chenodeoxycholic acid attenuates high-fat diet-induced obesity and hyperglycemia via the G protein-coupled bile acid receptor 1 and proliferator-activated receptor gamma pathway. Exp. Ther. Med. 2017, 14, 5305–5312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.J.; Zhao, J.F.; Gui, W.F.; Sun, D.; Dai, H.J.; Xiao, L.; Chu, H.K.; Du, F.; Zhu, Q.J.; Schnabl, B.; et al. Tauroursodeoxycholic acid inhibits intestinal inflammation and barrier disruption in mice with non-alcoholic fatty liver disease. Br. J. Pharmacol. 2018, 175, 469–484. [Google Scholar] [CrossRef] [PubMed]

| Item | Chr 1 | Plasmid 1 | Plasmid 2 |
|---|---|---|---|
| Genome Length | 1,723,288 bp | 204,234 bp | 26,769 bp |
| Number of scaffold(contig) | 1 | 1 | 1 |
| GC content | 33.00% | 31.90% | 32.79% |
| Num of CDS | 1587 | 199 | 29 |
| tRNA | 78 | ||
| rRNA | 22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Liao, R.; Huang, J.; Xiao, C.; Yang, Y.; Wang, H.; He, D.; Yan, H.; Yang, C. Lactobacillus salivarius SNK-6 Regulates Liver Lipid Metabolism Partly via the miR-130a-5p/MBOAT2 Pathway in a NAFLD Model of Laying Hens. Cells 2022, 11, 4133. https://doi.org/10.3390/cells11244133
Zhu L, Liao R, Huang J, Xiao C, Yang Y, Wang H, He D, Yan H, Yang C. Lactobacillus salivarius SNK-6 Regulates Liver Lipid Metabolism Partly via the miR-130a-5p/MBOAT2 Pathway in a NAFLD Model of Laying Hens. Cells. 2022; 11(24):4133. https://doi.org/10.3390/cells11244133
Chicago/Turabian StyleZhu, Lihui, Rongrong Liao, Jiwen Huang, Changfeng Xiao, Yunzhou Yang, Huiying Wang, Daqian He, Huaxiang Yan, and Changsuo Yang. 2022. "Lactobacillus salivarius SNK-6 Regulates Liver Lipid Metabolism Partly via the miR-130a-5p/MBOAT2 Pathway in a NAFLD Model of Laying Hens" Cells 11, no. 24: 4133. https://doi.org/10.3390/cells11244133
APA StyleZhu, L., Liao, R., Huang, J., Xiao, C., Yang, Y., Wang, H., He, D., Yan, H., & Yang, C. (2022). Lactobacillus salivarius SNK-6 Regulates Liver Lipid Metabolism Partly via the miR-130a-5p/MBOAT2 Pathway in a NAFLD Model of Laying Hens. Cells, 11(24), 4133. https://doi.org/10.3390/cells11244133







