A Leucine Zipper Dimerization Strategy to Generate Soluble T Cell Receptors Using the Escherichia coli Expression System
Abstract
:1. Introduction
2. Materials and Methods
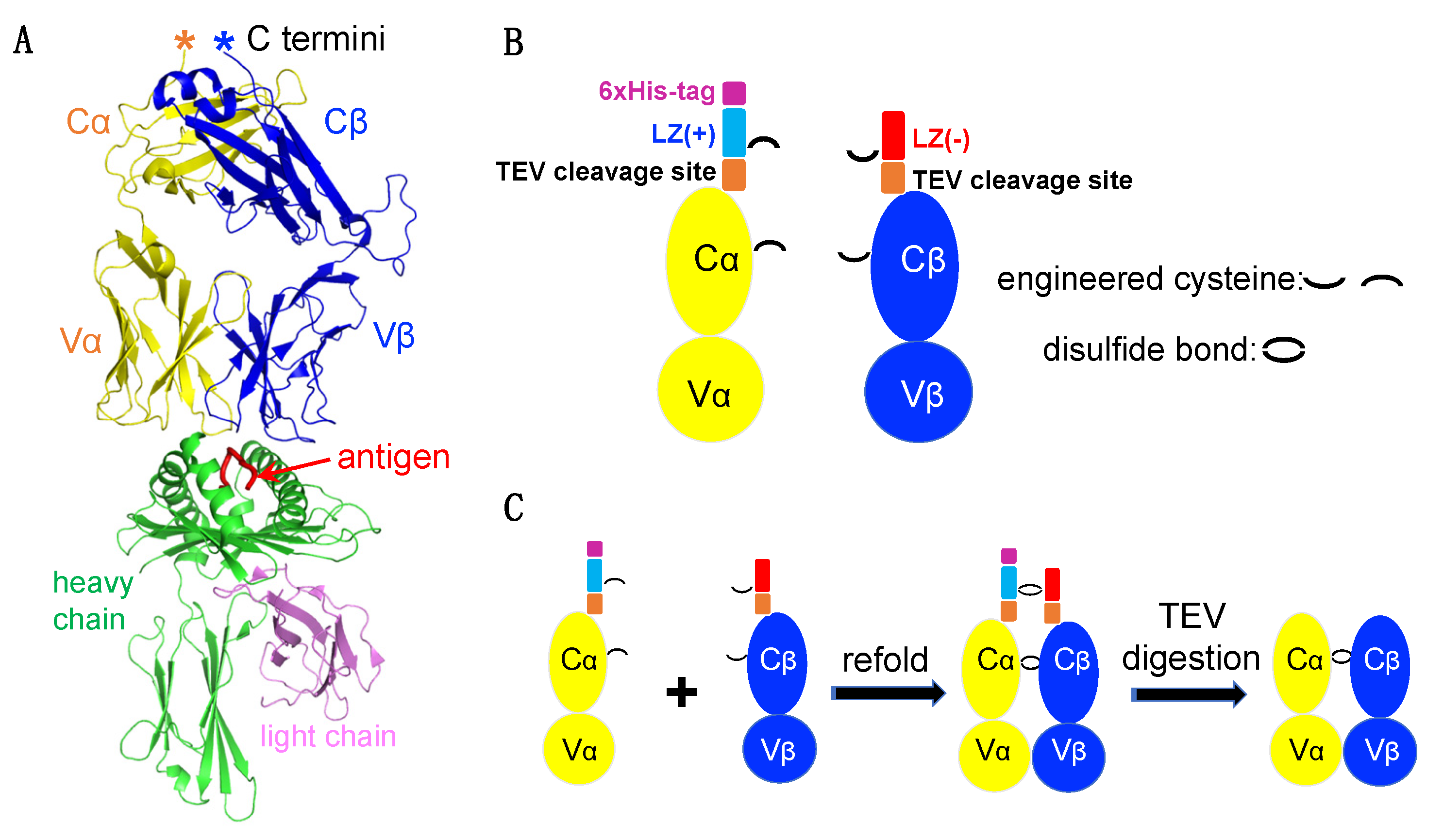
2.1. Construct Design
2.2. DNA Synthesis and Plasmid Construction
2.3. Inclusion Body Expression and Extraction
2.4. Soluble TCR Refolding
2.5. Protein Purification and Examination
2.6. Protein Digestion
2.7. Surface Plasmon Resonance
2.8. Software for Data Analysis
3. Results
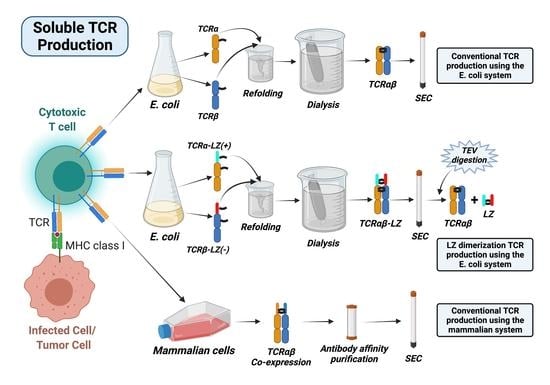
3.1. TCRα and TCRβ Chain Construct Design and Protein Production Workflow
3.2. Inclusion Bodies Expression, Purification and Solubilization
3.3. Refolding of Soluble C8 TCR
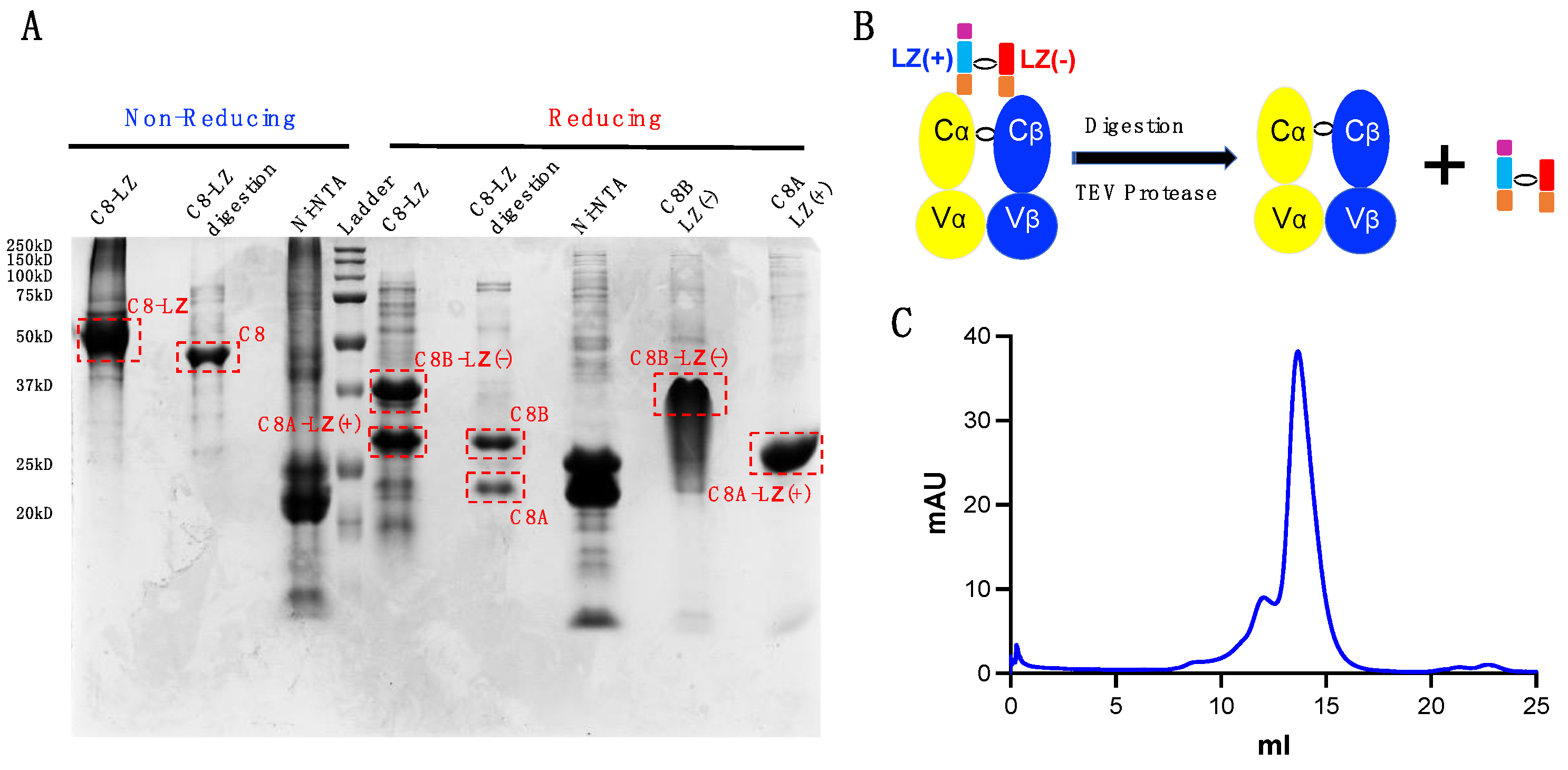
3.4. Soluble C8 TCR Purification
3.5. C8-LZ TCR and QW9-B*57 Binding Affinity Measurements
3.6. TEV Digestion for Leucine Zipper Removal
3.7. Application to TCRs beyond C8 TCR
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- La Gruta, N.L.; Gras, S.; Daley, S.R.; Thomas, P.G.; Rossjohn, J. Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 2018, 18, 467–478. [Google Scholar] [CrossRef]
- Wang, J.H.; Reinherz, E.L. The structural basis of αβ T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol. Rev. 2012, 250, 102–119. [Google Scholar] [CrossRef]
- von Andrian, U.H.; Mackay, C.R. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 2000, 343, 1020–1034. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mizsei, R.; Tan, K.; Mallis, R.J.; Duke-Cohan, J.S.; Akitsu, A.; Tetteh, P.W.; Dubey, A.; Hwang, W.; Wagner, G.; et al. Pre-T cell receptors topologically sample self-ligands during thymocyte β-selection. Science 2021, 371, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Teng, M.K.; Reinherz, E.L.; Wang, J.H. Strict Major Histocompatibility Complex Molecule Class-Specific Binding by Co-Receptors Enforces MHC-Restricted alphabeta TCR Recognition during T Lineage Subset Commitment. Front. Immunol. 2013, 4, 383. [Google Scholar] [CrossRef] [Green Version]
- Germain, R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002, 2, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Klein, L.; Kyewski, B.; Allen, P.M.; Hogquist, K.A. Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 2014, 14, 377–391. [Google Scholar] [CrossRef] [Green Version]
- Guermonprez, P.; Valladeau, J.; Zitvogel, L.; Thery, C.; Amigorena, S. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 2002, 20, 621–667. [Google Scholar] [CrossRef]
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459. [Google Scholar] [CrossRef]
- Wong, P.; Pamer, E.G. CD8 T cell responses to infectious pathogens. Annu. Rev. Immunol. 2003, 21, 29–70. [Google Scholar] [CrossRef]
- Schatz, D.G.; Oettinger, M.A.; Schlissel, M.S. V(D)J recombination: Molecular biology and regulation. Annu. Rev. Immunol. 1992, 10, 359–383. [Google Scholar] [CrossRef]
- Goldrath, A.W.; Bevan, M.J. Selecting and maintaining a diverse T-cell repertoire. Nature 1999, 402, 255–262. [Google Scholar] [CrossRef]
- Tonegawa, S. Nobel lecture in physiology or medicine—1987. Somatic generation of immune diversity. Vitr. Cell Dev. Biol. 1988, 24, 253–265. [Google Scholar] [CrossRef]
- Schatz, D.G.; Ji, Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 2011, 11, 251–263. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, Y.; Cheng, Y.; Glanville, J.; Zhang, D.; Lee, J.Y.; Olshen, R.A.; Weyand, C.M.; Boyd, S.D.; Goronzy, J.J. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl. Acad. Sci. USA 2014, 111, 13139–13144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Lian, X.; Gao, C.; Sun, X.; Einkauf, K.B.; Chevalier, J.M.; Chen, S.M.Y.; Hua, S.; Rhee, B.; Chang, K.; et al. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature 2020, 585, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Gaiha, G.D.; Rossin, E.J.; Urbach, J.; Landeros, C.; Collins, D.R.; Nwonu, C.; Muzhingi, I.; Anahtar, M.N.; Waring, O.M.; Piechocka-Trocha, A.; et al. Structural topology defines protective CD8(+) T cell epitopes in the HIV proteome. Science 2019, 364, 480–484. [Google Scholar] [CrossRef]
- Collins, D.R.; Gaiha, G.D.; Walker, B.D. CD8(+) T cells in HIV control, cure and prevention. Nat. Rev. Immunol. 2020, 20, 471–482. [Google Scholar] [CrossRef]
- Chen, H.; Ndhlovu, Z.M.; Liu, D.; Porter, L.C.; Fang, J.W.; Darko, S.; Brockman, M.A.; Miura, T.; Brumme, Z.L.; Schneidewind, A.; et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat. Immunol. 2012, 13, 691–700. [Google Scholar] [CrossRef] [Green Version]
- Rossjohn, J.; Gras, S.; Miles, J.J.; Turner, S.J.; Godfrey, D.I.; McCluskey, J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev. Immunol. 2015, 33, 169–200. [Google Scholar] [CrossRef] [PubMed]
- Sewell, A.K. Why must T cells be cross-reactive? Nat. Rev. Immunol. 2012, 12, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Garboczi, D.N.; Ghosh, P.; Utz, U.; Fan, Q.R.; Biddison, W.E.; Wiley, D.C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 1996, 384, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Boulter, J.M.; Glick, M.; Todorov, P.T.; Baston, E.; Sami, M.; Rizkallah, P.; Jakobsen, B.K. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 2003, 16, 707–711. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, E.K.; Rutkowski, R.; Stafford, W.F., 3rd; Kim, P.S. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science 1989, 245, 646–648. [Google Scholar] [CrossRef]
- Walseng, E.; Walchli, S.; Fallang, L.E.; Yang, W.; Vefferstad, A.; Areffard, A.; Olweus, J. Soluble T-cell receptors produced in human cells for targeted delivery. PLoS ONE 2015, 10, e0119559. [Google Scholar] [CrossRef]
- Chang, V.T.; Crispin, M.; Aricescu, A.R.; Harvey, D.J.; Nettleship, J.E.; Fennelly, J.A.; Yu, C.; Boles, K.S.; Evans, E.J.; Stuart, D.I.; et al. Glycoprotein structural genomics: Solving the glycosylation problem. Structure 2007, 15, 267–273. [Google Scholar] [CrossRef] [Green Version]
- Stewart-Jones, G.B.; McMichael, A.J.; Bell, J.I.; Stuart, D.I.; Jones, E.Y. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 2003, 4, 657–663. [Google Scholar] [CrossRef]
- Tan, N.Y.; Bailey, U.M.; Jamaluddin, M.F.; Mahmud, S.H.; Raman, S.C.; Schulz, B.L. Sequence-based protein stabilization in the absence of glycosylation. Nat. Commun. 2014, 5, 3099. [Google Scholar] [CrossRef] [Green Version]
- Tropea, J.E.; Cherry, S.; Waugh, D.S. Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol. 2009, 498, 297–307. [Google Scholar] [CrossRef]
- Glasel, J.A. Validity of nucleic acid purities monitored by 260 nm/280 nm absorbance ratios. Biotechniques 1995, 18, 62–63. [Google Scholar]
- Coutard, B.; Danchin, E.G.; Oubelaid, R.; Canard, B.; Bignon, C. Single pH buffer refolding screen for protein from inclusion bodies. Protein Expr. Purif. 2012, 82, 352–359. [Google Scholar] [CrossRef]
- Huang, J.; Zeng, X.; Sigal, N.; Lund, P.J.; Su, L.F.; Huang, H.; Chien, Y.H.; Davis, M.M. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc. Natl. Acad. Sci. USA 2016, 113, E1890–E1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niedzialkowska, E.; Gasiorowska, O.; Handing, K.B.; Majorek, K.A.; Porebski, P.J.; Shabalin, I.G.; Zasadzinska, E.; Cymborowski, M.; Minor, W. Protein purification and crystallization artifacts: The tale usually not told. Protein Sci. 2016, 25, 720–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudolph, M.G.; Wilson, I.A. The specificity of TCR/pMHC interaction. Curr. Opin. Immunol. 2002, 14, 52–65. [Google Scholar] [CrossRef]
- Baker, B.M.; Scott, D.R.; Blevins, S.J.; Hawse, W.F. Structural and dynamic control of T-cell receptor specificity, cross-reactivity, and binding mechanism. Immunol. Rev. 2012, 250, 10–31. [Google Scholar] [CrossRef]
- Poorebrahim, M.; Mohammadkhani, N.; Mahmoudi, R.; Gholizadeh, M.; Fakhr, E.; Cid-Arregui, A. TCR-like CARs and TCR-CARs targeting neoepitopes: An emerging potential. Cancer Gene Ther. 2021, 28, 581–589. [Google Scholar] [CrossRef]
- Douglass, J.; Hsiue, E.H.; Mog, B.J.; Hwang, M.S.; DiNapoli, S.R.; Pearlman, A.H.; Miller, M.S.; Wright, K.M.; Azurmendi, P.A.; Wang, Q.; et al. Bispecific antibodies targeting mutant RAS neoantigens. Sci. Immunol. 2021, 6, eabd5515. [Google Scholar] [CrossRef]
- Hsiue, E.H.; Wright, K.M.; Douglass, J.; Hwang, M.S.; Mog, B.J.; Pearlman, A.H.; Paul, S.; DiNapoli, S.R.; Konig, M.F.; Wang, Q.; et al. Targeting a neoantigen derived from a common TP53 mutation. Science 2021, 371, eabc8697. [Google Scholar] [CrossRef] [PubMed]
- Sermer, D.; Brentjens, R. CAR T-cell therapy: Full speed ahead. Hematol. Oncol. 2019, 37 (Suppl. S1), 95–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.F.; Wang, H.Y. Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res. 2017, 27, 11–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, T.N.; Kishton, R.J.; Restifo, N.P. Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat. Med. 2019, 25, 1488–1499. [Google Scholar] [CrossRef]
- Das, D.K.; Feng, Y.; Mallis, R.J.; Li, X.; Keskin, D.B.; Hussey, R.E.; Brady, S.K.; Wang, J.H.; Wagner, G.; Reinherz, E.L.; et al. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. USA 2015, 112, 1517–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Gutshall, L.; Jiang, H.; Baker, A.; Beil, E.; Obmolova, G.; Carton, J.; Taudte, S.; Amegadzie, B. Two routes for production and purification of Fab fragments in biopharmaceutical discovery research: Papain digestion of mAb and transient expression in mammalian cells. Protein Expr. Purif. 2009, 67, 182–189. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, A.; Piechocka-Trocha, A.; Li, X.; Walker, B.D. A Leucine Zipper Dimerization Strategy to Generate Soluble T Cell Receptors Using the Escherichia coli Expression System. Cells 2022, 11, 312. https://doi.org/10.3390/cells11030312
Zhang A, Piechocka-Trocha A, Li X, Walker BD. A Leucine Zipper Dimerization Strategy to Generate Soluble T Cell Receptors Using the Escherichia coli Expression System. Cells. 2022; 11(3):312. https://doi.org/10.3390/cells11030312
Chicago/Turabian StyleZhang, Angela, Alicja Piechocka-Trocha, Xiaolong Li, and Bruce D. Walker. 2022. "A Leucine Zipper Dimerization Strategy to Generate Soluble T Cell Receptors Using the Escherichia coli Expression System" Cells 11, no. 3: 312. https://doi.org/10.3390/cells11030312
APA StyleZhang, A., Piechocka-Trocha, A., Li, X., & Walker, B. D. (2022). A Leucine Zipper Dimerization Strategy to Generate Soluble T Cell Receptors Using the Escherichia coli Expression System. Cells, 11(3), 312. https://doi.org/10.3390/cells11030312