Exosome as a Delivery Vehicle for Cancer Therapy
Abstract
:1. Introduction
1.1. Drug Delivery Vehicles for Cancer Therapy
1.2. Basic Properties of Exosomes
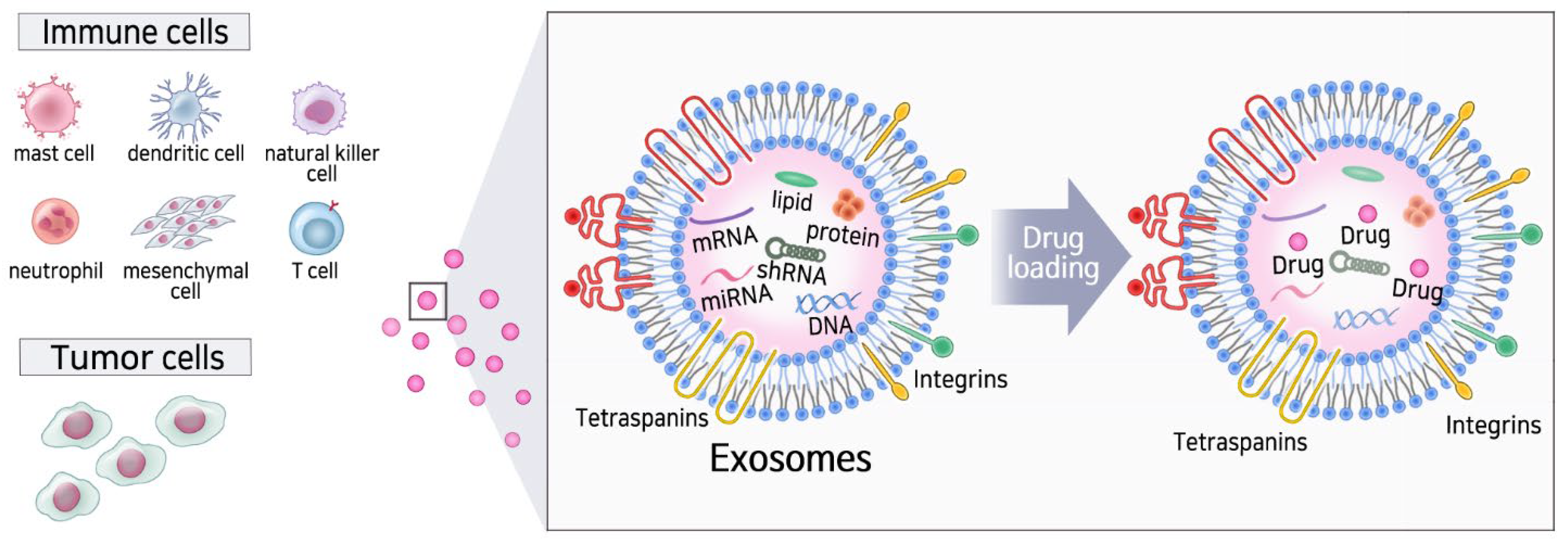
2. Natural Cell-Type Specific Exosomes
3. Artificial Exosomes as a Drug Delivery Vehicle
3.1. Limitations of Artificial Lipid Bilayer Nanoparticles
3.2. Advantages of Artificial Exosomes Compared to Artificial Lipid Bilayer Nanoparticles
3.3. Challenges Associated with Artificial Exosomes Compared to Lipid Bilayer Nanoparticles
4. Purification and Drug Loading of Exosomes
4.1. Approaches for the Isolation of Exosomes
4.2. Approaches for Drug Loading on Exosomes
5. Therapeutic Aspects of Exosomes as a DDS
5.1. Exosomes: The Natural Drug Delivery Vehicle
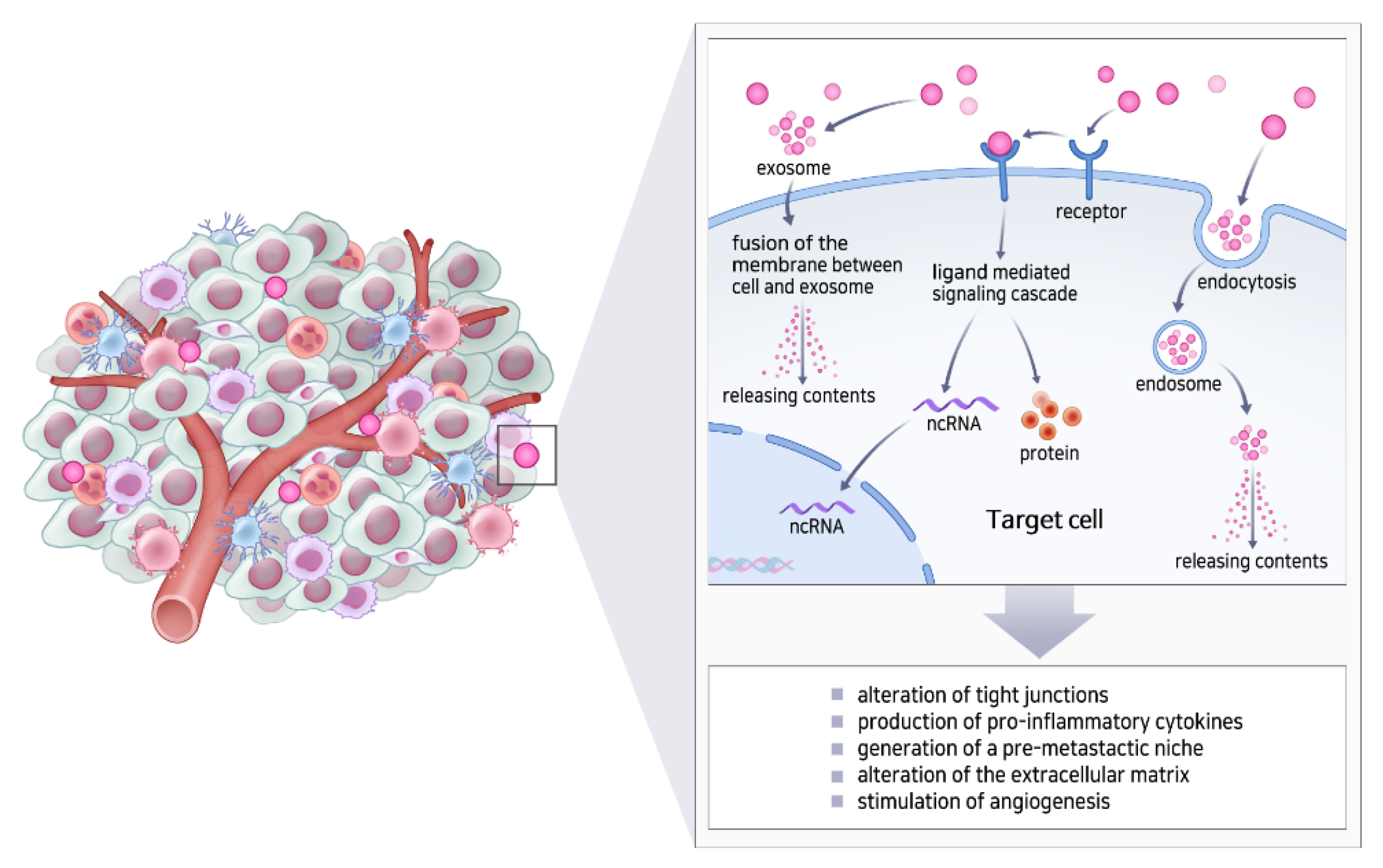
5.2. Exosomes in the Tumor Microenvironment (TME)
5.3. Engineering of Exosomes for Drug Delivery
5.4. Clinical Applications of Artificial Exosomes
6. Summary and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senapati, S.; Mahanta, A.K.; Kumar, S.; Maiti, P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct. Target. Ther. 2018, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, H.; Wu, J.; Sawa, T.; Matsumura, Y.; Hori, K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: A review. J. Control. Release 2000, 65, 271–284. [Google Scholar] [CrossRef]
- Koo, H.; Huh, M.S.; Sun, I.C.; Yuk, S.H.; Choi, K.; Kim, K.; Kwon, I.C. In vivo targeted delivery of nanoparticles for theranosis. Acc. Chem. Res. 2011, 44, 1018–1028. [Google Scholar] [CrossRef]
- Peer, D.; Karp, J.M.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef]
- Sun, W.; Gu, Z. Engineering DNA scaffolds for delivery of anticancer therapeutics. Biomater. Sci. 2015, 3, 1018–1024. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Sinha, D.; Roy, S.; Saha, P.; Chatterjee, N.; Bishayee, A. Trends in Research on Exosomes in Cancer Progression and Anticancer Therapy. Cancers 2021, 13, 326. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, W.; Hao, Y.; He, C.; Li, L.; Zhu, G. Exosome-orchestrated hypoxic tumor microenvironment. Mol. Cancer 2019, 18, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antimisiaris, S.G.; Mourtas, S.; Marazioti, A. Exosomes and Exosome-Inspired Vesicles for Targeted Drug Delivery. Pharmaceutics 2018, 10, 218. [Google Scholar] [CrossRef] [Green Version]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [Green Version]
- Fontana, S.; Saieva, L.; Taverna, S.; Alessandro, R. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: State of the art and new perspectives. Proteomics 2013, 13, 1581–1594. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Frank, S.; Aguirre, A.; Hescheler, J.; Kurian, L. A lncRNA Perspective into (Re)Building the Heart. Front. Cell Dev. Biol. 2016, 4, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Ljubimov, V.A.; Zhou, C.; Tong, Y.; Liang, J. Cell-free circulating tumor DNA in cancer. Chin. J. Cancer 2016, 35, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, R.C.; Yeo, R.W.; Padmanabhan, J.; Choo, A.; de Kleijn, D.P.; Lim, S.K. Isolation and Characterization of Exosome from Human Embryonic Stem Cell-Derived C-Myc-Immortalized Mesenchymal Stem Cells. Methods Mol. Biol. 2016, 1416, 477–494. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.W.; Lee, J.H.; Kim, S.Y.; Pack, C.G.; Ha, D.H.; Park, S.R.; Youn, J.; Cho, B.S. Advances in Analysis of Biodistribution of Exosomes by Molecular Imaging. Int. J. Mol. Sci. 2020, 21, 665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Justiz Vaillant, A.A.; Sabir, S.; Jan, A. Physiology, Immune Response; StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Jenjaroenpun, P.; Kremenska, Y.; Nair, V.M.; Kremenskoy, M.; Joseph, B.; Kurochkin, I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 2013, 1, e201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinman, R.M. Decisions about dendritic cells: Past, present, and future. Annu. Rev. Immunol. 2012, 30, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Chaput, N.; Taieb, J.; Schartz, N.E.; Andre, F.; Angevin, E.; Zitvogel, L. Exosome-based immunotherapy. Cancer Immunol. Immunother. 2004, 53, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Thery, C.; Ploix, S.; Tursz, T.; Lapierre, V.; Lantz, O.; Zitvogel, L.; Chaput, N. Dendritic cell-derived exosomes for cancer immunotherapy: What’s next? Cancer Res. 2010, 70, 1281–1285. [Google Scholar] [CrossRef] [Green Version]
- Hao, S.; Liu, Y.; Yuan, J.; Zhang, X.; He, T.; Wu, X.; Wei, Y.; Sun, D.; Xiang, J. Novel exosome-targeted CD4+ T cell vaccine counteracting CD4+25+ regulatory T cell-mediated immune suppression and stimulating efficient central memory CD8+ CTL responses. J. Immunol. 2007, 179, 2731–2740. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Gao, F.X.; Wang, C.; Qin, M.; Han, F.; Xu, T.; Hu, Z.; Long, Y.; He, X.M.; Deng, X.; et al. IL-6 and IL-8 secreted by tumour cells impair the function of NK cells via the STAT3 pathway in oesophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 321. [Google Scholar] [CrossRef]
- Fais, S. NK cell-released exosomes: Natural nanobullets against tumors. Oncoimmunology 2013, 2, e22337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Pace, A.L.; Tumino, N.; Besi, F.; Alicata, C.; Conti, L.A.; Munari, E.; Maggi, E.; Vacca, P.; Moretta, L. Characterization of Human NK Cell-Derived Exosomes: Role of DNAM1 Receptor in Exosome-Mediated Cytotoxicity Against Tumor. Cancers 2020, 12, 661. [Google Scholar] [CrossRef] [Green Version]
- Federici, C.; Shahaj, E.; Cecchetti, S.; Camerini, S.; Casella, M.; Iessi, E.; Camisaschi, C.; Paolino, G.; Calvieri, S.; Ferro, S.; et al. Natural-Killer-Derived Extracellular Vesicles: Immune Sensors and Interactors. Front. Immunol. 2020, 11, 262. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Kalimuthu, S.; Gangadaran, P.; Oh, J.M.; Lee, H.W.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. Exosomes Derived From Natural Killer Cells Exert Therapeutic Effect in Melanoma. Theranostics 2017, 7, 2732–2745. [Google Scholar] [CrossRef]
- Stone, K.D.; Prussin, C.; Metcalfe, D.D. IgE, mast cells, basophils, and eosinophils. J. Allergy Clin. Immunol. 2010, 125, S73–S80. [Google Scholar] [CrossRef] [PubMed]
- Ekstrom, K.; Valadi, H.; Sjostrand, M.; Malmhall, C.; Bossios, A.; Eldh, M.; Lotvall, J. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles 2012, 1, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Peng, J.; Wang, S.; Liu, X.; Zhang, K.; Zhang, Z.; Wang, C.; Jing, X.; Zhou, C.; Wang, Y. Applications of stem cell-derived exosomes in tissue engineering and neurological diseases. Rev. Neurosci. 2018, 29, 531–546. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Lin, L.; Wang, J.; Xiao, H.; Li, J.; Peng, X.; Dai, H.; Li, L. Mast Cell-Derived Exosomes Promote Th2 Cell Differentiation via OX40L-OX40 Ligation. J. Immunol. Res. 2016, 2016, 3623898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skokos, D.; Botros, H.G.; Demeure, C.; Morin, J.; Peronet, R.; Birkenmeier, G.; Boudaly, S.; Mecheri, S. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J. Immunol. 2003, 170, 3037–3045. [Google Scholar] [CrossRef] [Green Version]
- Vargas, A.; Roux-Dalvai, F.; Droit, A.; Lavoie, J.P. Neutrophil-Derived Exosomes: A New Mechanism Contributing to Airway Smooth Muscle Remodeling. Am. J. Respir. Cell Mol. Biol. 2016, 55, 450–461. [Google Scholar] [CrossRef]
- Shao, S.; Fang, H.; Zhang, J.; Jiang, M.; Xue, K.; Ma, J.; Zhang, J.; Lei, J.; Zhang, Y.; Li, B.; et al. Neutrophil exosomes enhance the skin autoinflammation in generalized pustular psoriasis via activating keratinocytes. FASEB J. 2019, 33, 6813–6828. [Google Scholar] [CrossRef] [PubMed]
- Brook, A.C.; Jenkins, R.H.; Clayton, A.; Kift-Morgan, A.; Raby, A.C.; Shephard, A.P.; Mariotti, B.; Cuff, S.M.; Bazzoni, F.; Bowen, T.; et al. Neutrophil-derived miR-223 as local biomarker of bacterial peritonitis. Sci. Rep. 2019, 9, 10136. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.R.; Lempke, S.L.; Miller, M.M.; Bush, D.M.; Braswell, B.G.; Estes, C.L.; Benedict, E.L.; Mahon, A.R.; Sabo, S.L.; Greenlee-Wacker, M.C. Effect of extracellular vesicles from S. aureus-challenged human neutrophils on macrophages. J. Leukoc. Biol. 2020, 108, 1841–1850. [Google Scholar] [CrossRef]
- Genschmer, K.R.; Russell, D.W.; Lal, C.; Szul, T.; Bratcher, P.E.; Noerager, B.D.; Abdul Roda, M.; Xu, X.; Rezonzew, G.; Viera, L.; et al. Activated PMN Exosomes: Pathogenic Entities Causing Matrix Destruction and Disease in the Lung. Cell 2019, 176, 113–126.e15. [Google Scholar] [CrossRef] [Green Version]
- Bianco, P.; Robey, P.G.; Simmons, P.J. Mesenchymal stem cells: Revisiting history, concepts, and assays. Cell Stem Cell 2008, 2, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, F.G.; Carvalho, M.M.; Sousa, N.; Salgado, A.J. Mesenchymal stem cells secretome: A new paradigm for central nervous system regeneration? Cell. Mol. Life Sci. 2013, 70, 3871–3882. [Google Scholar] [CrossRef]
- Gomari, H.; Forouzandeh Moghadam, M.; Soleimani, M. Targeted cancer therapy using engineered exosome as a natural drug delivery vehicle. OncoTargets Ther. 2018, 11, 5753–5762. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.J.; Wu, J.Y.; Liu, J.; Xu, W.; Qiu, X.; Huang, S.; Hu, X.B.; Xiang, D.X. Artificial exosomes for translational nanomedicine. J. Nanobiotechnol. 2021, 19, 242. [Google Scholar] [CrossRef]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Li, S.D.; Huang, L. Nanoparticles evading the reticuloendothelial system: Role of the supported bilayer. Biochim. Biophys. Acta 2009, 1788, 2259–2266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, M.; Abu Lila, A.S.; Shimizu, T.; Alaaeldin, E.; Hussein, A.; Sarhan, H.A.; Szebeni, J.; Ishida, T. PEGylated liposomes: Immunological responses. Sci. Technol. Adv. Mater. 2019, 20, 710–724. [Google Scholar] [CrossRef] [Green Version]
- Ishida, T.; Ichihara, M.; Wang, X.; Yamamoto, K.; Kimura, J.; Majima, E.; Kiwada, H. Injection of PEGylated liposomes in rats elicits PEG-specific IgM, which is responsible for rapid elimination of a second dose of PEGylated liposomes. J. Control. Release 2006, 112, 15–25. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dams, E.T.; Laverman, P.; Oyen, W.J.; Storm, G.; Scherphof, G.L.; van Der Meer, J.W.; Corstens, F.H.; Boerman, O.C. Accelerated blood clearance and altered biodistribution of repeated injections of sterically stabilized liposomes. J. Pharmacol. Exp. Ther. 2000, 292, 1071–1079. [Google Scholar]
- Semple, S.C.; Harasym, T.O.; Clow, K.A.; Ansell, S.M.; Klimuk, S.K.; Hope, M.J. Immunogenicity and rapid blood clearance of liposomes containing polyethylene glycol-lipid conjugates and nucleic Acid. J. Pharmacol. Exp. Ther. 2005, 312, 1020–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, P.L.; Harada, T.; Christian, D.A.; Pantano, D.A.; Tsai, R.K.; Discher, D.E. Minimal “Self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science 2013, 339, 971–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Segaliny, A.; et al. Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cell. Mol. Bioeng. 2016, 9, 509–529. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Lakhal, S.; Mager, I.; Wood, M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug. Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef]
- Rahman, M.M.; Shimizu, K.; Yamauchi, M.; Takase, H.; Ugawa, S.; Okada, A.; Inoshima, Y. Acidification effects on isolation of extracellular vesicles from bovine milk. PLoS ONE 2019, 14, e0222613. [Google Scholar] [CrossRef] [Green Version]
- Wasik, M.; Nazimek, K.; Nowak, B.; Askenase, P.W.; Bryniarski, K. Delayed-Type Hypersensitivity Underlying Casein Allergy Is Suppressed by Extracellular Vesicles Carrying miRNA-150. Nutrients 2019, 11, 907. [Google Scholar] [CrossRef] [Green Version]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef] [PubMed]
- Maroto, R.; Zhao, Y.; Jamaluddin, M.; Popov, V.L.; Wang, H.; Kalubowilage, M.; Zhang, Y.; Luisi, J.; Sun, H.; Culbertson, C.T.; et al. Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. J. Extracell. Vesicles 2017, 6, 1359478. [Google Scholar] [CrossRef] [Green Version]
- Trenkenschuh, E.; Richter, M.; Heinrich, E.; Koch, M.; Fuhrmann, G.; Friess, W. Enhancing the Stabilization Potential of Lyophilization for Extracellular Vesicles. Adv. Healthc. Mater. 2021, e2100538. [Google Scholar] [CrossRef] [PubMed]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Cheruvanky, A.; Zhou, H.; Pisitkun, T.; Kopp, J.B.; Knepper, M.A.; Yuen, P.S.; Star, R.A. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Ren. Physiol. 2007, 292, F1657–F1661. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.W.; Wieckowski, E.; Taylor, D.D.; Reichert, T.E.; Watkins, S.; Whiteside, T.L. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin. Cancer Res. 2005, 11, 1010–1020. [Google Scholar]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Corso, G.; Mager, I.; Lee, Y.; Gorgens, A.; Bultema, J.; Giebel, B.; Wood, M.J.A.; Nordin, J.Z.; Andaloussi, S.E. Reproducible and scalable purification of extracellular vesicles using combined bind-elute and size exclusion chromatography. Sci. Rep. 2017, 7, 11561. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.K.; Khan, M.A.; Zubair, H.; Srivastava, S.K.; Khushman, M.; Singh, S.; Singh, A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019, 9, 5335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanada, M.; Bachmann, M.H.; Hardy, J.W.; Frimannson, D.O.; Bronsart, L.; Wang, A.; Sylvester, M.D.; Schmidt, T.L.; Kaspar, R.L.; Butte, M.J.; et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA 2015, 112, E1433–E1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Familtseva, A.; Jeremic, N.; Tyagi, S.C. Exosomes: Cell-created drug delivery systems. Mol. Cell. Biochem. 2019, 459, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.; Busatto, S.; Pham, A.; Tian, M.; Suh, A.; Carson, K.; Quintero, A.; Lafrence, M.; Malik, H.; Santana, M.X.; et al. Extracellular vesicle-based drug delivery systems for cancer treatment. Theranostics 2019, 9, 8001–8017. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, L.; Peng, Y.; Yu, S.; Liu, J.; Wu, L.; Zhang, L.; Wu, Q.; Chang, X.; Yu, X.; et al. Dendritic cells loaded with tumor derived exosomes for cancer immunotherapy. Oncotarget 2018, 9, 2887–2894. [Google Scholar] [CrossRef] [Green Version]
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Ilkhani, K.; Bastami, M.; Delgir, S.; Safi, A.; Talebian, S.; Alivand, M.R. The Engaged Role of Tumor Microenvironment in Cancer Metabolism: Focusing on Cancer-Associated Fibroblast and Exosome Mediators. Anticancer Agents Med. Chem. 2021, 21, 254–266. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Ohlund, D.; Elyada, E.; Tuveson, D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 2014, 211, 1503–1523. [Google Scholar] [CrossRef]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S.; Nittner, D.; Burton, O.; Bassez, A.; Decaluwe, H.; Pircher, A.; Van den Eynde, K.; et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018, 24, 1277–1289. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Fernandes, A.R.; Baptista, P.V. Exosome in tumour microenvironment: Overview of the crosstalk between normal and cancer cells. Biomed. Res. Int. 2014, 2014, 179486. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raskov, H.; Orhan, A.; Gaggar, S.; Gogenur, I. Cancer-Associated Fibroblasts and Tumor-Associated Macrophages in Cancer and Cancer Immunotherapy. Front. Oncol. 2021, 11, 668731. [Google Scholar] [CrossRef]
- Eguchi, T.; Taha, E.A.; Calderwood, S.K.; Ono, K. A Novel Model of Cancer Drug Resistance: Oncosomal Release of Cytotoxic and Antibody-Based Drugs. Biology 2020, 9, 47. [Google Scholar] [CrossRef] [Green Version]
- Wyciszkiewicz, A.; Kalinowska-Lyszczarz, A.; Nowakowski, B.; Kazmierczak, K.; Osztynowicz, K.; Michalak, S. Expression of small heat shock proteins in exosomes from patients with gynecologic cancers. Sci. Rep. 2019, 9, 9817. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, P.; Haldar, M.K.; Karandish, F.; Confeld, M.; Hossain, R.; Borowicz, P.; Gange, K.; Xia, L.; Sarkar, K.; Mallik, S. Tissue-Penetrating, Hypoxia-Responsive Echogenic Polymersomes for Drug Delivery to Solid Tumors. Chemistry 2018, 24, 12490–12494. [Google Scholar] [CrossRef]
- Zuo, H.D.; Yao, W.W.; Chen, T.W.; Zhu, J.; Zhang, J.J.; Pu, Y.; Liu, G.; Zhang, X.M. The effect of superparamagnetic iron oxide with iRGD peptide on the labeling of pancreatic cancer cells in vitro: A preliminary study. Biomed. Res. Int. 2014, 2014, 852352. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Belhadj, Z.; He, B.; Deng, H.; Song, S.; Zhang, H.; Wang, X.; Dai, W.; Zhang, Q. A combined “eat me/don’t eat me” strategy based on extracellular vesicles for anticancer nanomedicine. J. Extracell. Vesicles 2020, 9, 1806444. [Google Scholar] [CrossRef] [PubMed]
- Fam, S.Y.; Chee, C.F.; Yong, C.Y.; Ho, K.L.; Mariatulqabtiah, A.R.; Tan, W.S. Stealth Coating of Nanoparticles in Drug-Delivery Systems. Nanomaterials 2020, 10, 787. [Google Scholar] [CrossRef] [Green Version]
- Shimoda, M.; Khokha, R. Metalloproteinases in extracellular vesicles. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xie, F.; Wang, L.; Zhang, L.; Zhang, S.; Fang, M.; Zhou, F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell. Mol. Immunol. 2020, 17, 323–334. [Google Scholar] [CrossRef]
- Marcus, M.E.; Leonard, J.N. FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharmaceuticals 2013, 6, 659–680. [Google Scholar] [CrossRef] [Green Version]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, W.; He, C.; Hao, Y.; Wang, L.; Li, L.; Zhu, G. Prospects and challenges of extracellular vesicle-based drug delivery system: Considering cell source. Drug. Deliv. 2020, 27, 585–598. [Google Scholar] [CrossRef] [Green Version]

| Method | Pros | Cons |
|---|---|---|
| Ultracentrifugation | Low cost, high purity, massive production | Time-consuming, mechanical damage, specialized equipment requirement |
| Ultrafiltration | Low cost, less time consuming, good portability | Moderate purity, mechanical damage, high cost |
| Immunological separation | High purity, no chemical contamination, simple | Small volume production, high cost |
| Loading Method | Extracellular Vesicle (EV) Source | Loading Content | Loading Measurement | Efficiency (Type, %) | |
|---|---|---|---|---|---|
| Sonication | Raw 264.7 macrophages (mouse) | Paclitaxel (PTX) | High-performance liquid chromatography (HPLC) | Loading capacity | 28.29 (SEM ± 1.38%) |
| Raw 264.7 macrophages (mouse) | Dox | Fluorescence of Dox | Encapsulation efficiency | 8.0–11.0% | |
| Raw 264.7 macrophages (mouse) | Catalase | Catalase enzymatic activity | Loading capacity | 26.1 (SEM ± 1.2%) | |
| Saponin permeabilization | Raw 264.7 macrophages (mouse) | Catalase | Catalase enzymatic activity | Loading capacity | 18.5 (SEM ± 1.3%) |
| Mixing | Raw 264.7 macrophages (mouse) | Paclitaxel (PTX) | High-performance liquid chromatography (HPLC) | Loading capacity | 1.4 (SEM ± 0.38%) |
| LNCaP and PC-3 (human) | PTX | Ultra-performance liquid chromatography (UPLC) | Encapsulation efficiency | 9.2% (SD ± 4.5%) | |
| Milk (bovine) | PTX | UPLC | Encapsulation efficiency | 7.9 ± 1.0% | |
| Raw 264.7 macrophages (mouse) | Catalase | Catalase enzymatic activity | Loading capacity | 4.9 (SEM ± 0.5%) | |
| Electroporation | Raw 264.7 macrophages (mouse) | Paclitaxel (PTX) | High-performance liquid chromatography (HPLC) | Loading capacity | 5.3 (SEM ± 0.48%) |
| Immature dendritic cells (mouse) | Doxorubicin (Dox) | Fluorescence of Dox | Encapsulation efficiency | <20% | |
| Primary immature dendritic cells (mouse) | Glyceraldehyde 3-phosphate dehydrogenase | qPCR analysis, fluorescence microscopy | Encapsulation efficiency | 10–38% | |
| Primary dendritic cells (mouse) | Vascular endothelial growth factor (VEGF) siRNA | qPCR analysis | Encapsulation efficiency | 3% | |
| Source of Exosomes | Disease Type | Drugs | Isolation Methods |
|---|---|---|---|
| Raw 264.7 macrophages (mouse) | Multi-drug resistant cancers (in vitro and mouse models) | Doxorubicin and paclitaxel | Low-speed centrifugation with precipitating reagents and purifying column |
| Primary dendritic cells (mouse) | Breast cancer (in vitro and mouse models) | VEGF siRNA | Differential centrifugation and UC |
| Neutrophils | Malignant glioma | Doxorubicin | Ultracentrifugation |
| MSC | Colorectal cancer | Doxorubicin | Ultracentrifugation |
| Milk (bovine) | Lung cancer (in vitro and mouse models) | Paclitaxel | Differential gradient centrifugation and UC |
| MCF-7 breast carcinoma cells (human) | Breast carcinoma (in vitro) | Doxorubicin | Differential gradient centrifugation |
| LNCaP and PC-3 prostate cancer cells (human) | Prostate cancer (in vitro) | Paclitaxel | Differential centrifugation |
| Lewis lung carcinoma cells (mouse) | Lung cancer (in vitro) | Methotrexate | Differential gradient centrifugation |
| Immature dendritic cells (mouse) | Breast cancer (in vitro and mouse models) | Doxorubicin | Ultrafiltration, UC, and gradient centrifugation |
| HeLa cervical cancer cells (human) | Cervical cancer (in vitro) | Dextran | Precipitating reagents (total exosome isolation kit, Invitrogen) |
| H22 hepatocarcinoma cells (mouse) | Hepatocarcinoma (in vitro and mouse models) | Cisplatin | Differential gradient centrifugation |
| Gastric cancer (SKBR-3) | Gastric cancer | Trastuzumab | Ultracentrifugation |
| EL-4 lymphoma cells (mouse) | Tumor-induced inflammation (in vitro and mouse models) | Curcumin | Sucrose gradient centrifugation |
| Bone-marrow-derived MSCs (human) | Lung cancer (in vitro) | TRAIL | Filtration |
| Pleural mesothelioma (in vitro) | TRAIL | Filtration | |
| Renal cancer (in vitro) | TRAIL | Filtration | |
| Breast adenocarcinoma (in vitro) | TRAIL | Filtration | |
| Neuroblastoma (in vitro) | TRAIL | Filtration | |
| B16-F10 melanoma cells (mouse) | Melanoma (in vitro) | Superparamagnetic iron oxide nanoparticles | Ultracentrifugation (UC) |
| B16BL6 melanoma cells (mouse) | Melanoma (in vitro and mouse models) | CpG DNA | Filtration and differential UC |
| ADR/MCF-7 breast carcinoma cells (human) | Breast carcinoma (in vitro) | Cisplatin | Differential gradient centrifugation |
| A549 lung carcinoma cells (human) | Lung carcinoma (in vitro, mouse models, and stage IV human patients) | Doxorubicin | Differential gradient centrifugation |
| M1 macrophage | Pancreatic cancer | Gemcitabine/Deferasirox | Ultracentrifugation |
| Human breast cancer cell line (EFM-192A) | Breast cancer | Trastuzumab | Ultracentrifugation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, B.; Chang, S. Exosome as a Delivery Vehicle for Cancer Therapy. Cells 2022, 11, 316. https://doi.org/10.3390/cells11030316
Moon B, Chang S. Exosome as a Delivery Vehicle for Cancer Therapy. Cells. 2022; 11(3):316. https://doi.org/10.3390/cells11030316
Chicago/Turabian StyleMoon, Bohyun, and Suhwan Chang. 2022. "Exosome as a Delivery Vehicle for Cancer Therapy" Cells 11, no. 3: 316. https://doi.org/10.3390/cells11030316
APA StyleMoon, B., & Chang, S. (2022). Exosome as a Delivery Vehicle for Cancer Therapy. Cells, 11(3), 316. https://doi.org/10.3390/cells11030316







