Fine-Tuning of Piezo1 Expression and Activity Ensures Efficient Myoblast Fusion during Skeletal Myogenesis
Abstract
:1. Introduction
2. Materials and Methods
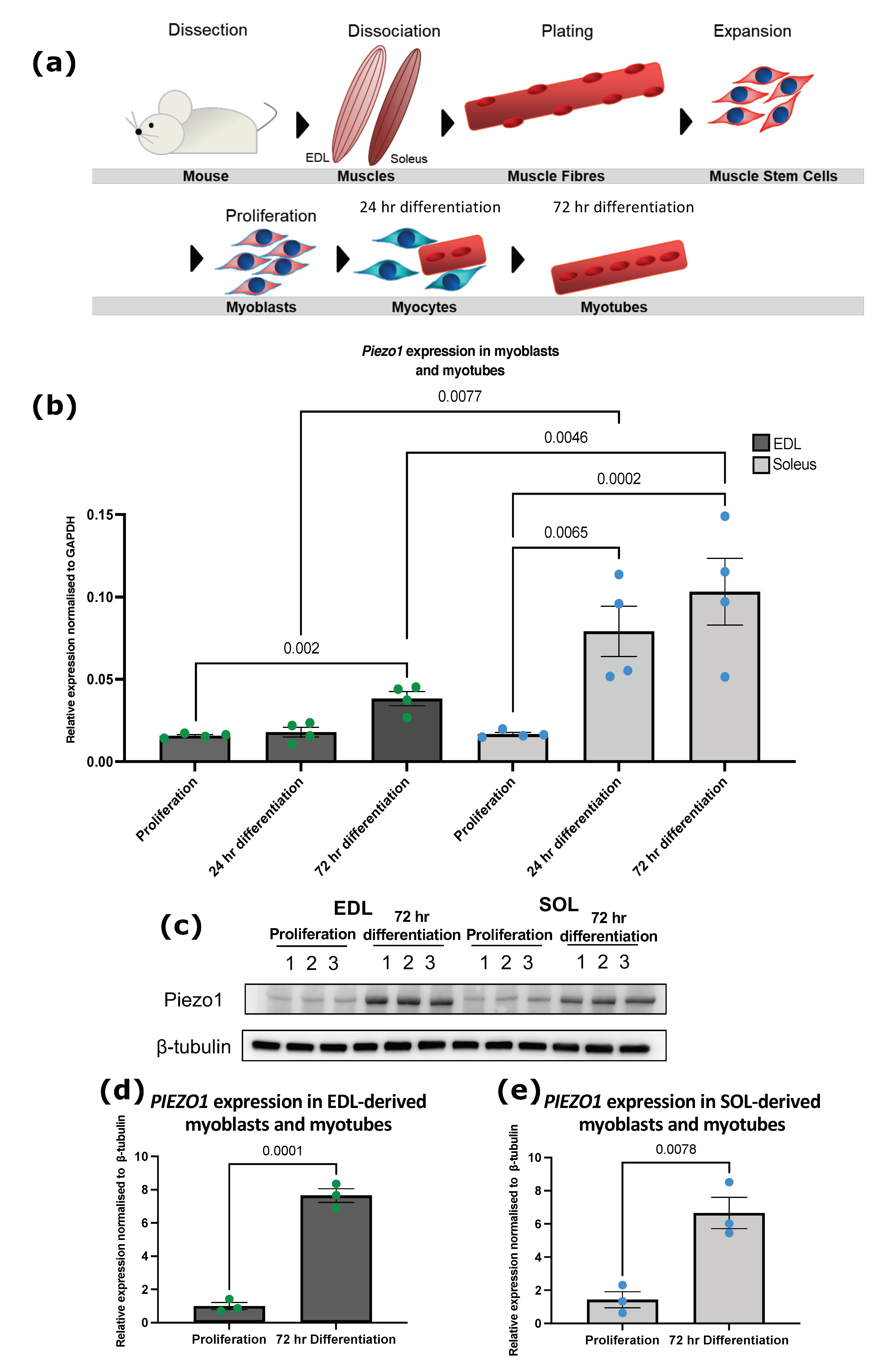
2.1. Primary Myoblast Cell Culture
2.2. siRNA Transfection
2.3. RNA Extraction and Reverse Transcription
2.4. Real-Time Quantitative PCR (RT-qPCR)
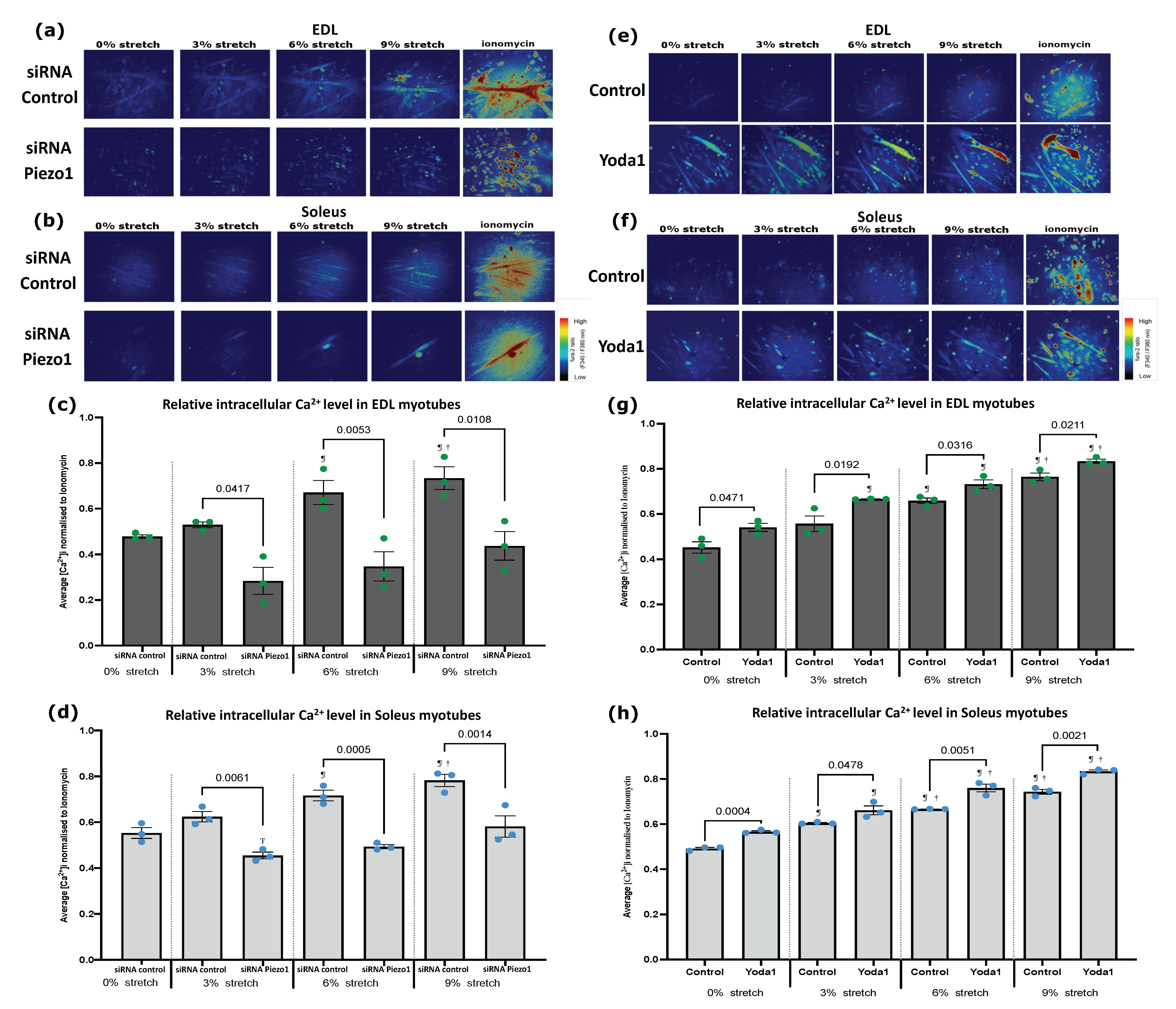
2.5. Piezo1 Activation
2.6. Immunolabelling
2.7. EdU Incorporation
2.8. Phalloidin Labelling
2.9. Image Analysis and Quantification
2.10. Stretch Experiments and Imaging of Intracellular Ca2+ Level
2.11. Immunoblotting
2.12. Statistical Analysis
3. Results
3.1. Piezo1 Is Upregulated during Myoblast Differentiation
3.2. Piezo1 Regulates Ca2+ Influx during Myogenesis
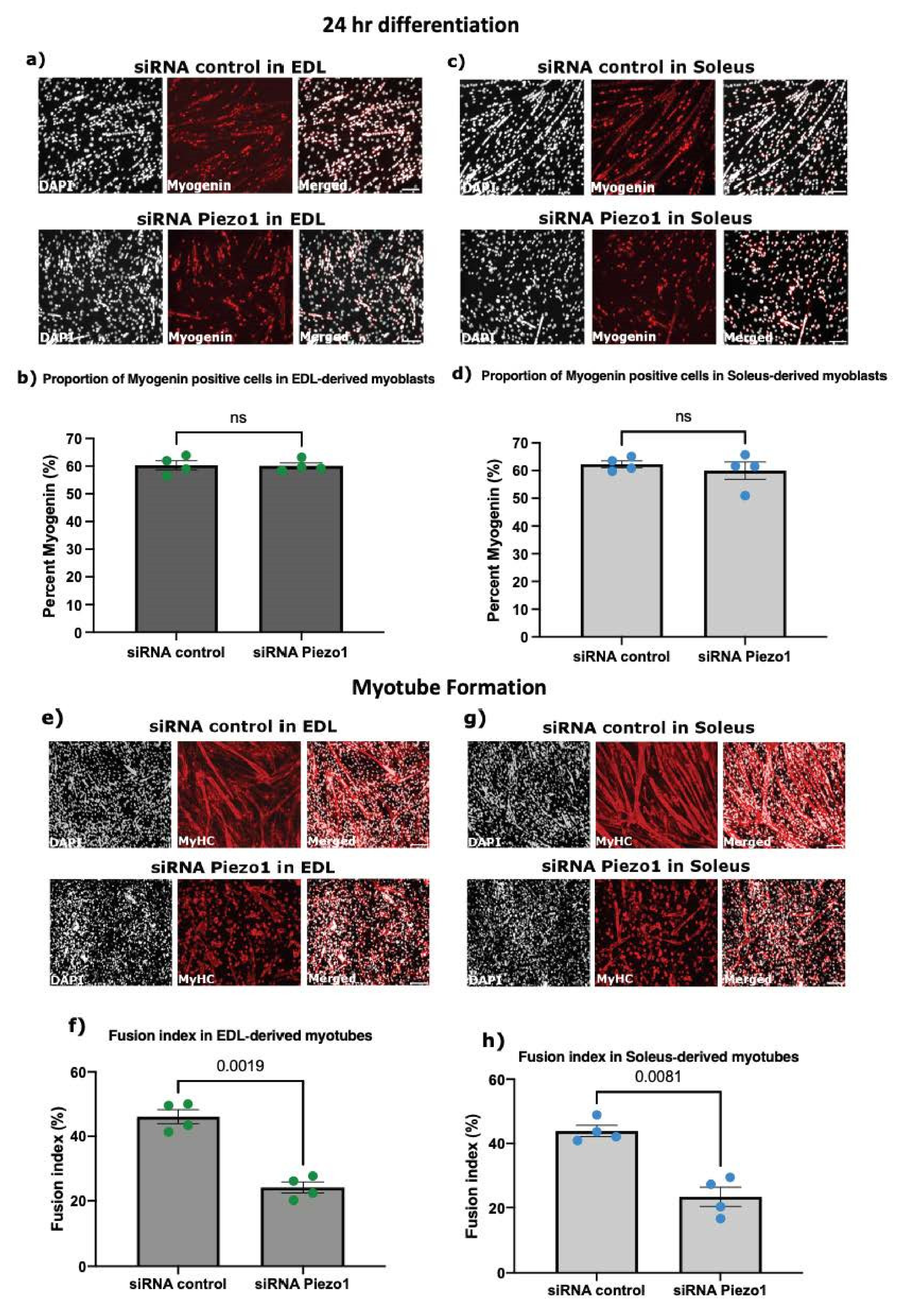
3.3. Piezo1 Is Dispensable for Myoblast Proliferation and Early Commitment to Differentiation
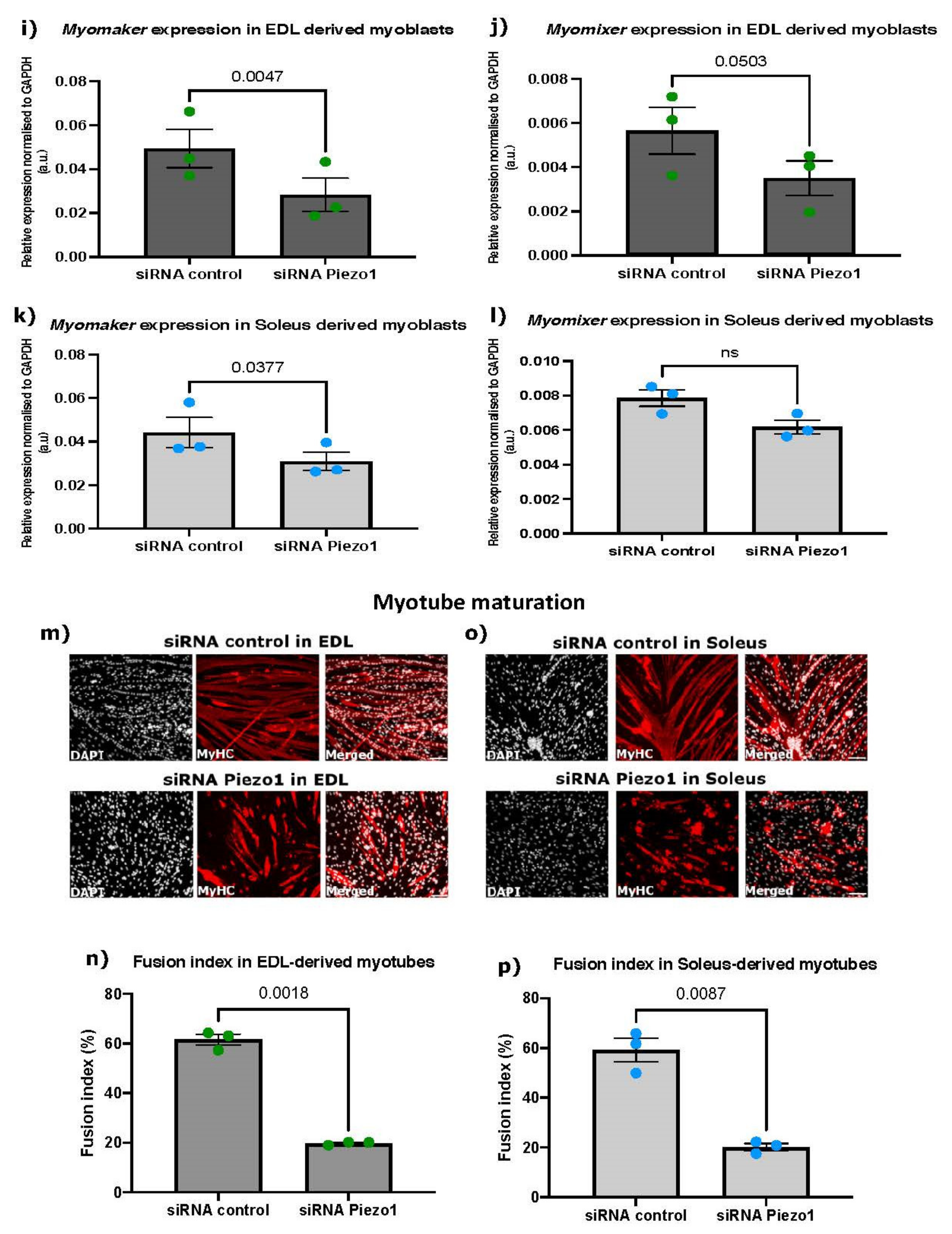
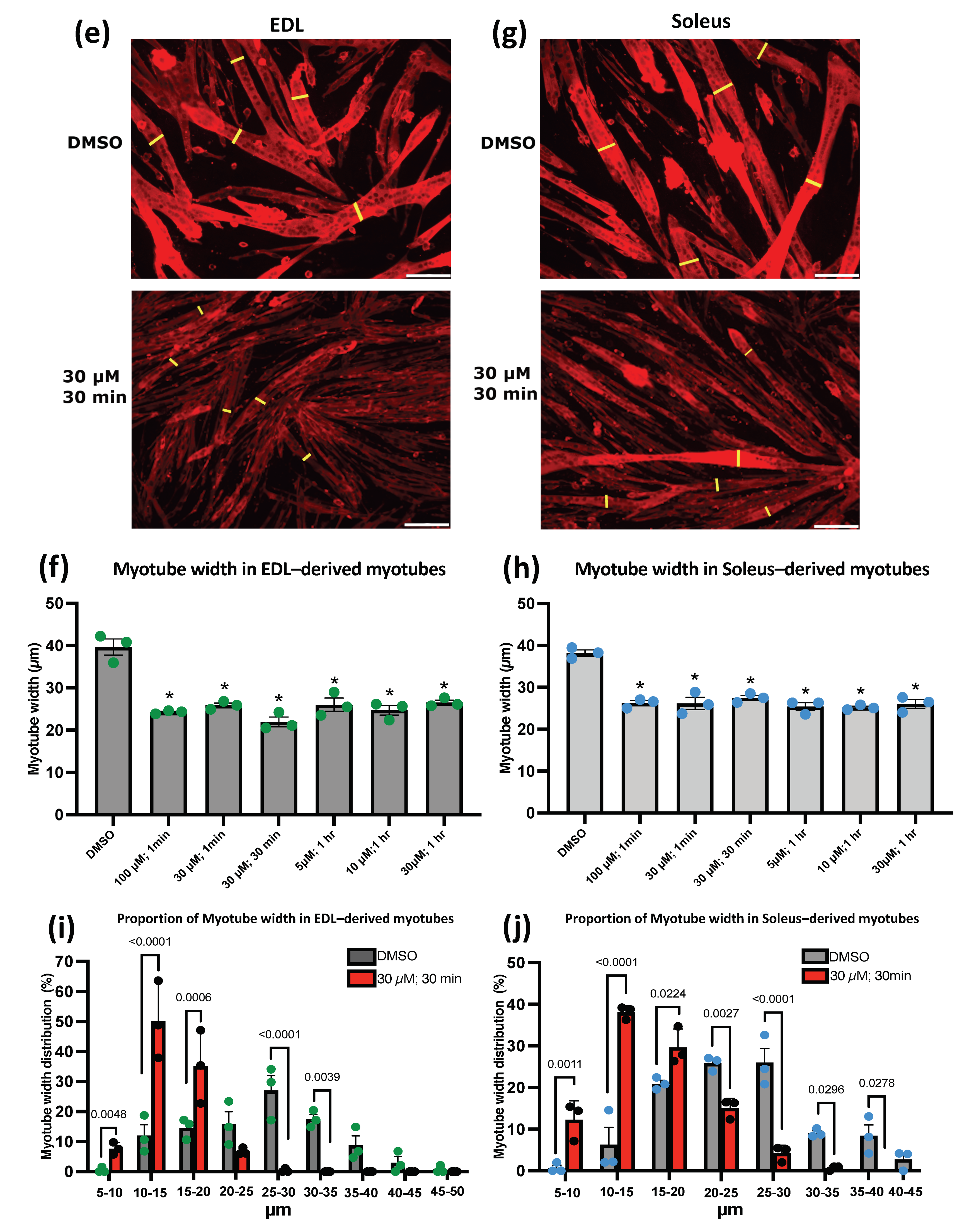
3.4. Piezo1 Activity Is Finely Regulated during Myoblast Fusion
3.5. Deregulation of Piezo1 Alters Cytoskeletal Remodelling during Differentiation
4. Discussion
4.1. Piezo1 in Myogenesis
4.2. Piezo1 Activation and Ca2+ Influx
4.3. Piezo1 and Muscular Dystrophies
4.4. Piezo1 and the Myoblast Fusion Machinery
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mauro, A. Satellite Cell of Skeletal Muscle Fibers. J. Cell Biol. 1961, 9, 493–495. [Google Scholar] [CrossRef] [PubMed]
- Zammit, P.S.; Relaix, F.; Nagata, Y.; Ruiz, A.P.; Collins, C.; Partridge, T.A.; Beauchamp, J. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006, 119, 1824–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zammit, P.S.; Partridge, T.A.; Yablonka-Reuveni, Z. The Skeletal Muscle Satellite Cell: The Stem Cell That Came in From the Cold. J. Histochem. Cytochem. 2006, 54, 1177–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relaix, F.; Zammit, P.S. Satellite cells are essential for skeletal muscle regeneration: The cell on the edge returns centre stage. Development 2012, 139, 2845–2856. [Google Scholar] [CrossRef] [Green Version]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 Are Essential Components of Distinct Mechanically Activated Cation Channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chi, S.; Guo, H.; Li, G.; Wang, L.; Zhao, Q.; Rao, Y.; Zu, L.; He, W.; Xiao, B. A lever-like transduction pathway for long-distance chemical- and mechano-gating of the mechanosensitive Piezo1 channel. Nat. Commun. 2018, 9, 1300. [Google Scholar] [CrossRef]
- Cinar, E.; Zhou, S.; DeCourcey, J.; Wang, Y.; Waugh, R.E.; Wan, J. Piezo1 regulates mechanotransductive release of ATP from human RBCs. Proc. Natl. Acad. Sci. USA 2015, 112, 11783–11788. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.; Li, W.; Zhao, Q.; Li, N.; Chen, M.; Zhi, P.; Li, R.; Gao, N.; Xiao, B.; Yang, M. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature 2015, 527, 64–69. [Google Scholar] [CrossRef]
- Sachs, F. Stretch-activated ion channels: What are they? Physiology 2010, 25, 50–56. [Google Scholar] [CrossRef]
- Saotome, K.; Murthy, S.; Kefauver, J.M.; Whitwam, T.; Patapoutian, A.; Ward, A.B. Structure of the mechanically activated ion channel Piezo1. Nature 2017, 554, 481–486. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, H.; Li, X.; Xiao, B. The mechanosensitive Piezo1 channel: A three-bladed propeller-like structure and a lever-like mechanogating mechanism. FEBS J. 2018, 286, 2461–2470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Zhou, H.; Chi, S.; Wang, Y.; Wang, J.; Geng, J.; Wu, K.; Liu, W.; Zhang, T.; Dong, M.-Q.; et al. Structure and mechanogating mechanism of the Piezo1 channel. Nature 2018, 554, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Coste, B.; Murthy, S.; Mathur, J.; Schmidt, M.; Mechioukhi, Y.; Delmas, P.; Patapoutian, A. Piezo1 ion channel pore properties are dictated by C-terminal region. Nat. Commun. 2015, 6, 7223. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Lansman, J.B. Calcium entry through stretch-in-activated ion channels in MDX myotubes. Nature 1990, 344, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Bönnemann, C.G.; Muntoni, F. Muscular dystrophies. Lancet 2019, 394, 2025–2038. [Google Scholar] [CrossRef]
- Collins, C.A.; Zammit, P.S. Isolation and Grafting of Single Muscle Fibres. In Stem Cells in Regenerative Medicine; Humana Press: Totowa, NJ, USA, 2009; Volume 482, pp. 319–330. [Google Scholar] [CrossRef]
- Lacroix, J.J.; Botello-Smith, W.M.; Luo, Y. Probing the gating mechanism of the mechanosensitive channel Piezo1 with the small molecule Yoda1. Nat. Commun. 2018, 9, 2029. [Google Scholar] [CrossRef] [Green Version]
- Berchtold, M.W.; Brinkmeier, H.; Müntener, M. Calcium Ion in Skeletal Muscle: Its Crucial Role for Muscle Function, Plasticity, and Disease. Physiol. Rev. 2000, 80, 1215–1265. [Google Scholar] [CrossRef] [Green Version]
- Gehlert, S.; Bloch, W.; Suhr, F. Ca2+-Dependent Regulations and Signaling in Skeletal Muscle: From Electro-Mechanical Coupling to Adaptation. Int. J. Mol. Sci. 2015, 16, 1066–1095. [Google Scholar] [CrossRef]
- Allen, D.G.; Lamb, G.D.; Westerblad, H. Skeletal Muscle Fatigue: Cellular Mechanisms. Physiol. Rev. 2008, 88, 287–332. [Google Scholar] [CrossRef] [Green Version]
- Nourse, J.L.; Pathak, M.M. How cells channel their stress: Interplay between Piezo1 and the cytoskeleton. Semin. Cell Dev. Biol. 2017, 71, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, T.; Mochizuki, T.; Nakagomi, H.; Kira, S.; Watanabe, M.; Takayama, Y.; Suzuki, Y.; Koizumi, S.; Takeda, M.; Tominaga, M. Functional Role for Piezo1 in Stretch-evoked Ca2+ Influx and ATP Release in Urothelial Cell Cultures. J. Biol. Chem. 2014, 289, 16565–16575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millay, D.P.; O’Rourke, J.R.; Sutherland, L.B.; Bezprozvannaya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 2013, 499, 301–305. [Google Scholar] [CrossRef] [Green Version]
- Albuisson, J.; Murthy, S.; Bandell, M.; Coste, B.; Louis-Dit-Picard, H.; Mathur, J.; Fénéant-Thibault, M.; Tertian, G.; De Jaureguiberry, J.-P.; Syfuss, P.-Y.; et al. Dehydrated hereditary stomatocytosis linked to gain-of-function mutations in mechanically activated PIEZO1 ion channels. Nat. Commun. 2013, 4, 1884. [Google Scholar] [CrossRef]
- McHugh, B.J.; Murdoch, A.; Haslett, C.; Sethi, T. Loss of the Integrin-Activating Transmembrane Protein Fam38A (Piezo1) Promotes a Switch to a Reduced Integrin-Dependent Mode of Cell Migration. PLoS ONE 2012, 7, e40346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Sun, Z.; Zhang, X.; Niu, K.; Wang, Y.; Zheng, J.; Li, H.; Liu, Y. Loss of stretch-activated channels, PIEZOs, accelerates non-small cell lung cancer progression and cell migration. Biosci. Rep. 2019, 39, BSR20181679. [Google Scholar] [CrossRef]
- Gottlieb, P.A.; Bae, C.; Sachs, F. Gating the mechanical channel Piezo1: A comparison between whole-cell and patch recording. Channels 2012, 6, 282–289. [Google Scholar] [CrossRef] [Green Version]
- Friedl, P. Prespecification and plasticity: Shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 2004, 16, 14–23. [Google Scholar] [CrossRef]
- Li, J.; Hou, B.; Tumova, S.; Muraki, K.; Bruns, A.; Ludlow, M.J.; Sedo, A.; Hyman, A.J.; McKeown, L.; Young, R.S.; et al. Piezo1 integration of vascular architecture with physiological force. Nature 2014, 515, 279–282. [Google Scholar] [CrossRef]
- Archer, N.M.; Shmukler, B.E.; Andolfo, I.; Vandorpe, D.H.; Gnanasambandam, R.; Higgins, J.M.; Rivera, A.; Fleming, M.; Sachs, F.; Gottlieb, P.; et al. Hereditary xerocytosis revisited. Am. J. Hematol. 2014, 89, 1142–1146. [Google Scholar] [CrossRef] [Green Version]
- Franco-Obregón, A.; Lansman, J.B. Mechanosensitive ion channels in skeletal muscle from normal and dystrophic mice. J. Physiol. 1994, 481, 299–309. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, M.; Hara, Y.; Okuda, M.; Itoh, K.; Nishioka, R.; Shiomi, A.; Nagao, K.; Mori, M.; Mori, Y.; Ikenouchi, J.; et al. Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nat. Commun. 2018, 9, 2049. [Google Scholar] [CrossRef] [PubMed]
- Ganassi, M.; Badodi, S.; Quiroga, H.P.O.; Zammit, P.S.; Hinits, Y.; Hughes, S.M. Myogenin promotes myocyte fusion to balance fibre number and size. Nat. Commun. 2018, 9, 4232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Cai, M.; Si, Y.; Zhang, J.; Du, S. Knockout of myomaker results in defective myoblast fusion, reduced muscle growth and increased adipocyte infiltration in zebrafish skeletal muscle. Hum. Mol. Genet. 2018, 27, 3542–3554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, Q.; Millay, D.P. Requirement of myomaker-mediated stem cell fusion for skeletal muscle hypertrophy. eLife 2017, 6, e20007. [Google Scholar] [CrossRef] [PubMed]
- Leikina, E.; Gamage, D.G.; Prasad, V.; Goykhberg, J.; Crowe, M.; Diao, J.; Kozlov, M.M.; Chernomordik, L.V.; Millay, D.P. Myomaker and Myomerger Work Independently to Control Distinct Steps of Membrane Remodeling during Myoblast Fusion. Dev. Cell 2018, 46, 767–780.e7. [Google Scholar] [CrossRef] [Green Version]
- Ganassi, M.; Muntoni, F.; Zammit, P.S. Defining and identifying satellite cell-opathies within muscular dystrophies and myopathies. Exp. Cell Res. 2021, 411, 112906. [Google Scholar] [CrossRef]

| Gene | Species | siRNA ID |
|---|---|---|
| scrambled non-targeting siRNA (All Stars Negative Control siRNA) | Mouse | Qiagen, 1027281 |
| Piezo1 | Mouse | Qiagen, S104420409 |
| Piezo1 | Mouse | Qiagen, S104420402 |
| Piezo1 | Mouse | Qiagen, S100814807 |
| Piezo1 | Mouse | Qiagen, S100814821 |
| Gene | Species | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Reference |
|---|---|---|---|---|
| Gapdh | Mouse | TGTGTCCGTGGATCTGA | TTGCTGTTGAAGTCGCAGGAG | Takara Bio, MA050371 |
| Piezo1 | Mouse | CTTTATCATGAAGTGCAGCCGAG | CCAGATGATGGCGATGAGGA | Takara Bio, MA125411 |
| Myomaker | Mouse | CATGCGCCGTGACATTCTG | AAGCATTGTGAAGGTCGATCTCTG | Takara Bio, MA131293 |
| Myomixer | Mouse | GAATCCACCGCAGGCAAA | ACCATCGGGAGCAATGGAAC | Takara Bio, MA101853 |
| Primary Antibody | Dilution | Reference |
|---|---|---|
| Monoclonal mouse—Myogenin | 1:10 | Development Studies Hybridoma Bank (DSHB), F5D-s |
| Monoclonal mouse—MF20 (myosin heavy chain) | 1:10 | Development Studies Hybridoma Bank (DSHB), MF20-s |
| Polyclonal rabbit—Piezo1 | 1:100 | Proteintech, 15939-1-AP |
| Secondary antibody | Dilution | Reference |
| Donkey anti-mouse IgG (H + L), Alexa Fluor® 555 | 1:500 | Life Technologies, A21203 |
| Donkey anti-rabbit IgG (H + L), Alexa Fluor® 488 | 1:500 | Life Technologies, A-21206 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortuste Quiroga, H.P.; Ganassi, M.; Yokoyama, S.; Nakamura, K.; Yamashita, T.; Raimbach, D.; Hagiwara, A.; Harrington, O.; Breach-Teji, J.; Asakura, A.; et al. Fine-Tuning of Piezo1 Expression and Activity Ensures Efficient Myoblast Fusion during Skeletal Myogenesis. Cells 2022, 11, 393. https://doi.org/10.3390/cells11030393
Ortuste Quiroga HP, Ganassi M, Yokoyama S, Nakamura K, Yamashita T, Raimbach D, Hagiwara A, Harrington O, Breach-Teji J, Asakura A, et al. Fine-Tuning of Piezo1 Expression and Activity Ensures Efficient Myoblast Fusion during Skeletal Myogenesis. Cells. 2022; 11(3):393. https://doi.org/10.3390/cells11030393
Chicago/Turabian StyleOrtuste Quiroga, Huascar Pedro, Massimo Ganassi, Shingo Yokoyama, Kodai Nakamura, Tomohiro Yamashita, Daniel Raimbach, Arisa Hagiwara, Oscar Harrington, Jodie Breach-Teji, Atsushi Asakura, and et al. 2022. "Fine-Tuning of Piezo1 Expression and Activity Ensures Efficient Myoblast Fusion during Skeletal Myogenesis" Cells 11, no. 3: 393. https://doi.org/10.3390/cells11030393






