Characterization of Gfat1 (zeppelin) and Gfat2, Essential Paralogous Genes Which Encode the Enzymes That Catalyze the Rate-Limiting Step in the Hexosamine Biosynthetic Pathway in Drosophila melanogaster
Abstract
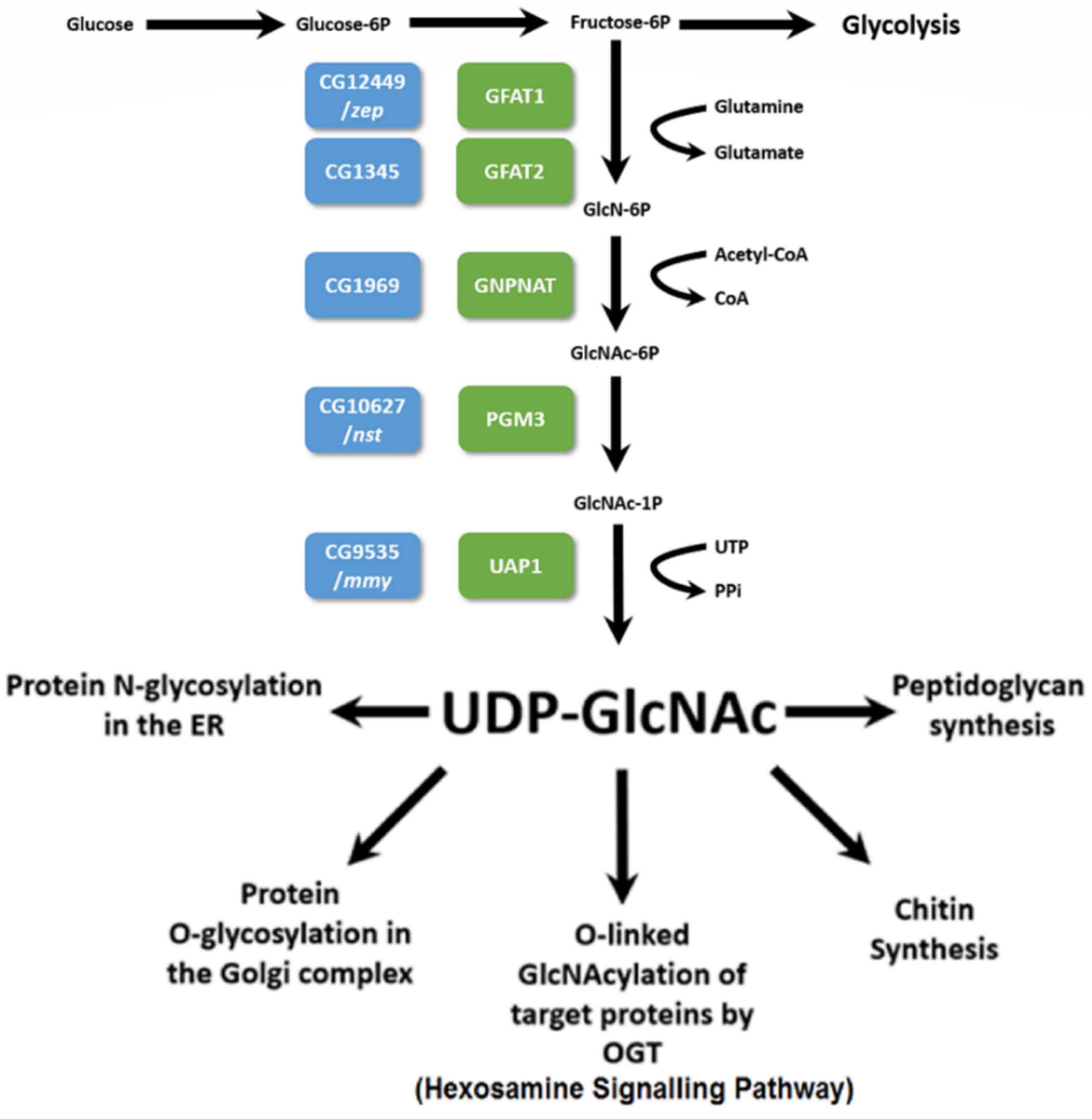
1. Introduction
2. Materials and Methods
2.1. Pre-Existing Drosophila Stocks, Routine Culture Conditions, Identification of New Zep/Gfat1 Alleles and Routine Genetic Crosses
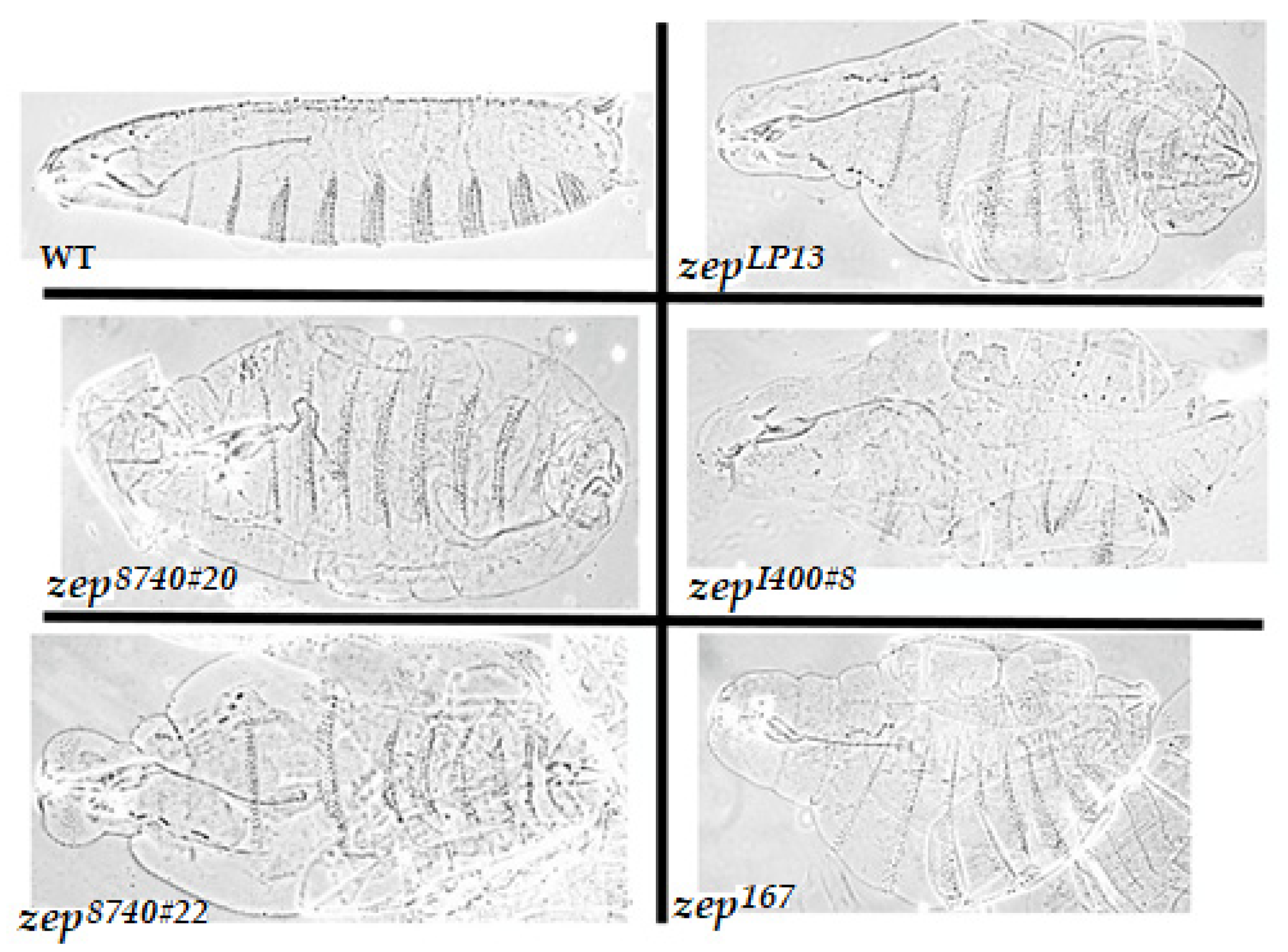
2.2. Cuticle Analysis of Newly-Isolated Zep Alleles
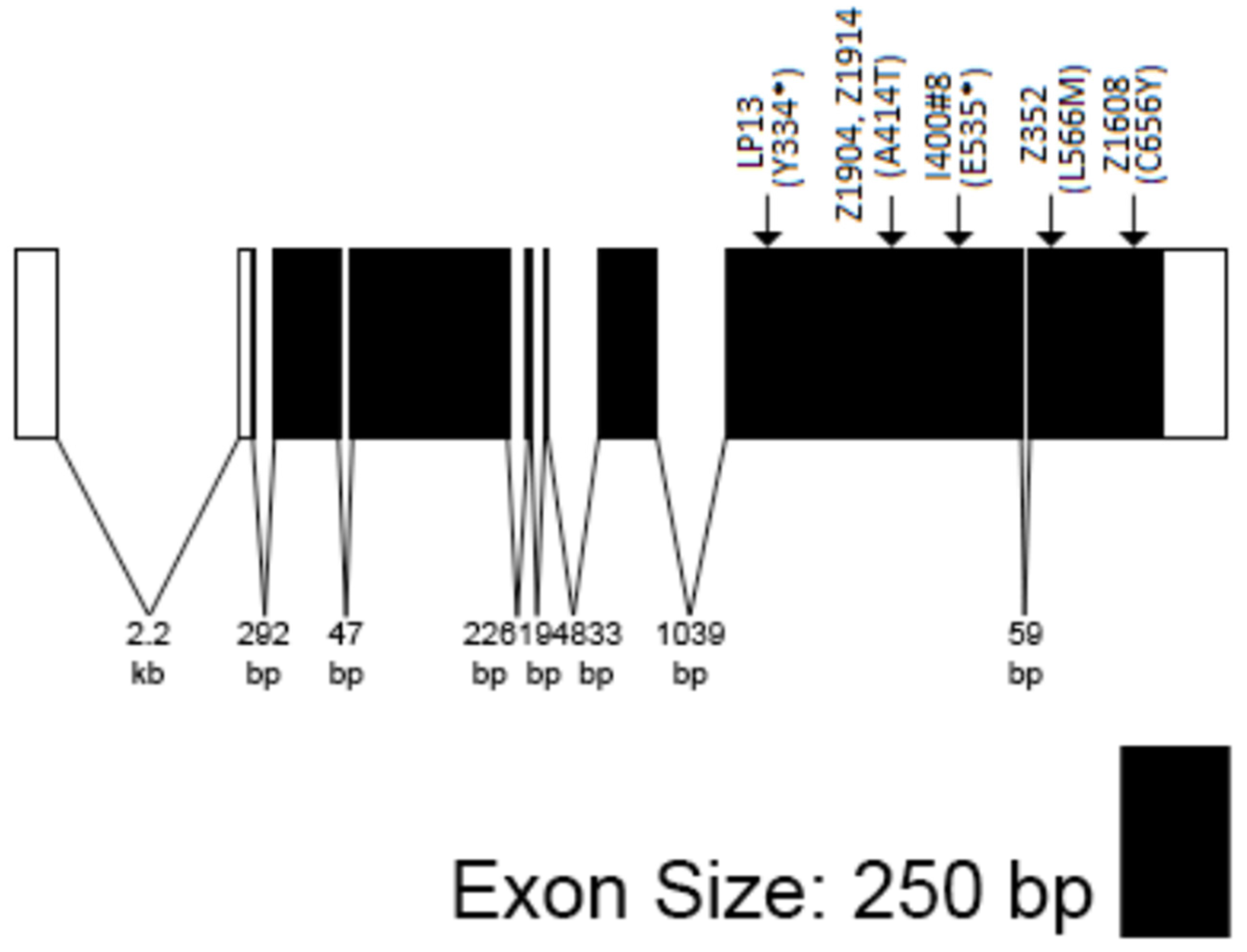
2.3. Sequence Analysis of Zep Alleles
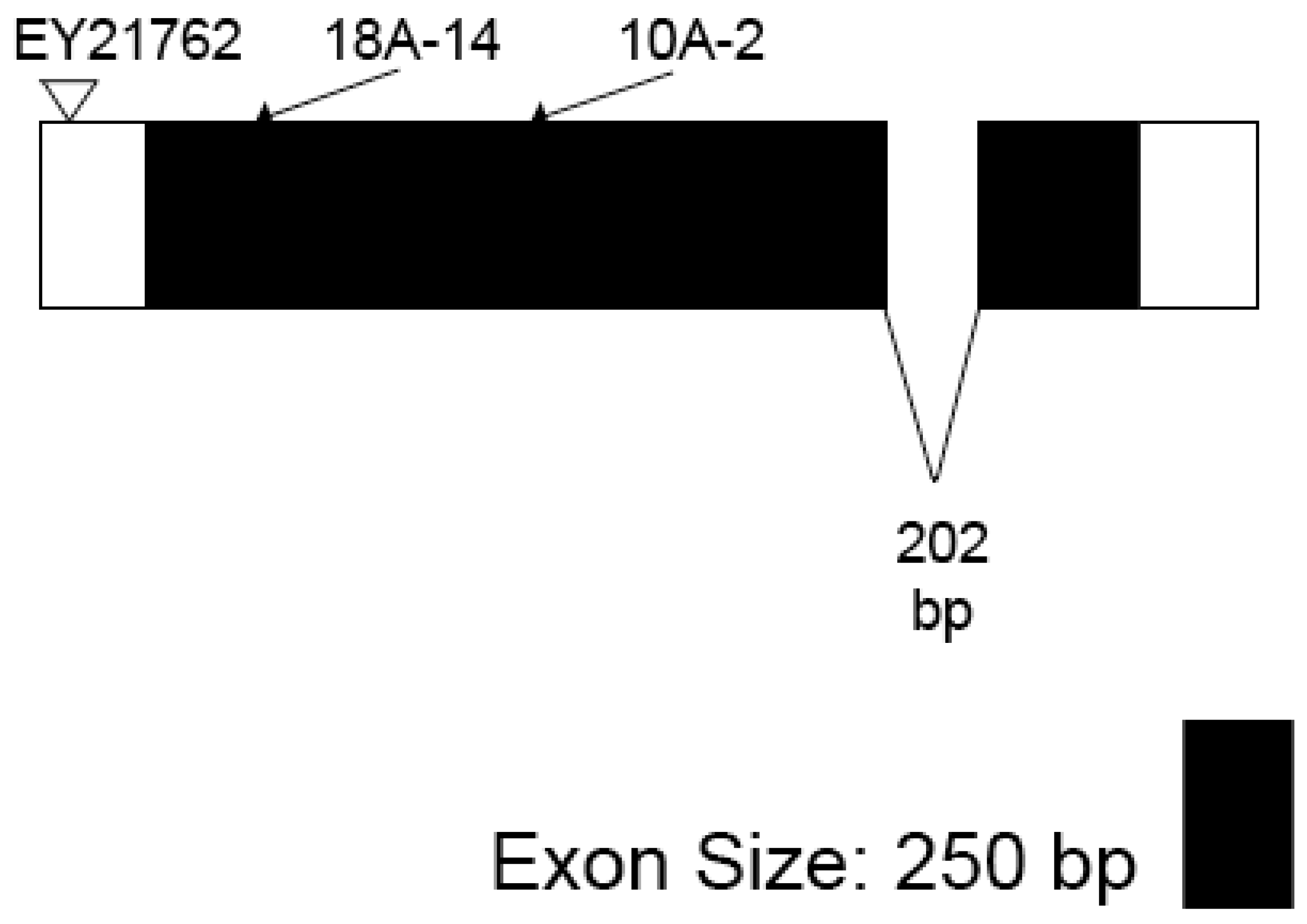
2.4. Isolation and Genetic and Molecular Characterization of Putative Gfat2 Excision Mutations
2.5. Sequence Analysis of Gfat2 Mutants
2.6. Lethal Phase Analysis of a Gfat2 Deletion Mutant
2.7. Generation of a Gfat1+ Rescue cDNA Construct
2.8. Gfat1 and Gfat2 Rescue Crosses
2.9. Gfat1 RNAi Stocks and Crosses
2.10. Real-Time qPCR
2.11. Tests for Essential Tissue-Specific Requirements for Gfat1 and Gfat2
2.12. Fluorescence In Situ Hybridization (FISH) Localization of Gfat1 and Gfat2 in Different Species of Drosophila
3. Results
3.1. The Zep Locus Corresponds to Gfat1
3.2. Isolation, Sequence and Lethal Phase Analysis of Gfat2 Mutant Alleles
3.3. Gfat1/Zep and Gfat2 Mutants Can Be Rescued by Ubiquitous Expression of Gfat1+ and Gfat2+ cDNA Transgenes
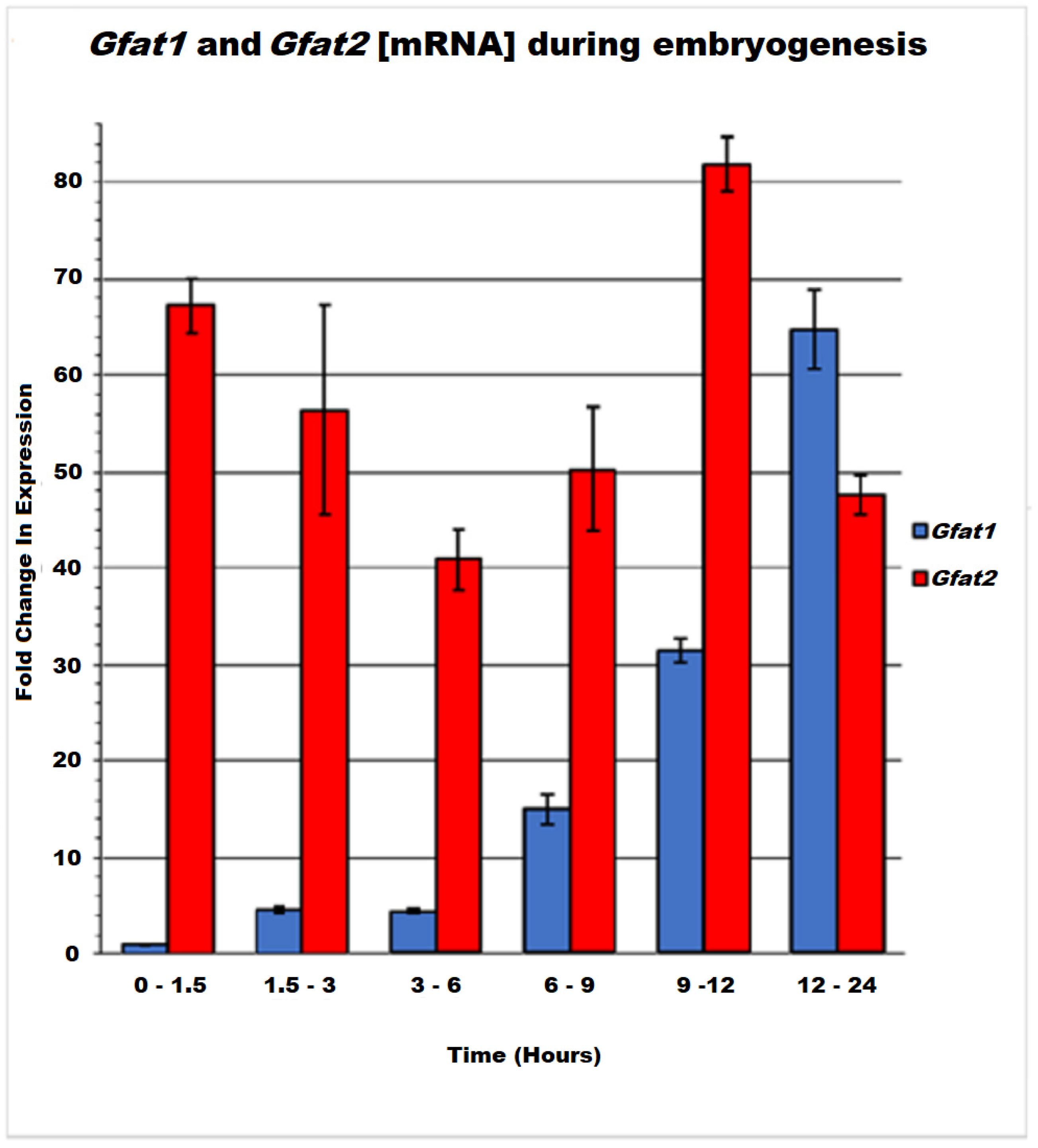
3.4. Gfat1 and Gfat2 Genes Exhibit Different Expression Patterns during Development
3.5. Evidence for Differences in Essential Tissue-Specific Requirements for Gfat1 and Gfat2
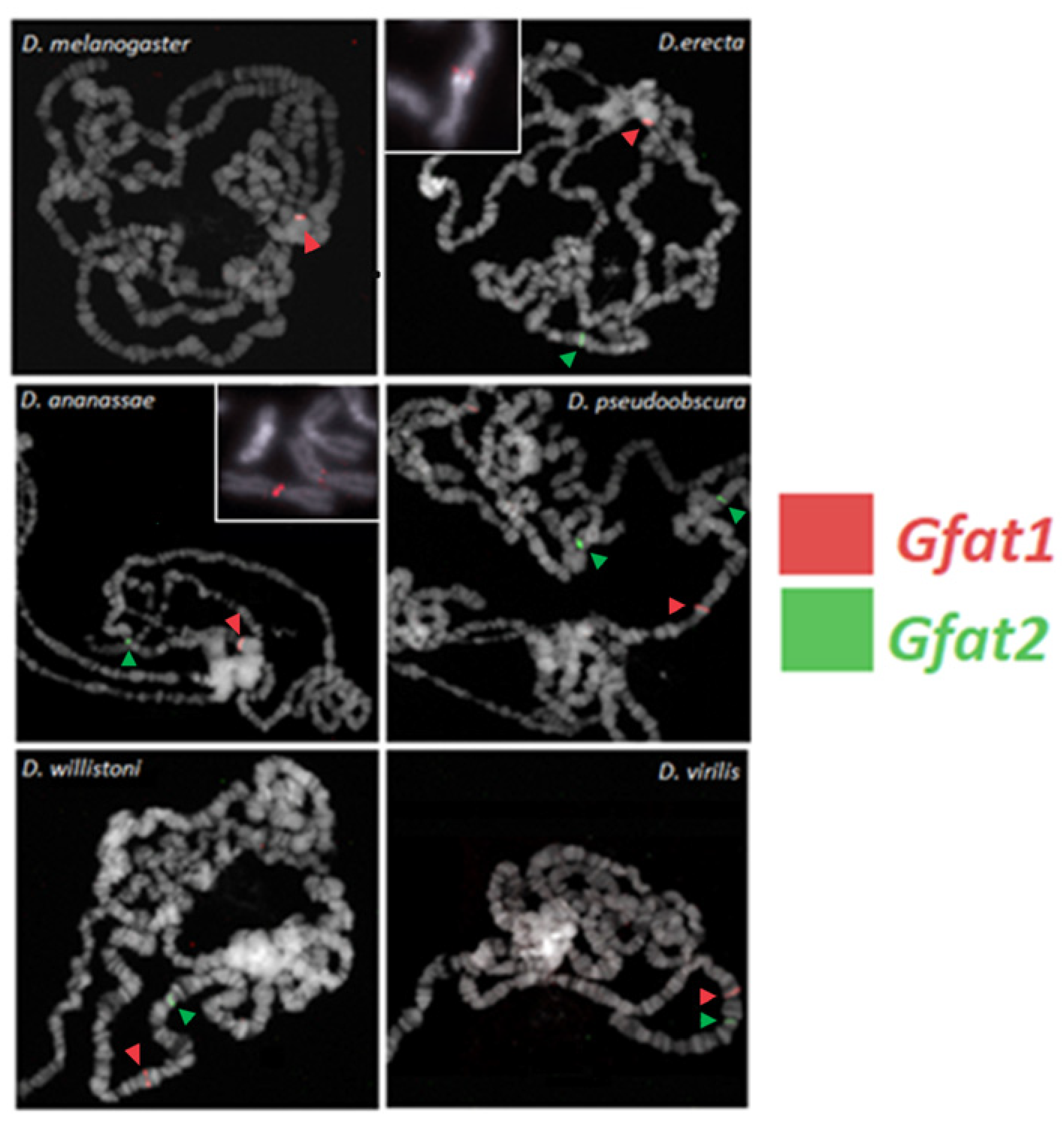
3.6. FISH Localization of the Gfat1 and Gfat2 Genes in Six Different Drosophila Species
3.7. Micro-Synteny of Gfat1 and Gfat2 and Flanking Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Darley-Usmar, M.V.; Ball, L.E.; Chatham, J.C. Protein O-linked β-N-acetylglucosamine: A novel effector of cardiomyocyte metabolism and function. J. Mol. Cell. Cardiol. 2012, 52, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Mattila, J.; Hietakangas, V. Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 2017, 207, 1231–1253. [Google Scholar] [CrossRef] [PubMed]
- Chiaradonna, F.; Ricciardiello, F.; Palorini, R. The nutrient-sensing hexosamine biosynthetic pathway as the hub of cancer metabolic rewiring. Cells 2018, 7, 53. [Google Scholar] [CrossRef]
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019, 17, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Graack, H.-R.; Cinque, U.; Kress, H. Functional regulation of glutamine:fructose-6-phosphate aminotransferase 1 (Gfat1) of Drosophila melanogaster in a UDP-N-acetylglucosamine and cAMP manner. Biochem. J. 2001, 360, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Denzel, M.S.; Antebi, A. Hexosamine pathway and (ER) protein quality control. Curr. Opin. Cell Biol. 2015, 33, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Oki, T.; Yamazaki, K.; Kuromitsu, J.; Okada, M.; Tanaka, I. cDNA cloning and mapping of a novel subtype of Glutamine:fructose-6-phosphate amidotransferase (Gfat2) in human and mouse. Genomics 1999, 57, 227–234. [Google Scholar] [CrossRef]
- Kato, N.; Dasgupta, R.; Smartt, C.T.; Christensen, B.M. Glucosamine:fructose-6-phosphate aminotransferase: Gene characterization, chitin biosynthesis and peritrophic matrix formation in Aedes aegypti. Insect Mol. Biol. 2002, 11, 207–216. [Google Scholar] [CrossRef]
- Smith, R.J.; Milewski, S.; Brown, A.J.P.; Gooday, G.W. Isolation and characterization of the GFA1 gene encoding glutamine:fructose-6-phosphate amidotransferase of Candida albicans. J. Bacteriol. 1996, 178, 2320–2327. [Google Scholar] [CrossRef][Green Version]
- Ram, A.F.J.; Arentshorst, M.; Damveld, R.A.; vanKuyk, P.A.; Klis, F.M.; van den Hondel, C.A. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine: Fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology 2004, 150, 3315–3326. [Google Scholar] [CrossRef]
- Luo, C.; Shao, W.; Li, X.; Chen, Z.; Liu, Y. Molecular cloning, sequencing and expression of a l-glutamine d-fructose 6-phosphate amidotransferase gene from Volvariella volvacea. Protein J. 2009, 28, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Huynh, Q.K.; Hoffman, R.T.; Crook, E.D.; Daniels, M.C.; Gulve, E.A.; McClain, D.A. Regulation of glutamine:fructose-6-phosphate amidotransferase by cAMP-dependent protein kinase. Diabetes 1998, 47, 1836–1840. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Riesland, L.; Paterson, A.J.; Kudlow, J.E. Phosphorylation of mouse glutamine:fructose-6-phosphate amidotransferase 2 (GFAT2) by cAMP-dependent protein kinase increases the enzyme activity. J. Biol. Chem. 2004, 279, 29988–29993. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Tsuji, N.; Miyoshi, T.; Motobu, M.; Islam, M.K.; Alim, M.A.; Fujisaki, K. Characterization of glutamine:Fructose-6-phosphate amino transferase from the ixodid tick, Haemaphysalis longicornis, and its critical role in host blood feeding. Int. J. Parasitol. 2007, 37, 383–392. [Google Scholar] [CrossRef]
- Denzel, M.S.; Storm, N.J.; Gutschmidt, A.; Baddi, R.; Hinze, Y.; Jarosch, E.; Sommer, T.; Hoppe, T.; Antebi, A. Hexosamine pathway metabolites enhance protein quality and prolong life. Cell 2014, 156, 1167–1178. [Google Scholar] [CrossRef]
- Wang, Z.V.; Deng, Y.; Gao, N.; Pedrozo, Z.; Li, D.L.; Morales, C.R.; Criollo, A.; Luo, X.; Tan, W.; Jiang, N. Spliced-box protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell 2014, 156, 1179–1192. [Google Scholar] [CrossRef]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell. Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef]
- Hanover, J.A.; Chen, W.; Bond, M.R. O-GlcNAc in cancer: An Oncometabolism-fueled vicious cycle. J. Bioenerg. Biomembr. 2018, 50, 155–173. [Google Scholar] [CrossRef]
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231. [Google Scholar] [CrossRef]
- Parker, M.P.; Peterson, K.R.; Slawson, C. O-GlcNAcylation and O-GlcNAc cycling regulate gene transcription: Emerging roles in cancer. Cancers 2021, 13, 1666. [Google Scholar] [CrossRef]
- Sun, L.; Lv, S.; Song, T. 2021 O-GlcNAcylation links oncogenic signals and cancer epigenetics. Discov. Oncol. 2021, 12, 54. [Google Scholar] [CrossRef]
- Moussian, B.; Schwarz, H.; Bartoszewski, S.; Nusslein-Volhard, C. Involvement of chitin in exoskeleton morphogenesis in Drosophila melanogaster. J. Morphol. 2005, 264, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Tonning, A.; Helms, S.; Schwarz, H.; Uv, A.E.; Moussian, B. Hormonal regulation of mummy is needed for extracellular formation and epithelial morphogenesis in Drosophila. Development 2006, 133, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, S.; Dierick, H.A.; Bejsovec, A. Genetic control of cuticle formation during embryonic development of Drosophila melanogaster. Genetics 2002, 161, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Syrzycka, M.; Hallson, G.; Fitzpatrick, K.A.; Kim, I.; Cotsworth, S.; Hollebakken, R.E.; Simonetto, K.; Yang, L.; Luongo, S.; Beja, K.; et al. Genetic and molecular analysis of essential genes in centromeric heterochromatin of the left arm of chromosome 3 in Drosophila melanogaster. Genes Genomics Genet. 2019, 7, 1581–1595. [Google Scholar] [CrossRef]
- Coulthard, A.B.; Alm, C.; Cealiac, I.; Sinclair, D.A.; Honda, B.M.; Rossi, F.; Dimitri, P.; Hilliker, A.J. Essential loci in centromeric heterochromatin of Drosophila melanogaster. I: The right arm of chromosome 2. Genetics 2010, 185, 479–495. [Google Scholar] [CrossRef]
- Yasuhara, J.C.; DeCrease, C.H.; Wakimoto, B.T. Evolution of heterochromatic genes of Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 10959–10963. [Google Scholar] [CrossRef]
- Thurmond, J.; Goodman, J.L.; Strelets, V.B.; Attrill, H.; Gramates, L.S.; Marygold, S.J.; Matthews, B.B.; Millburn, G.; Antonazzo, G.; Trovisco, V.; et al. FlyBase 2.0: The next generation. Nucleic Acids Res. 2019, 47, D759–D765. [Google Scholar] [CrossRef] [PubMed]
- Koundakjian, E.J.; Cowana, D.M.; Hardy, R.W.; Beckera, A.H. The Zuker Collection: A Resource for the Analysis of Autosomal Gene Function in Drosophila melanogaster. Genetics 2004, 167, 203–206. [Google Scholar] [CrossRef]
- Irion, U.; Leptin, M. Developmental and cell biological functions of the Drosophila DEAD-box protein abstrakt. Curr. Biol. 1999, 9, 1373–1381. [Google Scholar] [CrossRef]
- Marchant, G.E.; Holm, D.G. Genetic analysis of the heterochromatin of chromosome 3 in Drosophila melanogaster. I. Products of compound autosome detachment. Genetics 1988, 120, 503–517. [Google Scholar] [CrossRef]
- Marchant, G.E.; Holm, D.G. Genetic analysis of the heterochromatin of chromosome 3 in Drosophila melanogaster. II. Vital loci identified through EMS mutagenesis. Genetics 1988, 120, 519–532. [Google Scholar] [CrossRef]
- Koryakov, D.E.; Zhimulev, I.F.; Dimitri, P. Cytogenetic analysis of the third chromosome heterochromatin of Drosophila melanogaster. Genetics 2002, 160, 509–517. [Google Scholar] [CrossRef]
- Koryakov, D.E.; Domanitskaya, E.V.; Belyakin, S.N.; Zhimulev, E.F. Abnormal tissue-dependent polytenization of chromosome 3 pericentric heterochromatin in Drosophila melanogaster. J. Cell Sci. 2003, 116, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, K.A. Genetic and Molecular Characterization of Chromosome 3 Heterochromatin in Drosophila melanogaster. Ph.D.Thesis, W.A.C. Bennett Library QH 599 F584 2005, Simon Fraser University, Burnaby, BC, Canada, 2005. [Google Scholar]
- Jackson, C. Gfat1/Zeppelin is an Essential Gene Involved in Cuticle Formation in D. melanogaster. Master’s Thesis, W.A.C. Bennett Library QH 470 D7 J32, Simon Fraser University, Burnaby, BC, Canada, 2007. [Google Scholar]
- Stapleton, M.; Carlson, J.; Brokstein, P.; Yu, C.; Champe, M.; George, R.; Guarin, H.; Kronmiller, B.; Pacleb, J.; Park, S.; et al. A Drosophila full-length cDNA resource. Genome Biol. 2002, 3, 1–8. [Google Scholar] [CrossRef]
- Niccoli, T.; Cabecinha, M.; Tillmann, A.; Kerr, F.; Wong, C.T.; Cardenes, A.; Vincent, A.J.; Bettedi, L.; Li, L.; Grönke, S.; et al. Increased glucose transport into neurons rescues Abeta toxicity in Drosophila. Curr. Biol. 2016, 26, 2550. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 1, e45. [Google Scholar] [CrossRef] [PubMed]
- Pimpinelli, S.; Bonaccorsi, S.; Fanti, L.; Gatti, M. Preparation and Analysis of Drosophila mitotic Chromosomes, in Drosophila Protocols; Sullivan, W., Ashburner, M., Hawley, R.S., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2000; pp. 3–23. [Google Scholar]
- Fitzpatrick, K.A.; Sinclair, D.A.; Schulze, S.R.; Syrzycka, M.; Honda, B.M. A genetic and molecular profile of chromosome 3 centric heterochromatin in Drosophila melanogaster. Genome 2005, 48, 571–584. [Google Scholar] [CrossRef]
- Lindsley, D.L.; Sandler, L.; Baker, B.S.; Carpenter, A.T.C.; Denell, R.E.; Hall, J.C.; Jacobs, P.A.; Miklos, G.L.; Davis, B.K.; Gethmann, R.C.; et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 1972, 71, 157–184. [Google Scholar] [CrossRef]
- Tasaka, S.E.; Suzuki, D.T. Temperature-sensitive mutations in Drosophila melanogaster. XVII. Heat- and cold-sensitive lethals on chromosome 3. Genetics 1973, 74, 509–520. [Google Scholar] [CrossRef]
- Cotsworth, S.E.P. Gfat1 and Gfat2 Encode Functionally Equivalent Enzymes in Drosophila melanogaster: A Molecular, Genetic and Evolutionary Analysis. Master’s Thesis, W.A.C. Bennett Library Simon Fraser University, Burnaby, BC, Canada, 2018. [Google Scholar]
- Fisher, B.; Weiszmann, R.; Frise, E.; Hammonds, A.; Tomancak, P.; Beaton, A.; Berman, B.; Quan, E.; Shu, S.; Lewis, S.; et al. Patterns of Gene Expression in Drosophila embryogenesis. BDGP in Situ Homepage. 2012. Available online: https://insitu.fruitfly.org/cgi-bin/ex/insitu.pl (accessed on 20 January 2022).
- Liu, X.; Blaženović, I.; Contreras, A.J.; Pham, T.M.; Tabuloc, C.A.; Li, Y.H.; Ji, J.; Fiehn, O.; Chiu, J.C. Hexosamine biosynthetic pathway and O-GlcNAc-processing enzymes regulate daily rhythms in protein O-GlcNAcylation. Nat. Commun. 2021, 12, 4173. [Google Scholar] [CrossRef]
- Casas-Vila, N.; Bluhm, A.; Sayols, S.; Dinges, N.; Dejung, M.; Altenhein, T.; Kappei, D.; Altenhein, B.; Roignant, J.-Y.; Butter, F. The developmental proteome of Drosophila melanogaster. Genome Res. 2017, 27, 1273–1285. [Google Scholar] [CrossRef] [PubMed]
- Araujo, S.J.; Aslam, H.; Tear, G.; Casanova, J. Mummy/cystic encodes an enzyme required for chitin and glycan synthesis, involved in trachea, embryonic cuticle and CNS development—Analysis of its role in Drosophila morphogenesis. Dev. Biol. 2005, 288, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Schimmelpfeng, K.; Strunk, M.; Stork, T.; Klambt, C. mummy encodes an UDP-N-acetylglucosamine-diphosphorylase and is required during dorsal closure and nervous system development. Mech. Dev. 2006, 123, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, M.A.; Jayasinghe, V.R.; Grewal, R.; Bhat, K.M. The glycosylation pathway is required for the secretion of Slit receptor Robo on axons. Sci. Signal. 2017. [CrossRef]
- Sinclair, D.A.R.; Syrzycka, M.; Macauley, M.S.; Rastgardani, T.; Komljenovic, I.; Vocadlo, D.J.; Brock, H.W.; Honda, B.M. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. USA 2009, 106, 13427–13432. [Google Scholar] [CrossRef]
- Ozturk-Colak, A.; Moussian, B.; Araujo, S. Drosophila chitinous aECM and its cellular interactions during embryogenesis. Dev. Dyn. 2016, 245, 259–267. [Google Scholar] [CrossRef]
- Devine, W.P.; Lubarsky, B.; Shaw, K.; Luschnig, S.; Messina, L.; Krasnow, M.A. Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc. Nat. Acad. Sci. USA 2005, 102, 17014–17019. [Google Scholar] [CrossRef]
- Chen, P.; Visokay, S.; Abrams, J.M. Drosophila GFAT1 and GFAT2 enzymes encode obligate developmental functions. Fly 2020, 14, 9. [Google Scholar] [CrossRef]
- Mattila, J.; Kokki, K.; Hietakangas, V.; Boutros, M. Stem cell intrinsic hexosamine metabolism regulates intestinal adaptation to nutrient content. Dev. Cell 2018, 47, 112–121. [Google Scholar] [CrossRef]
- Oliveira, I.A.; Allonso, D.; Fernandes, T.V.A.; Lucena, D.M.S.; Ventura, G.T.; Dias, W.B.; Mohana-Borges, R.S.; Pascutti, P.G.; Todeschini, A.R. Enzymatic and structural properties of human glutamine:fructose-6-phosphate amidotransferase 2 (hGFAT2). J. Biol. Chem. 2020, 2020, 17. [Google Scholar] [CrossRef] [PubMed]
- Nabeebaccus, A.A.; Verma, S.; Zoccarato, A.; Emanuelli, G.; Santos, C.X.C.; Streckfuss-Bömeke, K.; Shah, A.M. Cardiomyocyte protein O-GlcNAcylation is regulated by GFAT1 not GFAT2. Biochem. Biophys. Res. Commun. 2021, 583, 121–127. [Google Scholar] [CrossRef]
- Chang, Q.; Su, K.; Baker, R.J.; Yang, X.; Paterson, A.J.; Kudlow, J.E. Phosphorylation of human glutamine:fructose-6-phosphate amidotransferase by cAMP-dependent protein kinase at Serine 205 blocks the enzyme activity. J. Biol. Chem. 2000, 275, 21981–21987. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, S.; Oshiro, N.; Miyamoto, T.; Yoshino, K.; Okamoto, S.; Ono, T.; Kikkawa, U.; Yonezawa, K. AMP-activated protein kinase phosphorylates glutamine: Fructose-6-phosphate amidotransferase 1 at Ser243 to modulate enzymatic activity. Genes Cells 2009, 14, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Schulze, S.R.; McAllister, B.F.; Sinclair, D.A.R.; Fitzpatrick, K.A.; Marchetti, M.; Pimpinelli, S.; Honda, B.M. Heterochromatic genes in Drosophila: A comparative analysis of two genes. Genetics 2006, 173, 1433–1445. [Google Scholar] [CrossRef][Green Version]
- Kulathinal, R.J.; Hartl, D.L. The latest buzz in comparative genomics. Genome Biol. 2005, 6, 201–203. [Google Scholar] [CrossRef][Green Version]
- Assis, R. Drosophila duplicate genes evolve new functions on the fly. Fly 2014, 8, 91–94. [Google Scholar] [CrossRef]
- Vilinsky, I.; Stewart, B.A.; Drummond, J.; Robinson, I.; Deitcher, D.L. A Drosophila SNAP-25 null mutant reveals context-dependent redundancy with SNAP-24 in neurotransmission. Genetics 2002, 162, 259–271. [Google Scholar] [CrossRef]
- Ono, H.; Rewitz, K.F.; Shinoda, T.; Itoyama, K.; Petryk, A.; Rybczynski, R.; Jarcho, M.; Warren, J.T.; Marqués, G.; Shimell, M.J.; et al. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 2006, 298, 555–570. [Google Scholar] [CrossRef]
- Syrzycka, M. Genetic and Molecular Characterization of Heterochromatic Genes in Drosophila melanogaster. Ph.D. Thesis, W.A.C. Bennett Library QH 5, Simon Fraser University, Burnaby, BC, Canada, 2009. [Google Scholar]
- De Renzis, S.; Elemento, O.; Tavazoie, S.; Wieschaus, E.F. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007, 5, e117. [Google Scholar] [CrossRef]
- Artieri, C.G.; Fraser, H.B. Transcript length mediates developmental timing expression across Drosophila. Mol. Biol. Evol. 2014, 31, 2879–2889. [Google Scholar] [CrossRef] [PubMed]

| Cross | Gfat210A2/Df(3R)BSC460 Offspring | ||
|---|---|---|---|
| Unhatched Embryos | First Instar Larvae | Second Instar Larvae | |
| Gfat210A2/TM3, Sb, Ser, Twi-GFP x Df(3R)BSC460/ TM3, Sb, Ser, Twi-GFP | 59 | 41 | 0 |
| Number of Relevant Progeny | |||||
|---|---|---|---|---|---|
| Mutant/TM3 or TM6B | Mutant/Deficiency or Mutant/Mutant | ||||
| Cross | Total | CyO/Gfat1+ or Gfat2+ | Act5C-GAL4/Gfat1+ or Gfat2+ | Cy/Gfat1+ or Gfat2+ | Act5C-GAL4/Gfat1+ or Gfat2+ |
| UAS-Gfat1+/CyRoi; Gfat1LP13/TM3, Sb | |||||
| X | 310 | 139 | 120 | 0 | 51 |
| Actin5C-GAL4/CyO; Df(3R)7B-90, e/TM3, Sb | |||||
| UAS-Gfat1+/CyRoi; Gfat210A−2/TM3 Ser | |||||
| X | 179 | 115 | 49 | 0 | 15 |
| Actin5C-GAL4/CyO; Df(3R)BSC567/TM3, Ser | |||||
| UAS-Gfat2+/CyRoi; Df(3R)BSC460/TM6B | |||||
| X | 177 | 95 | 50 | 0 | 32 |
| Actin5C-GAL4/CyO; Gfat210A−2/TM3, Ser | |||||
| UAS-Gfat2+/CyRoi; Gfat1z−1904/TM6B | |||||
| X | 156 | 84 | 54 | 0 | 18 |
| Actin5C-GAL4/CyO; Gfat1I400#8/TM3, Ser | |||||
| RNAi | Number of Progeny | Total | Comments on RNAi | |
|---|---|---|---|---|
| Gfat RNAi | Control (Balancer) | |||
| Gfat2/CyO (v105129) | 123 | 159 | 282 | male semi-lethality ** frequent unfurled wings |
| Gfat2/TM3, Sb (B34740) | 0 | 218 | 218 | lethal |
| Gfat1/TM3, Sb (HL) | 175 | 111 | 286 | viable |
| Gfat1/CyO (B42892) | 258 | 260 | 518 | viable |
| Gfat1 (v24539) | 539 | - | 539 | viable |
| Ogt/CyO (B50909) | 0 | 275 | 275 | lethal |
| RNAi | Number of Progeny | Total | Comments on RNAi | |
|---|---|---|---|---|
| Gfat RNAi | Control (Balancer) | |||
| Gfat2/CyO (v105129) | 0 | 148 | 148 | lethal |
| Gfat2/TM3, Sb (B34740) | 0 | 60 | 60 | lethal |
| Gfat1/TM3, Sb (HL) | 227 | 165 | 392 | viable |
| Gfat1/CyO (B42892) | 250 | 252 | 502 | viable |
| Gfat1 (v24539) | 60 | - | 60 | viable |
| Ogt/CyO (B50909) | 0 | 240 | 240 | lethal |
| Control or RNAi | Number of Progeny | Total | Comments on RNAi | |
|---|---|---|---|---|
| Gfat RNAi | Control (Balancer) | |||
| w1118 control | 92 | 116 | 208 | NA |
| Gfat2/CyO (v105129) | 40 | 132 | 172 | weak semi-lethality ** |
| Gfat2/TM3, Sb (B34740) | 0 | 269 | 269 | lethal |
| Gfat1/CyO (B42892) | 27 | 89 | 116 | weak semi-lethality ** |
| Gfat1 (v24539) | 0 | 202 | 202 | lethal |
| Species and Designation of Gfat1 Gene | Sequence Homology of Orthologous Gfat1 Proteins Versus D. m. Gfat1 * | CG42402 or Ortholog 5′ to Gfat1 ** | 5′ to 3′ Orientation Relative to That of Gfat1 | CG40198 or Ortholog 3′ to Gfat1 | 5′ to 3′ Orientation Relative to That of Gfat1 |
|---|---|---|---|---|---|
| melanogaster CG12449 | - | yes (22) | same | yes (23) | same |
| erecta GG12143 | 98/98 | yes (31) | same | yes (3) | opposite |
| ananassae GF23135 | 95/97 | yes (35) | same | yes (12) | same |
| pseudoobscura GA26267 | 97/98 | yes (35) | same | yes (6) | opposite |
| willistoni GK12920 | 96/98 | yes (38) | same | yes (8) | opposite |
| virilis GJ24380 | 96/98 | yes (28) | same | yes (28) | opposite |
| Species and Designation of Gfat2 Gene | Sequence Homology of Orthologous Gfat2 Protein Versus D. m. Gfat2 * | Moca-cyp or Ortholog 5′ to Gfat2 ** | 5′ to 3′ Orientation Relative to That of Gfat2 | larp or Ortholog 3′ to Gfat2 | 5′ to 3′ Orientation Relative to That of Gfat2 |
|---|---|---|---|---|---|
| melanogaster CG1345 | - | yes (0.5) | opposite | yes (0.6) | same |
| erecta GG12070 | 98/99 | yes (0.4) | opposite | yes (1.3) | same |
| ananassae GF16128 | 95/97 | yes (0.9) | opposite | yes (0.7) | same |
| pseudoobscura GA12297 | 92/95 | yes (2) | opposite | yes (6) | same |
| willistoni GK12142 | 92/95 | located 3′ (0.3) | same | 5′ gene: GK12141 (3) | same |
| virilis GJ22773 | 91/94 | located 3′ (0.4) | same | located 5′ (0.3) | opposite |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cotsworth, S.; Jackson, C.J.; Hallson, G.; Fitzpatrick, K.A.; Syrzycka, M.; Coulthard, A.B.; Bejsovec, A.; Marchetti, M.; Pimpinelli, S.; Wang, S.J.H.; et al. Characterization of Gfat1 (zeppelin) and Gfat2, Essential Paralogous Genes Which Encode the Enzymes That Catalyze the Rate-Limiting Step in the Hexosamine Biosynthetic Pathway in Drosophila melanogaster. Cells 2022, 11, 448. https://doi.org/10.3390/cells11030448
Cotsworth S, Jackson CJ, Hallson G, Fitzpatrick KA, Syrzycka M, Coulthard AB, Bejsovec A, Marchetti M, Pimpinelli S, Wang SJH, et al. Characterization of Gfat1 (zeppelin) and Gfat2, Essential Paralogous Genes Which Encode the Enzymes That Catalyze the Rate-Limiting Step in the Hexosamine Biosynthetic Pathway in Drosophila melanogaster. Cells. 2022; 11(3):448. https://doi.org/10.3390/cells11030448
Chicago/Turabian StyleCotsworth, Shawn, Catherine J. Jackson, Graham Hallson, Kathleen A. Fitzpatrick, Monika Syrzycka, Alistair B. Coulthard, Amy Bejsovec, Marcella Marchetti, Sergio Pimpinelli, Simon J. H. Wang, and et al. 2022. "Characterization of Gfat1 (zeppelin) and Gfat2, Essential Paralogous Genes Which Encode the Enzymes That Catalyze the Rate-Limiting Step in the Hexosamine Biosynthetic Pathway in Drosophila melanogaster" Cells 11, no. 3: 448. https://doi.org/10.3390/cells11030448
APA StyleCotsworth, S., Jackson, C. J., Hallson, G., Fitzpatrick, K. A., Syrzycka, M., Coulthard, A. B., Bejsovec, A., Marchetti, M., Pimpinelli, S., Wang, S. J. H., Camfield, R. G., Verheyen, E. M., Sinclair, D. A., Honda, B. M., & Hilliker, A. J. (2022). Characterization of Gfat1 (zeppelin) and Gfat2, Essential Paralogous Genes Which Encode the Enzymes That Catalyze the Rate-Limiting Step in the Hexosamine Biosynthetic Pathway in Drosophila melanogaster. Cells, 11(3), 448. https://doi.org/10.3390/cells11030448






