The Effects of Mechanical Loading Variations on the Hypertrophic, Anti-Apoptotic, and Anti-Inflammatory Responses of Differentiated Cardiomyocyte-like H9C2 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. H9C2 Cell Culture
2.2. Cardiomyocyte Mechanical Loading
2.3. Cell Lysis and RNA Extraction
2.4. Reverse Transcription and Real-Time PCR
2.5. Protein Extraction and Immunoblotting Analysis
2.6. Immunofluorescence
2.7. Cell Cycle Analysis
2.8. Statistical Analysis
3. Results
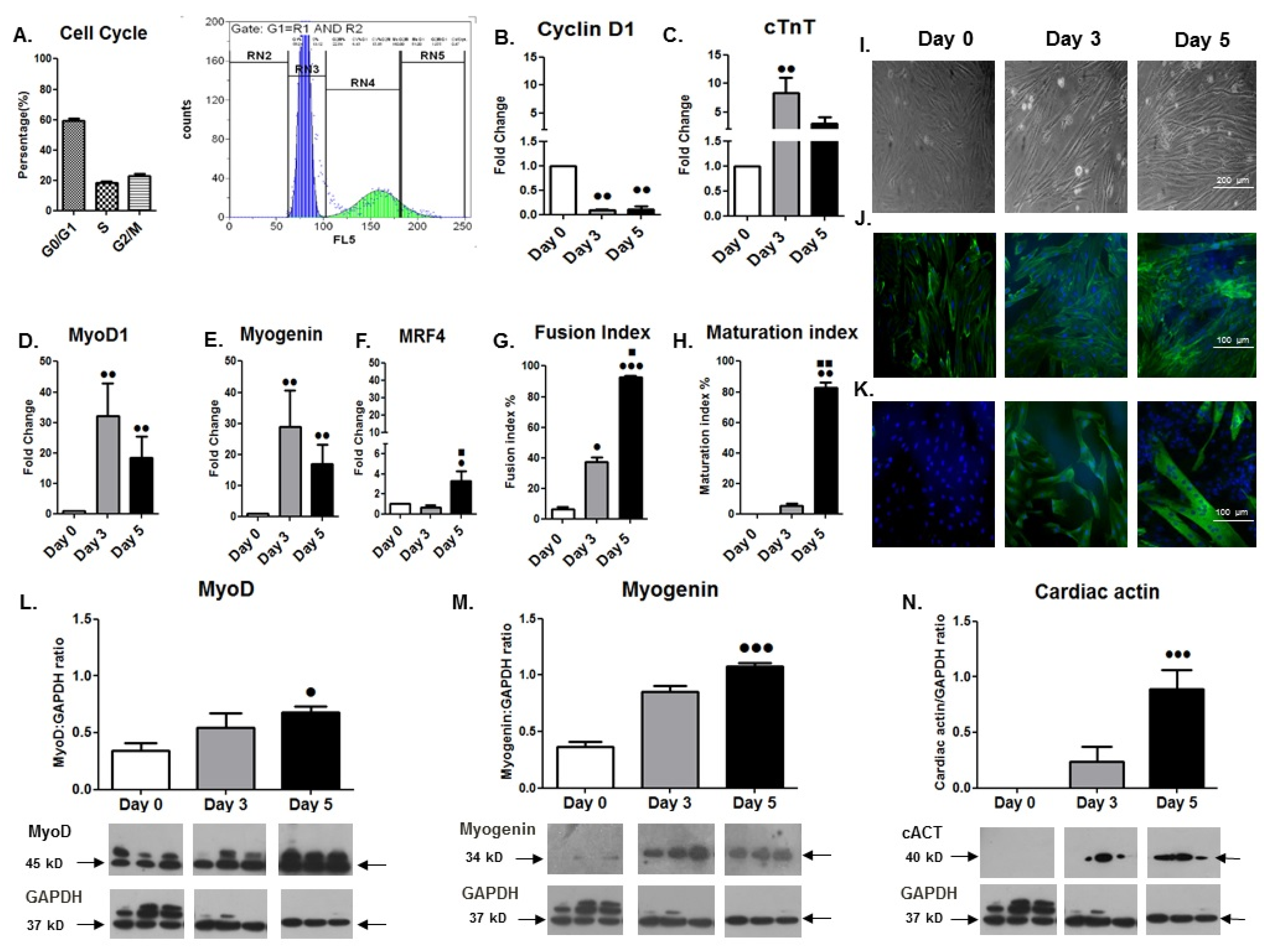
3.1. Differentiation Phenotype of Cardiomyocyte-like H9C2 Cells
3.1.1. Cell Cycle and Morphological Analyses
3.1.2. Gene Expression Changes during Differentiation
3.1.3. Changes in Signalling Pathways Activation during Differentiation
3.2. Responses of Differentiated Cardiomyocyte-like H9C2 Cells to Various Mechanical Loading Protocols
3.2.1. Myogenic Regulatory Factors
3.2.2. Muscle Hypertrophy/Atrophy Factors
3.2.3. Pro-Apoptotic Factors
3.2.4. Inflammation-Related Factors
3.2.5. Changes in the Activation of Key Intracellular Signalling Mediators
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hescheler, J.; Meyer, R.; Plant, S.; Krautwurst, D.; Rosenthal, W.; Schultz, G. Morphological, biochemical, and electrophysiological characterization of a clonal cell (H9c2) line from rat heart. Circ. Res. 1991, 69, 1476–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenco, J.; Lencova-Popelova, O.; Link, M.; Jirkovska, A.; Tambor, V.; Potuckova, E.; Stulik, J.; Simunek, T.; Sterba, M. Proteomic investigation of embryonic rat heart-derived H9c2 cell line sheds new light on the molecular phenotype of the popular cell model. Exp. Cell Res. 2015, 339, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Branco, A.F.; Pereira, S.P.; Gonzalez, S.; Gusev, O.; Rizvanov, A.A.; Oliveira, P.J. Gene expression profiling of H9c2 myoblast differentiation towards a cardiac-like phenotype. PLoS ONE 2015, 10, e0129303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCain, M.L.; Parker, K.K. Mechanotransduction: The role of mechanical stress, myocyte shape, and cytoskeletal architecture on cardiac function. Pflug. Arch 2011, 462, 89–104. [Google Scholar] [CrossRef]
- Takahashi, K.; Kakimoto, Y.; Toda, K.; Naruse, K. Mechanobiology in cardiac physiology and diseases. J. Cell. Mol. Med. 2013, 17, 225–232. [Google Scholar] [CrossRef]
- Palmieri, E.A.; Benincasa, G.; Di Rella, F.; Casaburi, C.; Monti, M.G.; De Simone, G.; Chiariotti, L.; Palombini, L.; Bruni, C.B.; Sacca, L.; et al. Differential expression of TNF-alpha, IL-6, and IGF-1 by graded mechanical stress in normal rat myocardium. Am. J. Physiol. Heart Circ. Physiol. 2002, 282, H926–H934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, I.L.; Hool, L.; Choi, Y.S. A review of in vitro platforms for understanding cardiomyocyte mechanobiology. Front. Bioeng. Biotechnol. 2019, 7, 133. [Google Scholar] [CrossRef] [Green Version]
- Martino, F.; Perestrelo, A.R.; Vinarsky, V.; Pagliari, S.; Forte, G. Cellular mechanotransduction: From tension to function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Saucerman, J.J.; Tan, P.M.; Buchholz, K.S.; McCulloch, A.D.; Omens, J.H. Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat. Rev. Cardiol. 2019, 16, 361–378. [Google Scholar] [CrossRef]
- Huang, H.; Kamm, R.D.; Lee, R.T. Cell mechanics and mechanotransduction: Pathways, probes, and physiology. Am. J. Physiol. Cell Physiol. 2004, 287, C1–C11. [Google Scholar] [CrossRef]
- Teng, E.L.; Engler, A.J. Mechanical influences on cardiovascular differentiation and disease modeling. Exp. Cell Res. 2019, 377, 103–108. [Google Scholar] [CrossRef]
- Jacot, J.G.; McCulloch, A.D.; Omens, J.H. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys. J. 2008, 95, 3479–3487. [Google Scholar] [CrossRef] [Green Version]
- Jacot, J.G.; Kita-Matsuo, H.; Wei, K.A.; Chen, H.S.; Omens, J.H.; Mercola, M.; McCulloch, A.D. Cardiac myocyte force development during differentiation and maturation. Ann. N. Y. Acad. Sci. 2010, 1188, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rysa, J.; Tokola, H.; Ruskoaho, H. Mechanical stretch induced transcriptomic profiles in cardiac myocytes. Sci. Rep. 2018, 8, 4733. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cheng, G.; Jin, R.; Afzal, M.R.; Samanta, A.; Xuan, Y.T.; Girgis, M.; Elias, H.K.; Zhu, Y.; Davani, A.; et al. Deletion of Interleukin-6 attenuates pressure overload-induced left ventricular hypertrophy and dysfunction. Circ. Res. 2016, 118, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Tzanis, G.; Philippou, A.; Karatzanos, E.; Dimopoulos, S.; Kaldara, E.; Nana, E.; Pitsolis, T.; Rontogianni, D.; Koutsilieris, M.; Nanas, S. Effects of high-intensity interval exercise training on skeletal myopathy of chronic heart failure. J. Card. Fail. 2017, 23, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Halapas, A.; Papalois, A.; Stauropoulou, A.; Philippou, A.; Pissimissis, N.; Chatzigeorgiou, A.; Kamper, E.; Koutsilieris, M. In vivo models for heart failure research. In Vivo 2008, 22, 767–780. [Google Scholar] [PubMed]
- Philippou, A.; Tryfonos, A.; Theos, A.; Nezos, A.; Halapas, A.; Maridaki, M.; Koutsilieris, M. Expression of tissue remodelling, inflammation- and angiogenesis-related factors after eccentric exercise in humans. Mol. Biol. Rep. 2021, 48, 4047–4054. [Google Scholar] [CrossRef]
- Tryfonos, A.; Tzanis, G.; Pitsolis, T.; Karatzanos, E.; Koutsilieris, M.; Nanas, S.; Philippou, A. Exercise training enhances angiogenesis-related gene responses in skeletal muscle of patients with chronic heart failure. Cells 2021, 10, 1915. [Google Scholar] [CrossRef]
- Krueger, W.; Bender, N.; Haeusler, M.; Henneberg, M. The role of mechanotransduction in heart failure pathobiology-a concise review. Heart Fail. Rev. 2021, 26, 981–995. [Google Scholar] [CrossRef]
- Branco, A.F.; Pereira, S.L.; Moreira, A.C.; Holy, J.; Sardao, V.A.; Oliveira, P.J. Isoproterenol cytotoxicity is dependent on the differentiation state of the cardiomyoblast H9c2 cell line. Cardiovasc. Toxicol. 2011, 11, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Asfour, H.A.; Allouh, M.Z.; Said, R.S. Myogenic regulatory factors: The orchestrators of myogenesis after 30 years of discovery. Exp. Biol. Med. 2018, 243, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Kopantseva, E.E.; Belyavsky, A.V. Key regulators of skeletal myogenesis. Mol. Biol. 2016, 50, 195–222. [Google Scholar] [CrossRef]
- Karalaki, M.; Fili, S.; Philippou, A.; Koutsilieris, M. Muscle regeneration: Cellular and molecular events. In Vivo 2009, 23, 779–796. [Google Scholar] [PubMed]
- Mutlak, M.; Kehat, I. Extracellular signal-regulated kinases 1/2 as regulators of cardiac hypertrophy. Front. Pharmacol. 2015, 6, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stavropoulou, A.; Halapas, A.; Sourla, A.; Philippou, A.; Papageorgiou, P.; Papalois, A.; Koutsilieris, M. IGF-1 expression in infarcted myocardium and MGF E peptide actions in rat cardiomyocytes in vitro. Mol. Med. 2009, 15, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Honsho, S.; Nishikawa, S.; Amano, K.; Zen, K.; Adachi, Y.; Kishita, E.; Matsui, A.; Katsume, A.; Yamaguchi, S.; Nishikawa, K.; et al. Pressure-mediated hypertrophy and mechanical stretch induces IL-1 release and subsequent IGF-1 generation to maintain compensative hypertrophy by affecting Akt and JNK pathways. Circ. Res. 2009, 105, 1149–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippou, A.; Halapas, A.; Maridaki, M.; Koutsilieris, M. Type I insulin-like growth factor receptor signaling in skeletal muscle regeneration and hypertrophy. J. Musculoskelet. Neuronal Interact. 2007, 7, 208–218. [Google Scholar]
- Zevolis, E.; Philippou, A.; Moustogiannis, A.; Chatzigeorgiou, A.; Koutsilieris, M. Optimizing mechanical stretching protocols for hypertrophic and anti-apoptotic responses in cardiomyocyte-like H9C2 cells. Mol. Biol. Rep. 2021, 48, 645–655. [Google Scholar] [CrossRef]
- Sardao, V.A.; Oliveira, P.J.; Holy, J.; Oliveira, C.R.; Wallace, K.B. Doxorubicin-induced mitochondrial dysfunction is secondary to nuclear p53 activation in H9c2 cardiomyoblasts. Cancer Chemother. Pharm. 2009, 64, 811–827. [Google Scholar] [CrossRef]
- Glass, D.J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell Biol. 2003, 5, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.P.; Wang, B.W.; Lo, H.M.; Shyu, K.G. Mechanical stretch induces apoptosis regulator TRB3 in cultured cardiomyocytes and volume-overloaded heart. PLoS ONE 2015, 10, e0123235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, N.; Hadden, T.J.; Rishi, A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 2011, 1813, 1978–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moustogiannis, A.; Philippou, A.; Taso, O.; Zevolis, E.; Pappa, M.; Chatzigeorgiou, A.; Koutsilieris, M. The effects of muscle cell aging on myogenesis. Int. J. Mol. Sci. 2021, 22, 3721. [Google Scholar] [CrossRef] [PubMed]
- Lindner, D.; Zietsch, C.; Tank, J.; Sossalla, S.; Fluschnik, N.; Hinrichs, S.; Maier, L.; Poller, W.; Blankenberg, S.; Schultheiss, H.P.; et al. Cardiac fibroblasts support cardiac inflammation in heart failure. Basic Res. Cardiol. 2014, 109, 428. [Google Scholar] [CrossRef]
- Shyni, G.L.; Renjitha, J.; Somappa, B.S.; Raghu, K.G. Zerumin A attenuates the inflammatory responses in LPS-stimulated H9c2 cardiomyoblasts. J. Biochem. Mol. Toxicol. 2021, 35, 1–11. [Google Scholar] [CrossRef]
- Moustogiannis, A.; Philippou, A.; Zevolis, E.; Taso, O.; Chatzigeorgiou, A.; Koutsilieris, M. Characterization of optimal strain, frequency and duration of mechanical loading on skeletal myotubes’ biological responses. In Vivo 2020, 34, 1779–1788. [Google Scholar] [CrossRef]
- Philippou, A.; Papageorgiou, E.; Bogdanis, G.; Halapas, A.; Sourla, A.; Maridaki, M.; Pissimissis, N.; Koutsilieris, M. Expression of IGF-1 isoforms after exercise-induced muscle damage in humans: Characterization of the MGF E peptide actions in vitro. In Vivo 2009, 23, 567–575. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Wu, F.; Gao, A.; Liu, J.; Shen, Y.; Xu, P.; Meng, J.; Wen, T.; Xu, L.; Xu, H. High modulus conductive hydrogels enhance in vitro maturation and contractile function of primary cardiomyocytes for uses in drug screening. Adv. Healthc. Mater. 2018, 7, e1800990. [Google Scholar] [CrossRef]
- Mei, C.; Chao, C.W.; Lin, C.W.; Li, S.T.; Wu, K.H.; Yang, K.C.; Yu, J. Three-dimensional spherical gelatin bubble-based scaffold improves the myotube formation of H9c2 myoblasts. Biotechnol. Bioeng. 2019, 116, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Engel, F.B.; Schebesta, M.; Duong, M.T.; Lu, G.; Ren, S.; Madwed, J.B.; Jiang, H.; Wang, Y.; Keating, M.T. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005, 19, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kankeu, C.; Clarke, K.; Van Haver, D.; Gevaert, K.; Impens, F.; Dittrich, A.; Roderick, H.L.; Passante, E.; Huber, H.J. Quantitative proteomics and systems analysis of cultured H9C2 cardiomyoblasts during differentiation over time supports a ‘function follows form’ model of differentiation. Mol. Omics 2018, 14, 181–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberston, M.J.; Raghunathan, S.; Potaman, V.N.; Zhang, F.; Stewart, M.D.; McConnell, B.K.; Schwartz, R.J. CRISPR-Cas9-induced IGF1 gene activation as a tool for enhancing muscle differentiation via multiple isoform expression. FASEB J. 2020, 34, 555–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinciguerra, M.; Santini, M.P.; Claycomb, W.C.; Ladurner, A.G.; Rosenthal, N. Local IGF-1 isoform protects cardiomyocytes from hypertrophic and oxidative stresses via SirT1 activity. Aging 2009, 2, 43–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippou, A.; Maridaki, M.; Halapas, A.; Koutsilieris, M. The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In Vivo 2007, 21, 45–54. [Google Scholar]
- Philippou, A.; Maridaki, M.; Pneumaticos, S.; Koutsilieris, M. The complexity of the IGF1 gene splicing, posttranslational modification and bioactivity. Mol. Med. 2014, 20, 202–214. [Google Scholar] [CrossRef]
- Papageorgiou, E.; Philippou, A.; Armakolas, A.; Christopoulos, P.F.; Dimakakos, A.; Koutsilieris, M. The human Ec peptide: The active core of a progression growth factor with species-specific mode of action. Hormones 2016, 15, 423–434. [Google Scholar] [CrossRef]
- Philippou, A.; Armakolas, A.; Panteleakou, Z.; Pissimissis, N.; Nezos, A.; Theos, A.; Kaparelou, M.; Armakolas, N.; Pneumaticos, S.G.; Koutsilieris, M. IGF1Ec expression in MG-63 human osteoblast-like osteosarcoma cells. Anticancer Res. 2011, 31, 4259–4265. [Google Scholar]
- Philippou, A.; Armakolas, A.; Koutsilieris, M. Evidence for the possible biological significance of the igf-1 gene alternative splicing in prostate cancer. Front. Endocrinol. 2013, 4, 31. [Google Scholar] [CrossRef] [Green Version]
- Armakolas, A.; Philippou, A.; Panteleakou, Z.; Nezos, A.; Sourla, A.; Petraki, C.; Koutsilieris, M. Preferential expression of IGF-1Ec (MGF) transcript in cancerous tissues of human prostate: Evidence for a novel and autonomous growth factor activity of MGF E peptide in human prostate cancer cells. Prostate 2010, 70, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Milingos, D.S.; Philippou, A.; Armakolas, A.; Papageorgiou, E.; Sourla, A.; Protopapas, A.; Liapi, A.; Antsaklis, A.; Mastrominas, M.; Koutsilieris, M. Insulinlike growth factor-1Ec (MGF) expression in eutopic and ectopic endometrium: Characterization of the MGF E-peptide actions in vitro. Mol. Med. 2011, 17, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Vassilakos, G.; Philippou, A.; Tsakiroglou, P.; Koutsilieris, M. Biological activity of the e domain of the IGF-1Ec as addressed by synthetic peptides. Hormones 2014, 13, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Vassilakos, G.; Philippou, A.; Koutsilieris, M. Identification of the IGF-1 processing product human Ec/rodent Eb peptide in various tissues: Evidence for its differential regulation after exercise-induced muscle damage in humans. Growth Horm. IGF Res. 2017, 32, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Jogo, M.; Shiraishi, S.; Tamura, T.A. Identification of MAFbx as a myogenin-engaged F-box protein in SCF ubiquitin ligase. FEBS Lett. 2009, 583, 2715–2719. [Google Scholar] [CrossRef] [Green Version]
- Abe, S.; Rhee, S.; Iwanuma, O.; Hiroki, E.; Yanagisawa, N.; Sakiyama, K.; Ide, Y. Effect of mechanical stretching on expressions of muscle specific transcription factors MyoD, Myf-5, myogenin and MRF4 in proliferated myoblasts. Anat. Histol. Embryol. 2009, 38, 305–310. [Google Scholar] [CrossRef]
- Sharples, A.P.; Stewart, C.E. Myoblast models of skeletal muscle hypertrophy and atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Langley, B.; Thomas, M.; Bishop, A.; Sharma, M.; Gilmour, S.; Kambadur, R. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J. Biol. Chem. 2002, 277, 49831–49840. [Google Scholar] [CrossRef] [Green Version]
- Philippou, A.; Maridaki, M.; Karatzas, T.; Koutsilieris, M. The multiple actions of the insulin-like growth factor-I signaling in the myocardium. In Introduction to Translational Cardiovascular Research; Springer: Cham, Switzerland, 2014; pp. 187–204. [Google Scholar]
- Gumucio, J.P.; Mendias, C.L. Atrogin-1, MuRF-1, and sarcopenia. Endocrine 2013, 43, 12–21. [Google Scholar] [CrossRef]
- Yang, Y.; Jemiolo, B.; Trappe, S. Proteolytic mRNA expression in response to acute resistance exercise in human single skeletal muscle fibers. J. Appl. Physiol. 2006, 101, 1442–1450. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Kitamura, Y.I.; Funahashi, Y.; Shawber, C.J.; Castrillon, D.H.; Kollipara, R.; DePinho, R.A.; Kitajewski, J.; Accili, D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Investig. 2007, 117, 2477–2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.J.; Broz, D.K.; Noderer, W.L.; Ferreira, J.P.; Overton, K.W.; Spencer, S.L.; Meyer, T.; Tapscott, S.J.; Attardi, L.D.; Wang, C.L. p53 suppresses muscle differentiation at the myogenin step in response to genotoxic stress. Cell Death Differ. 2015, 22, 560–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, P.M.; Buchholz, K.S.; Omens, J.H.; McCulloch, A.D.; Saucerman, J.J. Predictive model identifies key network regulators of cardiomyocyte mechano-signaling. PLoS Comput. Biol. 2017, 13, e1005854. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.X.; Zhang, W.M.; Zhang, H.J.; Li, T.T.; Wang, Y.L.; Qin, Y.W.; Gu, H.; Du, J. Mechanical stretch-induced endoplasmic reticulum stress, apoptosis and inflammation contribute to thoracic aortic aneurysm and dissection. J. Pathol. 2015, 236, 373–383. [Google Scholar] [CrossRef] [Green Version]






| Target Gene | 5′-3′ (Forward) Primer Sequence | 5′-3′ (Reverse) Primer Sequence |
|---|---|---|
| GAPDH | CAA CTC CCT CAA GAT TGT CAG CAA | GGC ATG GAC TGT GGT CAT GA |
| MYOD | TGC TCC TTT GAG ACA GCA GA | AGT AGG GAA GTG TGC GTG CT |
| MYOGENIN | AGG AGA GAA AGA TGG AGT CCA GAG | TAA CAA AAG AAG TCA CCC CAA GAG |
| MRF4 | AGG GCT CTC CTT TGT ATC CAG | TGG AAG AAA GGC GCT GAA GA |
| cTnT | GCG GAA GAG TGG GAA GAG ACA | CCA CAG CTC CTT GGC CTT CT |
| CYCLIN D1 | TCA AGT GTG ACC CGG ACT G | ATG TCC ACA TCT CGC ACG TC |
| IGF-1Ea | GTG GAC GCT CTT CAG TTC GT | GCT TCC TTT TCT TGT GTG TCG ATA G |
| IGF-1Eb | GTC CCC AGC ACA CAT CGC G | TCT TTT GTG CAA AAT AAG GCG TA |
| p53 | GAG AGA CCG CCG TAC AGA AG | AGC AGT TTG GGC TTT CCT CC |
| FoxO1 | AGT GGA TGG TGA AGA GCG TG | GAA GGG ACA GAT TGT GGC GA |
| TNF-α | CTC TTC TGC CTG CTG CAG TTG | ATG GGC TAC AGG CTT GTC ACT C |
| NF-kB | ATA GGC ACT GTC TTC TTT CAC CTC | ATA GGC ACT GTC TTC TTT CAC CTC |
| IL-6 | CCT TCC TAC CCC AAT TTC CAA T | AAC GCA CTA GGT TTG CCG AGT A |
| IL-1β | ATC CCA AGC AAT ACC CAA AG | GTG CTG ATG TAC CAG TTG GG |
| ATROGIN-1 | AAC AAG GAG GTA TAC AGT AAG G | AAT TGT TCA TGA AGT TCT TTT G |
| MYOSTATIN | CTG TAA CCT TCC CAG GAC CA | GCA GTC AAG CCC AAA GTC TC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zevolis, E.; Philippou, A.; Moustogiannis, A.; Chatzigeorgiou, A.; Koutsilieris, M. The Effects of Mechanical Loading Variations on the Hypertrophic, Anti-Apoptotic, and Anti-Inflammatory Responses of Differentiated Cardiomyocyte-like H9C2 Cells. Cells 2022, 11, 473. https://doi.org/10.3390/cells11030473
Zevolis E, Philippou A, Moustogiannis A, Chatzigeorgiou A, Koutsilieris M. The Effects of Mechanical Loading Variations on the Hypertrophic, Anti-Apoptotic, and Anti-Inflammatory Responses of Differentiated Cardiomyocyte-like H9C2 Cells. Cells. 2022; 11(3):473. https://doi.org/10.3390/cells11030473
Chicago/Turabian StyleZevolis, Evangelos, Anastassios Philippou, Athanasios Moustogiannis, Antonios Chatzigeorgiou, and Michael Koutsilieris. 2022. "The Effects of Mechanical Loading Variations on the Hypertrophic, Anti-Apoptotic, and Anti-Inflammatory Responses of Differentiated Cardiomyocyte-like H9C2 Cells" Cells 11, no. 3: 473. https://doi.org/10.3390/cells11030473








