Autophagy and Lysosomal Functionality in CMT2B Fibroblasts Carrying the RAB7K126R Mutation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibodies and Reagents
2.2. Cell Lines
2.3. Plasmids and Transfection
2.4. Western Blot Analysis
2.5. Oil-Red O Staining
2.6. Immunofluorescence, Confocal Microscopy and Live Imaging
2.7. DQBSA (Self-Quenched BODIPY Dye Conjugates of Bovine Serum Albumin) Assay
2.8. Flow Cytometry
2.9. Statistical Analysis
3. Results
3.1. Altered Autophagic Flux in CMT2B Fibroblasts
3.2. Transfection of CMT2B-Causing RAB7K126R Mutants Induces Impairment of the Autophagic Process
3.3. RAB7K126R Mutant Protein Affects the Lipophagy Mechanism
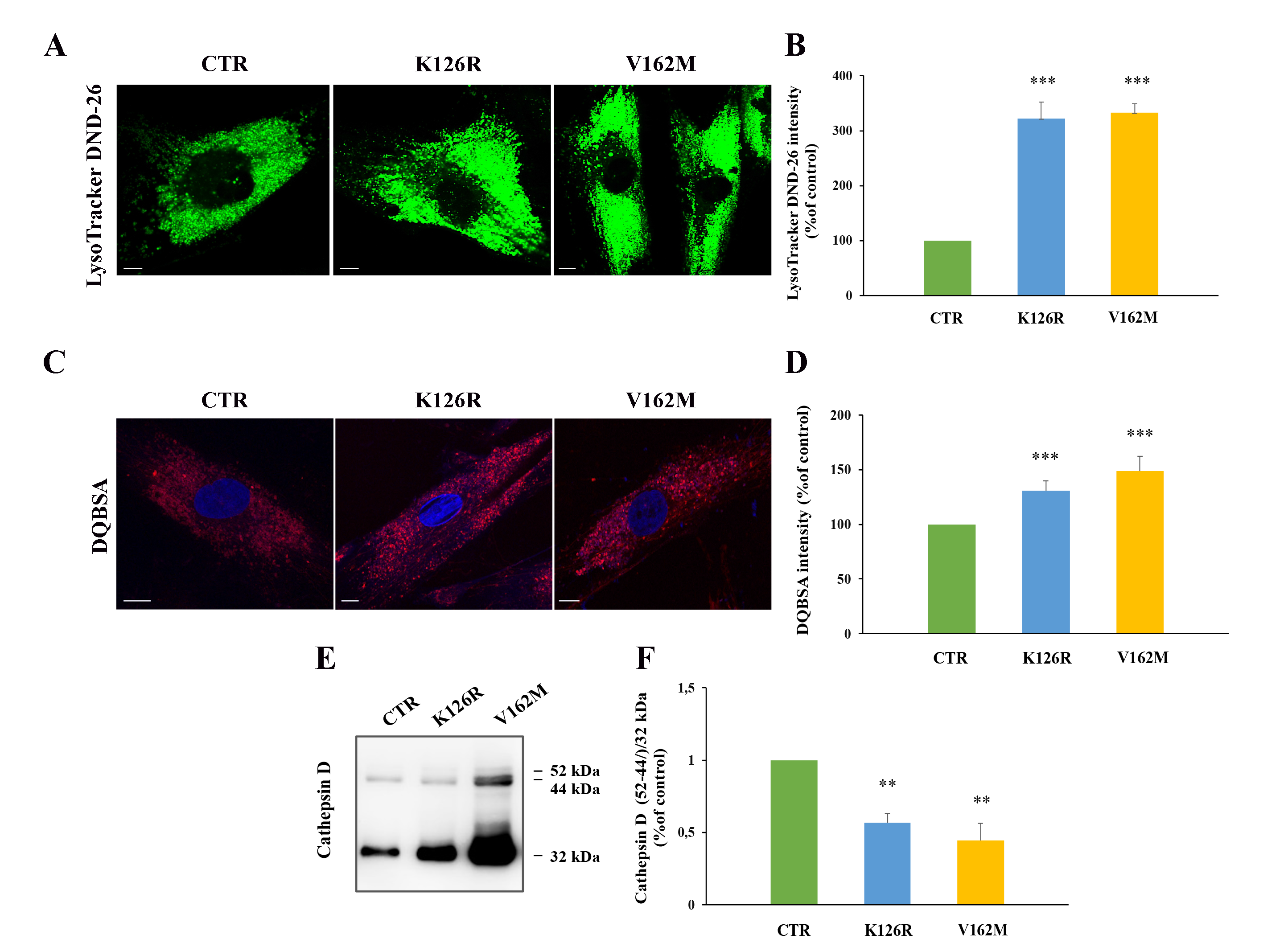
3.4. Lysosomal Activity Was Increased in Patient’s Fibroblasts Carrying the CMT2B-Causing RAB7K126R Mutation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bucci, C.; Bakke, O.; Progida, C. Charcot-marie-tooth disease and intracellular traffic. Prog. Neurobiol. 2012, 99, 191–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tazir, M.; Hamadouche, T.; Nouioua, S.; Mathis, S.; Vallat, J.M. Hereditary motor and sensory neuropathies or charcot-marie-tooth diseases: An update. J. Neurol. Sci. 2014, 347, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Manganelli, F.; Pisciotta, C.; Provitera, V.; Taioli, F.; Iodice, R.; Topa, A.; Fabrizi, G.M.; Nolano, M.; Santoro, L. Autonomic nervous system involvement in a new cmt2b family. J. Peripher. Nerv. Syst. 2012, 17, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Han, C.; Liu, W.; Wang, P.; Zhang, X. A novel rab7 mutation in a chinese family with charcot-marie-tooth type 2b disease. Gene 2014, 534, 431–434. [Google Scholar] [CrossRef]
- Houlden, H.; King, R.H.; Muddle, J.R.; Warner, T.T.; Reilly, M.M.; Orrell, R.W.; Ginsberg, L. A novel rab7 mutation associated with ulcero-mutilating neuropathy. Ann. Neurol. 2004, 56, 586–590. [Google Scholar] [CrossRef]
- Verhoeven, K.; De Jonghe, P.; Coen, K.; Verpoorten, N.; Auer-Grumbach, M.; Kwon, J.M.; FitzPatrick, D.; Schmedding, E.; De Vriendt, E.; Jacobs, A.; et al. Mutations in the small gtp-ase late endosomal protein rab7 cause charcot-marie-tooth type 2b neuropathy. Am. J. Hum. Genet. 2003, 72, 722–727. [Google Scholar] [CrossRef] [Green Version]
- Meggouh, F.; Bienfait, H.M.; Weterman, M.A.; de Visser, M.; Baas, F. Charcot-marie-tooth disease due to a de novo mutation of the rab7 gene. Neurology 2006, 67, 1476–1478. [Google Scholar] [CrossRef]
- Guerra, F.; Bucci, C. Multiple roles of the small gtpase rab7. Cells 2016, 5, 34. [Google Scholar] [CrossRef]
- Luzio, J.P.; Pryor, P.R.; Bright, N.A. Lysosomes: Fusion and function. Nat. Rev. Mol. Cell Biol. 2007, 8, 622–632. [Google Scholar] [CrossRef]
- Bucci, C.; Thomsen, P.; Nicoziani, P.; McCarthy, J.; van Deurs, B. Rab7: A key to lysosome biogenesis. Mol. Biol. Cell 2000, 11, 467–480. [Google Scholar] [CrossRef]
- Jager, S.; Bucci, C.; Tanida, I.; Ueno, T.; Kominami, E.; Saftig, P.; Eskelinen, E.L. Role for rab7 in maturation of late autophagic vacuoles. J. Cell. Sci. 2004, 117, 4837–4848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, M.; Munafó, D.; Berón, W.; Colombo, M. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell. Sci. 2004, 117, 2687–2697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, R.E.; Bucci, C.; Vieira, O.V.; Schroer, T.A.; Grinstein, S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: Role of rab7 and rilp. Mol. Cell Biol. 2003, 23, 6494–6506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamano, K.; Fogel, A.I.; Wang, C.; van der Bliek, A.M.; Youle, R.J. Mitochondrial rab gaps govern autophagosome biogenesis during mitophagy. eLife 2014, 3, e01612. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.; Schulze, R.J.; Weller, S.G.; Sletten, A.C.; Casey, C.A.; McNiven, M.A. The small gtpase rab7 as a central regulator of hepatocellular lipophagy. Hepatology 2015, 61, 1896–1907. [Google Scholar] [CrossRef] [Green Version]
- Rojas, R.; van Vlijmen, T.; Mardones, G.A.; Prabhu, Y.; Rojas, A.L.; Mohammed, S.; Heck, A.J.; Raposo, G.; van der Sluijs, P.; Bonifacino, J.S. Regulation of retromer recruitment to endosomes by sequential action of rab5 and rab7. J. Cell. Biol. 2008, 183, 513–526. [Google Scholar] [CrossRef] [Green Version]
- Seebohm, G.; Strutz-Seebohm, N.; Birkin, R.; Dell, G.; Bucci, C.; Spinosa, M.R.; Baltaev, R.; Mack, A.F.; Korniychuk, G.; Choudhury, A.; et al. Regulation of endocytic recycling of kcnq1/kcne1 potassium channels. Circ. Res. 2007, 100, 686–692. [Google Scholar] [CrossRef] [Green Version]
- Edinger, A.L.; Cinalli, R.M.; Thompson, C.B. Rab7 prevents growth factor-independent survival by inhibiting cell-autonomous nutrient transporter expression. Dev. Cell 2003, 5, 571–582. [Google Scholar] [CrossRef] [Green Version]
- Deinhardt, K.; Salinas, S.; Verastegui, C.; Watson, R.; Worth, D.; Hanrahan, S.; Bucci, C.; Schiavo, G. Rab5 and rab7 control endocytic sorting along the axonal retrograde transport pathway. Neuron 2006, 52, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Saxena, S.; Bucci, C.; Weis, J.; Kruttgen, A. The small gtpase rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor trka. J. Neurosci. 2005, 25, 10930–10940. [Google Scholar] [CrossRef]
- Kawauchi, T.; Sekine, K.; Shikanai, M.; Chihama, K.; Tomita, K.; Kubo, K.; Nakajima, K.; Nabeshima, Y.; Hoshino, M. Rab gtpases-dependent endocytic pathways regulate neuronal migration and maturation through n-cadherin trafficking. Neuron 2010, 67, 588–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saveri, P.; De Luca, M.; Nisi, V.; Pisciotta, C.; Romano, R.; Piscosquito, G.; Reilly, M.M.; Polke, J.M.; Cavallaro, T.; Fabrizi, G.M.; et al. Charcot-marie-tooth type 2b: A new phenotype associated with a novel rab7a mutation and inhibited egfr degradation. Cells 2020, 9, 1028. [Google Scholar] [CrossRef] [PubMed]
- Pareyson, D.; Saveri, P.; Piscosquito, G. Charcot-marie-tooth disease and related hereditary neuropathies: From gene function to associated phenotypes. Curr. Mol. Med. 2014, 14, 1009–1103. [Google Scholar] [CrossRef] [PubMed]
- Spinosa, M.R.; Progida, C.; De Luca, A.; Colucci, A.M.R.; Alifano, P.; Bucci, C. Functional characterization of rab7 mutant proteins associated with charcot-marie-tooth type 2b disease. J. Neurosci. 2008, 28, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Progida, C.; Spinosa, M.R.; Alifano, P.; Bucci, C. Characterization of the rab7k157n mutant protein associated with charcot-marie-tooth type 2b. Biochem. Biophys. Res. Commun. 2008, 372, 283–287. [Google Scholar] [CrossRef]
- McCray, B.A.; Skordalakes, E.; Taylor, J.P. Disease mutations in rab7 result in unregulated nucleotide exchange and inappropriate activation. Hum. Mol. Genet. 2010, 19, 1033–1047. [Google Scholar] [CrossRef] [Green Version]
- Cogli, L.; Progida, C.; Lecci, R.; Bramato, R.; Kruttgen, A.; Bucci, C. Cmt2b-associated rab7 mutants inhibit neurite outgrowth. Acta Neuropathol. 2010, 120, 491–501. [Google Scholar] [CrossRef]
- Yamauchi, J.; Torii, T.; Kusakawa, S.; Sanbe, A.; Nakamura, K.; Takashima, S.; Hamasaki, H.; Kawaguchi, S.; Miyamoto, Y.; Tanoue, A. The mood stabilizer valproic acid improves defective neurite formation caused by charcot-marie-tooth disease-associated mutant rab7 through the jnk signaling pathway. J. Neurosci. Res. 2010, 88, 3189–3197. [Google Scholar] [CrossRef]
- Mahar, M.; Cavalli, V. Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 2018, 19, 323–337. [Google Scholar] [CrossRef]
- Cogli, L.; Progida, C.; Thomas, C.L.; Spencer-Dene, B.; Donno, C.; Schiavo, G.; Bucci, C. Charcot-marie-tooth type 2b disease-causing rab7a mutant proteins show altered interaction with the neuronal intermediate filament peripherin. Acta Neuropathol. 2013, 125, 257–272. [Google Scholar] [CrossRef] [Green Version]
- Kirkcaldie, M.T.K.; Dwyer, S.T. The third wave: Intermediate filaments in the maturing nervous system. Mol. Cell Neurosci. 2017, 84, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Parlakian, A.; Paulin, D.; Izmiryan, A.; Xue, Z.; Li, Z. Intermediate filaments in peripheral nervous system: Their expression, dysfunction and diseases. Rev. Neurol. 2016, 172, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.; Rivellini, C.; De Luca, M.; Tonlorenzi, R.; Beli, R.; Manganelli, F.; Nolano, M.; Santoro, L.; Eskelinen, E.L.; Previtali, S.C.; et al. Alteration of the late endocytic pathway in charcot-marie-tooth type 2b disease. Cell Mol. Life Sci. 2020, 78, 351–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colecchia, D.; Stasi, M.; Leonardi, M.; Manganelli, F.; Nolano, M.; Veneziani, B.M.; Santoro, L.; Eskelinen, E.L.; Chiariello, M.; Bucci, C. Alterations of autophagy in the peripheral neuropathy charcot-marie-tooth type 2b. Autophagy 2018, 14, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Giudetti, A.M.; Guerra, F.; Longo, S.; Beli, R.; Romano, R.; Manganelli, F.; Nolano, M.; Mangini, V.; Santoro, L.; Bucci, C. An altered lipid metabolism characterizes charcot-marie-tooth type 2b peripheral neuropathy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 1588805. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.; Calcagnile, M.; Margiotta, A.; Franci, L.; Chiariello, M.; Alifano, P.; Bucci, C. Rab7a regulates vimentin phosphorylation through akt and pak. Cancers 2021, 13, 2220. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.; Girolimetti, G.; Beli, R.; Mitruccio, M.; Pacelli, C.; Ferretta, A.; Gasparre, G.; Cocco, T.; Bucci, C. Synergistic effect of mitochondrial and lysosomal dysfunction in parkinson’s disease. Cells 2019, 8, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, M.; Romano, R.; Bucci, C. Role of the v1g1 subunit of v-atpase in breast cancer cell migration. Sci. Rep. 2021, 11, 4615. [Google Scholar] [CrossRef]
- Gump, J.M.; Thorburn, A. Sorting cells for basal and induced autophagic flux by quantitative ratiometric flow cytometry. Autophagy 2014, 10, 1327–1334. [Google Scholar] [CrossRef]
- Marwaha, R.; Sharma, M. Dq-red bsa trafficking assay in cultured cells to assess cargo delivery to lysosomes. Bio. Protoc. 2017, 7, e2571. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar]
- Voss, E.W., Jr.; Workman, C.J.; Mummert, M.E. Detection of protease activity using a fluorescence-enhancement globular substrate. Biotechniques 1996, 20, 286–291. [Google Scholar] [CrossRef]
- Richo, G.; Conner, G.E. Proteolytic activation of human procathepsin d. Adv. Exp. Med. Biol. 1991, 306, 289–296. [Google Scholar]
- Bucci, C.; Parton, R.G.; Mather, I.H.; Stunnenberg, H.; Simons, K.; Hoflack, B.; Zerial, M. The small gtpase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 1992, 70, 715–728. [Google Scholar] [CrossRef] [Green Version]
- Gorvel, J.P.; Chavrier, P.; Zerial, M.; Gruenberg, J. Rab5 controls early endosome fusion in vitro. Cell 1991, 64, 915–925. [Google Scholar] [CrossRef]
- Van der Sluijs, P.; Hull, M.; Webster, P.; Mâle, P.; Goud, B.; Mellman, I. The small gtp-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 1992, 70, 729–740. [Google Scholar] [CrossRef]
- Gruenberg, J.; Stenmark, H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004, 5, 317–323. [Google Scholar] [CrossRef]
- Mallet, W.G.; Maxfield, F.R. Chimeric forms of furin and tgn38 are transported with the plasma membrane in the trans-golgi network via distinct endosomal pathways. J. Cell. Biol. 1999, 146, 345–359. [Google Scholar] [CrossRef] [Green Version]
- Banting, G.; Ponnambalam, S. Tgn38 and its orthologues: Roles in post-tgn vesicle formation and maintenance of tgn morphology. Biochim. Biophys. Acta 1997, 1355, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.L.; Urbe, S. The emerging shape of the escrt machinery. Nat. Rev. Mol. Cell Biol. 2007, 8, 355–368. [Google Scholar] [CrossRef]
- Progida, C.; Spinosa, M.R.; De Luca, A.; Bucci, C. Rilp interacts with the vps22 component of the escrt-ii complex. Biochem. Biophys. Res. Commun. 2006, 347, 1074–1079. [Google Scholar] [CrossRef]
- Maday, S. Mechanisms of neuronal homeostasis: Autophagy in the axon. Brain Res. 2016, 1649, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Boland, B.; Nixon, R.A. Neuronal macroautophagy: From development to degeneration. Mol. Aspects Med. 2006, 27, 503–519. [Google Scholar] [CrossRef]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef]
- Menzies, F.M.; Fleming, A.; Rubinsztein, D.C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015, 16, 345–357. [Google Scholar] [CrossRef]
- Wang, D.; Chan, C.C.; Cherry, S.; Hiesinger, P.R. Membrane trafficking in neuronal maintenance and degeneration. Cell Mol. Life Sci. 2013, 70, 2919–2934. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Coleman, M.; Zhang, L.; Zheng, X.; Yue, Z. Autophagy in axonal and dendritic degeneration. Trends Neurosci. 2013, 36, 418–428. [Google Scholar] [CrossRef] [Green Version]
- Kosacka, J.; Nowicki, M.; Blüher, M.; Baum, P.; Stockinger, M.; Toyka, K.V.; Klöting, I.; Stumvoll, M.; Serke, H.; Bechmann, I.; et al. Increased autophagy in peripheral nerv.ves may protect wistar ottawa karlsburg w rats against neuropathy. Exp. Neurol. 2013, 250, 125–135. [Google Scholar] [CrossRef]
- Ribas, V.T.; Schnepf, B.; Challagundla, M.; Koch, J.C.; Bähr, M.; Lingor, P. Early and sustained activation of autophagy in degenerating axons after spinal cord injury. Brain Pathol. 2015, 25, 157–170. [Google Scholar] [CrossRef]
- Launay, N.; Aguado, C.; Fourcade, S.; Ruiz, M.; Grau, L.; Riera, J.; Guilera, C.; Giròs, M.; Ferrer, I.; Knecht, E.; et al. Autophagy induction halts axonal degeneration in a mouse model of x-adrenoleukodystrophy. Acta Neuropathol. 2015, 129, 399–415. [Google Scholar] [CrossRef] [Green Version]
- Casas, C.; Isus, L.; Herrando-Grabulosa, M.; Mancuso, F.M.; Borrás, E.; Sabidó, E.; Forés, J.; Aloy, P. Network-based proteomic approaches reveal the neurodegenerative, neuroprotective and pain-related mechanisms involved after retrograde axonal damage. Sci. Rep. 2015, 5, 9185. [Google Scholar] [CrossRef] [Green Version]
- Hou, H.; Zhang, L.; Zhang, L.; Liu, D.; Xiong, Q.; Du, H.; Tang, P. Acute spinal cord injury could cause activation of autophagy in dorsal root ganglia. Spinal Cord 2013, 51, 679–682. [Google Scholar] [CrossRef] [Green Version]
- Connolly, G.P. Fibroblast models of neurological disorders: Fluorescence measurement studies. Trends Pharmacol. Sci. 1998, 19, 171–177. [Google Scholar] [CrossRef]
- Gasparini, L.; Racchi, M.; Binetti, G.; Trabucchi, M.; Solerte, S.B.; Alkon, D.; Etcheberrigaray, R.; Gibson, G.; Blass, J.; Paoletti, R.; et al. Peripheral markers in testing pathophysiological hypotheses and diagnosing alzheimer’s disease. FASEB J. 1998, 12, 17–34. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.M.; Martins, R.; Gandy, S.; Etcheberrigaray, R.; Ito, E.; Alkon, D.L.; Blass, J.; Gibson, G. Use of cultured fibroblasts in elucidating the pathophysiology and diagnosis of alzheimer’s disease. Ann. NY Acad. Sci. 1994, 747, 225–244. [Google Scholar] [CrossRef]
- Tesco, G.; Vergelli, M.; Amaducci, L.; Sorbi, S. Growth properties of familial alzheimer skin fibroblasts during in vitro aging. Exp. Gerontol. 1993, 28, 51–58. [Google Scholar] [CrossRef]
- Dreesmann, L.; Mittnacht, U.; Lietz, M.; Schlosshauer, B. Nerve fibroblast impact on schwann cell behavior. Eur. J. Cell Biol. 2009, 88, 285–300. [Google Scholar]
- Walther, T.C.; Farese, R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012, 81, 687–714. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Czaja, M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013, 20, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Missaglia, S.; Coleman, R.A.; Mordente, A.; Tavian, D. Neutral lipid storage diseases as cellular model to study lipid droplet function. Cells 2019, 8, 187. [Google Scholar] [CrossRef] [Green Version]
- Ganley, I.G.; Wong, P.M.; Jiang, X. Thapsigargin distinguishes membrane fusion in the late stages of endocytosis and autophagy. Autophagy 2011, 7, 1397–1399. [Google Scholar] [CrossRef] [Green Version]
- Ganley, I.G.; Wong, P.M.; Gammoh, N.; Jiang, X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell 2011, 42, 731–743. [Google Scholar] [CrossRef] [Green Version]
- Carew, J.S.; Espitia, C.M.; Esquivel 2nd, J.A.; Mahalingam, D.; Kelly, K.R.; Reddy, G.; Giles, F.J.; Nawrocki, S.T. Lucanthone is a novel inhibitor of autophagy that induces cathepsin d-mediated apoptosis. J. Biol. Chem. 2010, 286, 6602–6613. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Li, X.; Lu, C.; Bai, A.; Bielawski, J.; Bielawska, A.; Marshall, B.; Schoenlein, P.V.; Lebedyeva, I.O.; Liu, K. Ceramide activates lysosomal cathepsin b and cathepsin d to attenuate autophagy and induces er stress to suppress myeloid-derived suppressor cells. Oncotarget 2016, 7, 83907–83925. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Lambert, P.F.; Rapraeger, A.C.; Anderson, R.A. Stress-induced egfr trafficking: Mechanisms, functions, and therapeutic implications. Trends Cell Biol. 2016, 5, 352–366. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.; Simpson, J.; Fontana, R.; Kishi-Itakura, C.; Ktistakis, N.T.; Gammoh, N. Targeting of early endosomes by autophagy facilitates egfr recycling and signalling. EMBO Rep. 2019, 20, e47734. [Google Scholar] [CrossRef]





| Growth Condition | N° Lipid Droplets/Cell | Average Size (µ2) | Colocalization Rate vs. CD63 | Colocalization Rate vs. LAMP-1 | |
|---|---|---|---|---|---|
| CTR | FM | 118 ± 18 | 0,020 ± 0,007 | 0,993 ± 0,052 | 0,997 ± 0,0005 |
| ST | 112 ± 3 | 0,0048 ± 0,009 ** | 0,495 ± 0,06 * | 0,589 ± 0,05 ** | |
| K126R | FM | 275 ± 49 * | 0,022 ± 0,006 | 0,432 ± 0,1 * | 0,344 ± 0,03 *** |
| ST | 332 ± 35 *** | 0,027 ± 0,008 * | 0,215 ± 0,01 * | 0,285 ± 0,07 * | |
| V162M | FM | 395 ± 59 ** | 0,039 ± 0,001 * | 0,274 ± 0,03 *** | 0,355 ± 0,12 * |
| ST | 271 ± 57 * | 0,058 ± 0,01 ** | 0,173 ± 0,1 ** | 0,18 ± 0,03 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, R.; Del Fiore, V.S.; Saveri, P.; Palamà, I.E.; Pisciotta, C.; Pareyson, D.; Bucci, C.; Guerra, F. Autophagy and Lysosomal Functionality in CMT2B Fibroblasts Carrying the RAB7K126R Mutation. Cells 2022, 11, 496. https://doi.org/10.3390/cells11030496
Romano R, Del Fiore VS, Saveri P, Palamà IE, Pisciotta C, Pareyson D, Bucci C, Guerra F. Autophagy and Lysosomal Functionality in CMT2B Fibroblasts Carrying the RAB7K126R Mutation. Cells. 2022; 11(3):496. https://doi.org/10.3390/cells11030496
Chicago/Turabian StyleRomano, Roberta, Victoria Stefania Del Fiore, Paola Saveri, Ilaria Elena Palamà, Chiara Pisciotta, Davide Pareyson, Cecilia Bucci, and Flora Guerra. 2022. "Autophagy and Lysosomal Functionality in CMT2B Fibroblasts Carrying the RAB7K126R Mutation" Cells 11, no. 3: 496. https://doi.org/10.3390/cells11030496
APA StyleRomano, R., Del Fiore, V. S., Saveri, P., Palamà, I. E., Pisciotta, C., Pareyson, D., Bucci, C., & Guerra, F. (2022). Autophagy and Lysosomal Functionality in CMT2B Fibroblasts Carrying the RAB7K126R Mutation. Cells, 11(3), 496. https://doi.org/10.3390/cells11030496








