βIII-Tubulin Structural Domains Regulate Mitochondrial Network Architecture in an Isotype-Specific Manner
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
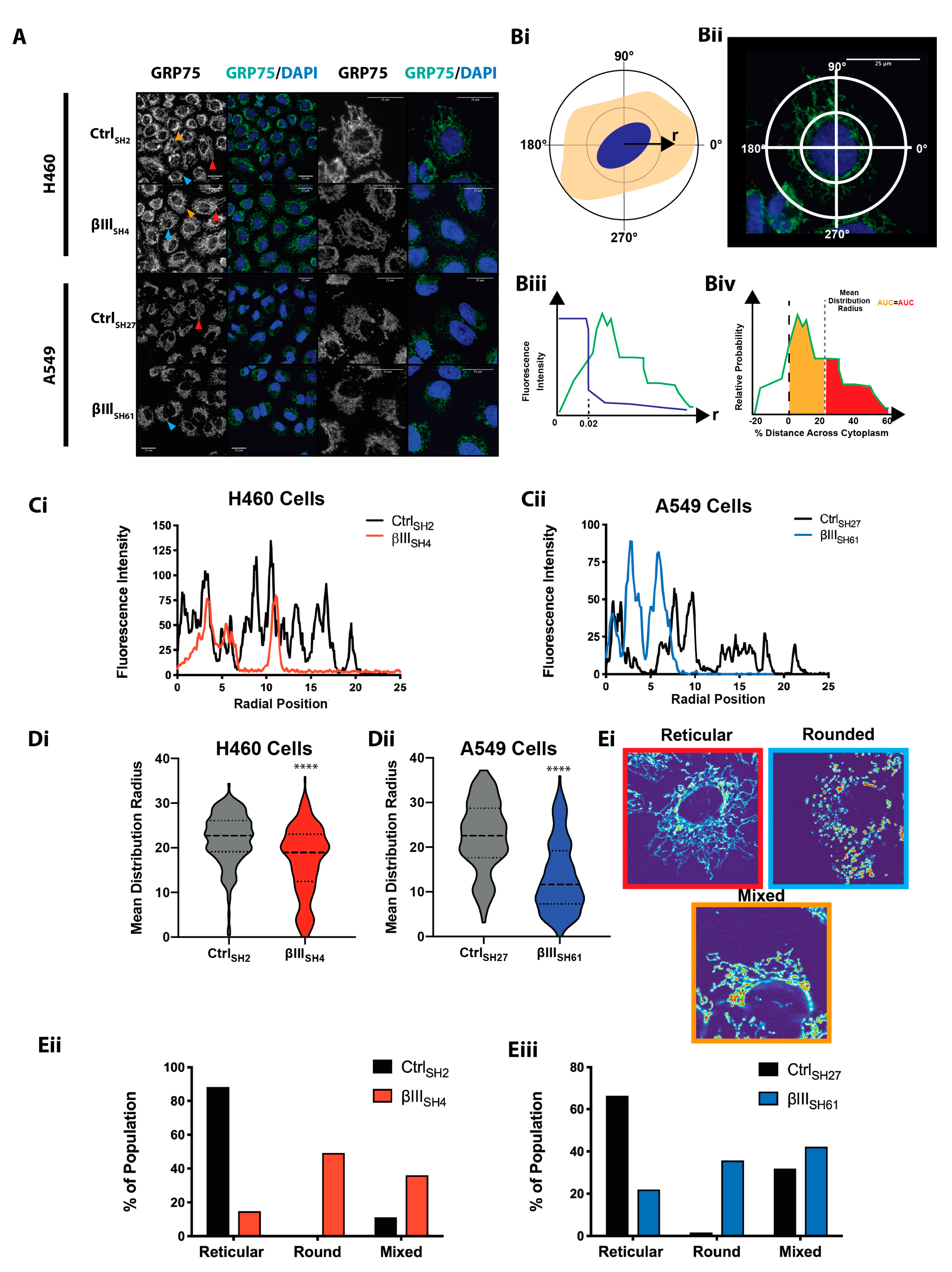
2.2. Mitochondrial Network Immunofluorescence
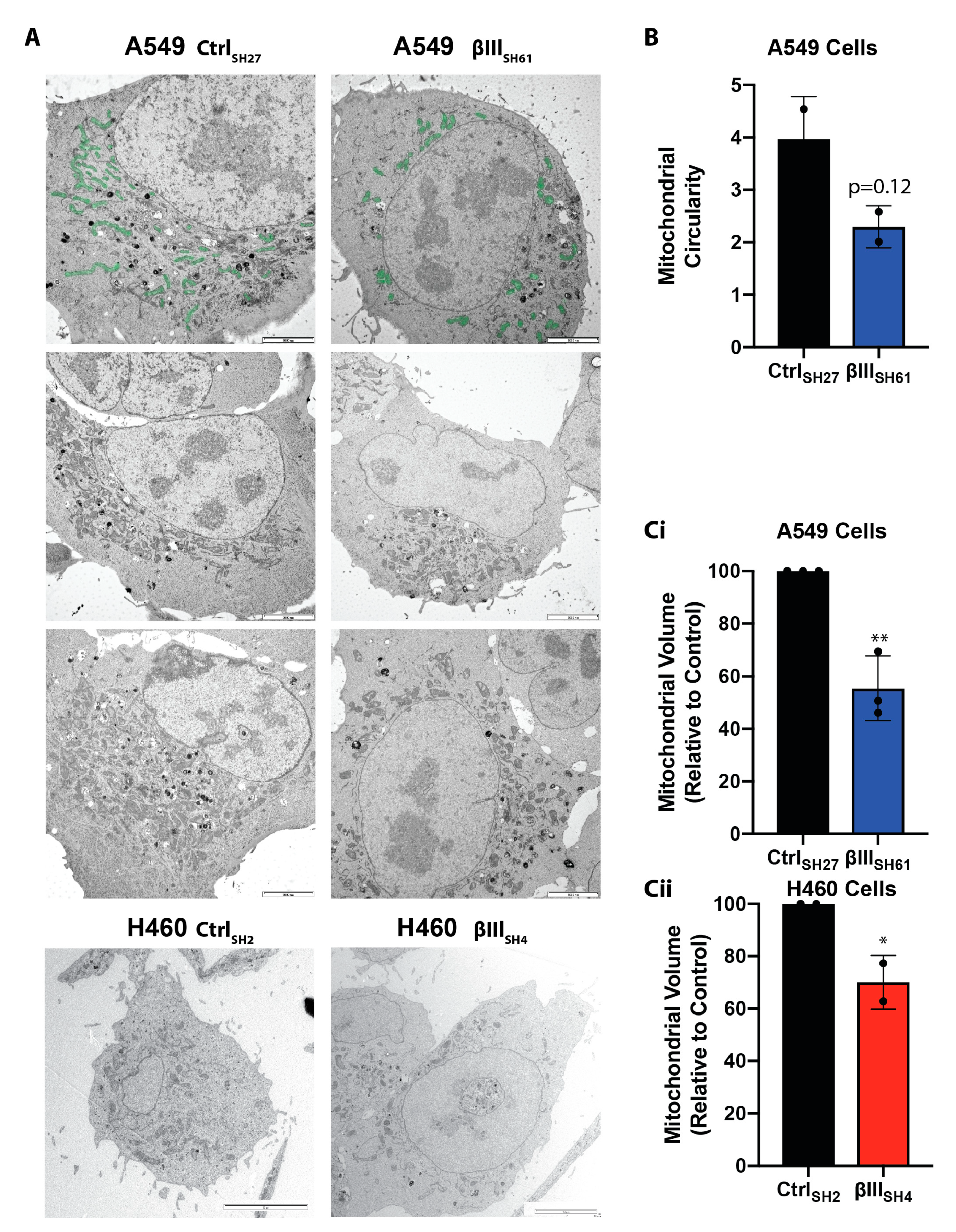
2.3. Transmission Electron Microscopy
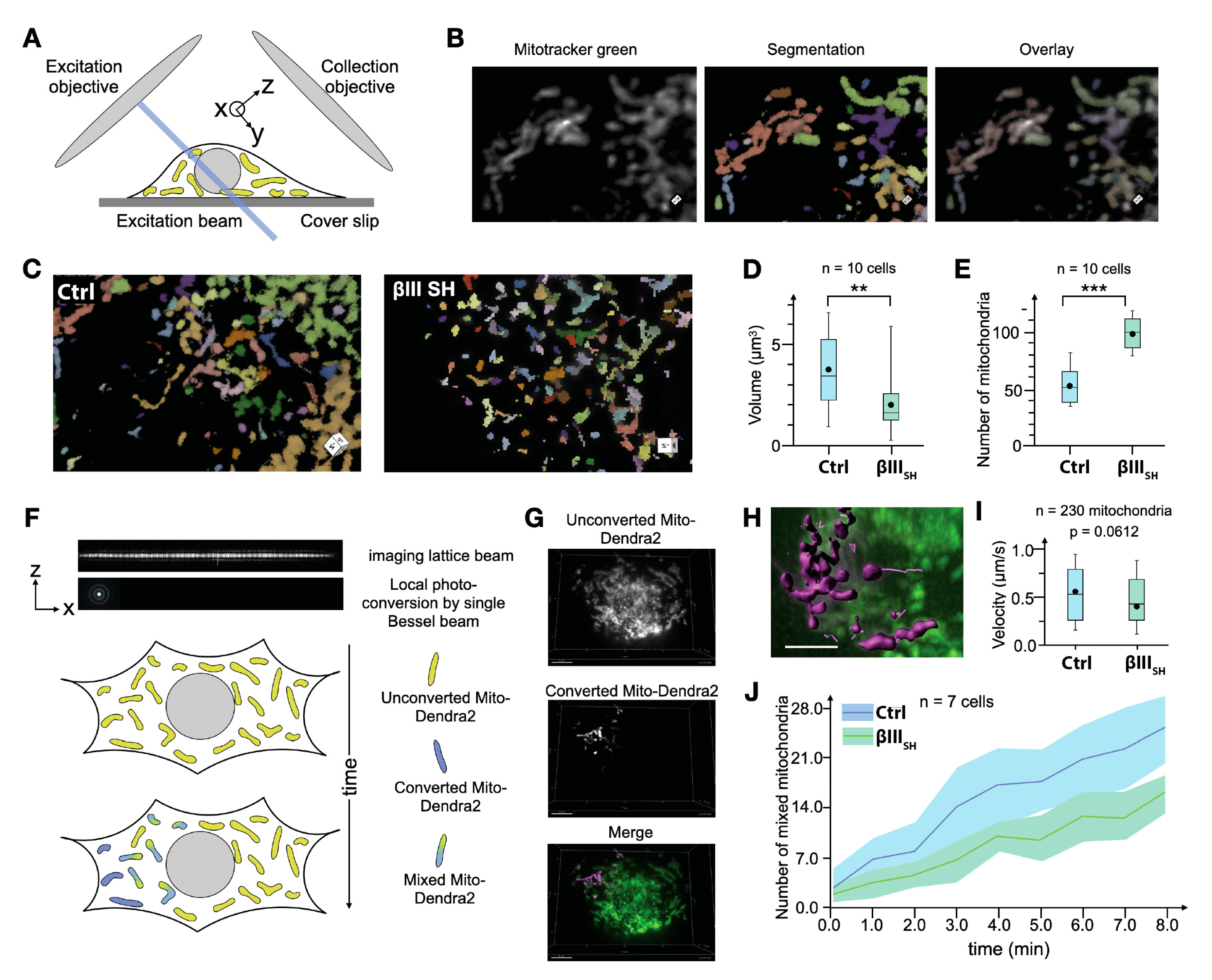
2.4. Preparation of NCI-H460 cells for Lattice Light-Sheet Imaging of Mitochondria
2.5. Lattice-Light Sheet Imaging Mitochondrial Dynamics
2.6. Analysis of Lattice-Light Sheet Imaging of Mitochondrial Dynamics
2.7. Code Availability
2.8. Statistical Analysis
3. Results
3.1. βIII-Tubulin Regulates the Mitochondrial Network Structure and Morphology
3.2. βIII-Tubulin Regulates Mitochondrial Volume
3.3. The βIII-Tubulin C-Terminal Tail Regulates the Mitochondrial Network Structure and Morphology
3.4. βIII-Tubulin Promotes a Dynamic Mitochondrial Network
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopata, M.A.; Cleveland, D. In vivo microtubules are copolymers of available beta-tubulin isotypes: Localization of each of six vertebrate beta-tubulin isotypes using polyclonal antibodies elicited by synthetic peptide antigens. J. Cell Biol. 1987, 105, 1707–1720. [Google Scholar] [CrossRef]
- Parker, A.L.; Ekavallaris, M.; McCarroll, J.A. Microtubules and Their Role in Cellular Stress in Cancer. Front. Oncol. 2014, 4, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, A.L.; Teo, W.S.; Pandzic, E.; Vicente, J.J.; McCarroll, J.A.; Wordeman, L.; Kavallaris, M. β-Tubulin carboxy-terminal tails exhibit isotype-specific effects on microtubule dynamics in human gene-edited cells. Life Sci. Alliance 2018, 1, e201800059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, A.L.; Turner, N.; McCarroll, J.; Kavallaris, M. βIII-Tubulin alters glucose metabolism and stress response signaling to promote cell survival and proliferation in glucose-starved non-small cell lung cancer cells. Carcinogenesis 2016, 37, 787–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarroll, J.; Gan, P.P.; Erlich, R.B.; Liu, M.; Dwarte, T.; Sagnella, S.S.; Akerfeldt, M.C.; Yang, L.; Parker, A.L.; Chang, M.; et al. TUBB3/βIII-Tubulin Acts through the PTEN/AKT Signaling Axis to Promote Tumorigenesis and Anoikis Resistance in Non–Small Cell Lung Cancer. Cancer Res. 2014, 75, 415–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Carré, M.; André, N.; Carles, G.; Borghi, H.; Brichese, L.; Briand, C.; Braguer, D. Tubulin Is an Inherent Component of Mitochondrial Membranes That Interacts with the Voltage-dependent Anion Channel. J. Biol. Chem. 2002, 277, 33664–33669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appaix, F.; Kuznetsov, A.V.; Usson, Y.; Kay, L.; Andrienko, T.; Olivares, J.; Kaambre, T.; Sikk, P.; Margreiter, R.; Saks, V. Possible Role of Cytoskeleton in Intracellular Arrangement and Regulation of Mitochondria. Exp. Physiol. 2003, 88, 175–190. [Google Scholar] [CrossRef] [Green Version]
- Anesti, V.; Scorrano, L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim. Biophys. Acta 2006, 1757, 692–699. [Google Scholar] [CrossRef]
- Pareyson, D.; Saveri, P.; Sagnelli, A.; Piscosquito, G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci. Lett. 2015, 596, 66–77. [Google Scholar] [CrossRef]
- Ren, Y.; Jiang, H.; Yang, F.; Nakaso, K.; Feng, J. Parkin Protects Dopaminergic Neurons against Microtubule-depolymerizing Toxins by Attenuating Microtubule-associated Protein Kinase Activation. J. Biol. Chem. 2009, 284, 4009–4017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Y.; Zhao, J.; Feng, J. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J. Neurosci. 2003, 23, 3316–3324. [Google Scholar] [CrossRef] [Green Version]
- Silva, D.F.; Esteves, A.R.; Arduino, D.M.; Oliveira, C.R.; Cardoso, S.M. Amyloid-β-Induced Mitochondrial Dysfunction Impairs the Autophagic Lysosomal Pathway in a Tubulin Dependent Pathway. J. Alzheimer’s Dis. 2011, 26, 565–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, R.; Wang, F.; McKeehan, W.L.; Liu, L. Autophagy Enhanced by Microtubule- and Mitochondrion-Associated MAP1S Suppresses Genome Instability and Hepatocarcinogenesis. Cancer Res. 2011, 71, 7537–7546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.-Y.; Sheng, Z.-H. Regulation of mitochondrial transport in neurons. Exp. Cell Res. 2015, 334, 35–44. [Google Scholar] [CrossRef] [Green Version]
- MacAskill, A.F.; Kittler, J.T. Control of mitochondrial transport and localization in neurons. Trends Cell Biol. 2010, 20, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.-H. Mitochondrial trafficking and anchoring in neurons: New insight and implications. J. Cell Biol. 2014, 204, 1087–1098. [Google Scholar] [CrossRef] [Green Version]
- Vemu, A.; Atherton, J.; Spector, J.O.; Moores, C.A.; Roll-Mecak, A. Tubulin isoform composition tunes microtubule dynamics. Mol. Biol. Cell 2017, 28, 3564–3572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, Y.; Tsuchiya, K.; Sumiyoshi, E.; Haruta, N.; Sugimoto, A. Tubulin isotype substitution revealed that isotype composition modulates microtubule dynamics in C. elegans embryos. J. Cell Sci. 2017, 130, 1652–1661. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Obara, C.J.; Szczesna, E.; Nixon-Abell, J.; Mahalingan, K.K.; Roll-Mecak, A.; Lippincott-Schwartz, J.; Blackstone, C. ER proteins decipher the tubulin code to regulate organelle distribution. Nature 2021, 601, 132–138. [Google Scholar] [CrossRef]
- Panda, D.; Miller, H.P.; Banerjee, A.; Luduena, R.F.; Wilson, L. Microtubule dynamics in vitro are regulated by the tubulin isotype composition. Proc. Natl. Acad. Sci. USA 1994, 91, 11358–11362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamath, K.; Wilson, L.; Cabral, F.; Jordan, M.A. βIII-Tubulin Induces Paclitaxel Resistance in Association with Reduced Effects on Microtubule Dynamic Instability. J. Biol. Chem. 2005, 280, 12902–12907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, P.P.; McCarroll, J.; Po’Uha, S.T.; Kamath, K.; Jordan, M.A.; Kavallaris, M. Microtubule Dynamics, Mitotic Arrest, and Apoptosis: Drug-Induced Differential Effects of βIII-Tubulin. Mol. Cancer Ther. 2010, 9, 1339–1348. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.; Jain, D.; Costa, J.; Velve-Casquillas, G.; Tran, P.T. mmb1p Binds Mitochondria to Dynamic Microtubules. Curr. Biol. 2011, 21, 1431–1439. [Google Scholar] [CrossRef] [Green Version]
- Mehta, K.; Chacko, L.A.; Chug, M.K.; Jhunjhunwala, S.; Ananthanarayanan, V. Association of mitochondria with microtubules inhibits mitochondrial fission by precluding assembly of the fission protein Dnm1. J. Biol. Chem. 2019, 294, 3385–3396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaffe, M.P.; Stuurman, N.; Vale, R.D. Mitochondrial positioning in fission yeast is driven by association with dynamic microtubules and mitotic spindle poles. Proc. Natl. Acad. Sci. USA 2003, 100, 11424–11428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McVicker, D.P.; Chrin, L.R.; Berger, C.L. The Nucleotide-binding State of Microtubules Modulates Kinesin Processivity and the Ability of Tau to Inhibit Kinesin-mediated Transport. J. Biol. Chem. 2011, 286, 42873–42880. [Google Scholar] [CrossRef] [Green Version]
- Feizabadi, M.S. The Contribution of the C-Terminal Tails of Microtubules in Altering the Force Production Specifications of Multiple Kinesin-1. Cell Biophys. 2016, 74, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Hirose, K.; Löwe, J.; Alonso, M.; Cross, R.; Amos, L. 3D Electron Microscopy of the Interaction of Kinesin with Tubulin. Cell Struct. Funct. 1999, 24, 277–284. [Google Scholar] [CrossRef] [Green Version]
- Sirajuddin, M.; Rice, L.M.; Vale, R.D. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 2014, 16, 335–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, Y.; Kanai, Y.; Okada, Y.; Nonaka, S.; Takeda, S.; Harada, A.; Hirokawa, N. Targeted Disruption of Mouse Conventional Kinesin Heavy Chain kif5B, Results in Abnormal Perinuclear Clustering of Mitochondria. Cell 1998, 93, 1147–1158. [Google Scholar] [CrossRef] [Green Version]
- Pilling, A.D.; Horiuchi, D.; Lively, C.M.; Saxton, W.M. Kinesin-1 and Dynein Are the Primary Motors for Fast Transport of Mitochondria inDrosophilaMotor Axons. Mol. Biol. Cell 2006, 17, 2057–2068. [Google Scholar] [CrossRef] [Green Version]
- Rostovtseva, T.K.; Gurnev, P.A.; Hoogerheide, D.P.; Rovini, A.; Sirajuddin, M.; Bezrukov, S.M.; Rostovtseva, T.K.; Gurnev, P.A.; Hoogerheide, D.P.; Rovini, A.; et al. Sequence diversity of tubulin isotypes in regulation of the mitochondrial voltage-dependent anion channel. J. Biol. Chem. 2018, 293, 10949–10962. [Google Scholar] [CrossRef] [Green Version]
- Rostovtseva, T.K.; Sheldon, K.L.; Hassanzadeh, E.; Monge, C.; Saks, V.; Bezrukov, S.M.; Sackett, D.L. Tubulin binding blocks mitochondrial voltage-dependent anion channel and regulates respiration. Proc. Natl. Acad. Sci. USA 2008, 105, 18746–18751. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, K.L.; Maldonado, E.N.; Lemasters, J.J.; Rostovtseva, T.K.; Bezrukov, S.M. Phosphorylation of Voltage-Dependent Anion Channel by Serine/Threonine Kinases Governs Its Interaction with Tubulin. PLoS ONE 2011, 6, e25539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, D.; Filliol, D.; Miehe, M.; Rendón, A. Interaction of brain mitochondria with microtubules reconstituted from brain tubulin and MAP2 or TAU. Cell Motil. Cytoskelet. 1993, 24, 245–255. [Google Scholar] [CrossRef]
- Cicchillitti, L.; Penci, R.; Di Michele, M.; Filippetti, F.; Rotilio, D.; Donati, M.B.; Scambia, G.; Ferlini, C. Proteomic characterization of cytoskeletal and mitochondrial class III β-tubulin. Mol. Cancer Ther. 2008, 7, 2070–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovini, A.; Savry, A.; Braguer, D.; Carré, M. Microtubule-targeted agents: When mitochondria become essential to chemotherapy. Biochim. et Biophys. Acta 2011, 1807, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Bernier-Valentin, F.; Aunis, D.; Rousset, B. Evidence for tubulin-binding sites on cellular membranes: Plasma membranes, mitochondrial membranes, and secretory granule membranes. J. Cell Biol. 1983, 97, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Tischfield, M.A.; Engle, E.C. Distinct α- and β-tubulin isotypes are required for the positioning, differentiation and survival of neurons: New support for the ‘multi-tubulin’ hypothesis. Biosci. Rep. 2010, 30, 319–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tischfield, M.A.; Baris, H.N.; Wu, C.; Rudolph, G.; Van Maldergem, L.; He, W.; Chan, W.-M.; Andrews, C.; Demer, J.L.; Robertson, R.L.; et al. Human TUBB3 Mutations Perturb Microtubule Dynamics, Kinesin Interactions, and Axon Guidance. Cell 2010, 140, 74–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, S.; Balasubramanian, R.; Chan, W.-M.; Kang, P.; Andrews, C.; Webb, B.D.; MacKinnon, S.E.; Oystreck, D.T.; Rankin, J.; Crawford, T.O.; et al. A novel syndrome caused by the E410K amino acid substitution in the neuronal β-tubulin isotype 3. Brain 2013, 136, 522–535. [Google Scholar] [CrossRef] [Green Version]
- Niwa, S.; Takahashi, H.; Hirokawa, N. β-Tubulin mutations that cause severe neuropathies disrupt axonal transport. EMBO J. 2013, 32, 1352–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarroll, J.A.; Gan, P.P.; Liu, M.; Kavallaris, M. βIII-Tubulin Is a Multifunctional Protein Involved in Drug Sensitivity and Tumorigenesis in Non–Small Cell Lung Cancer. Cancer Res. 2010, 70, 4995–5003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariotti, N.; Wu, Y.; Okano, S.; Gambin, Y.; Follett, J.; Rae, J.; Ferguson, C.; Teasdale, R.D.; Alexandrov, K.; Meunier, F.A.; et al. An inverted CAV1 (caveolin 1) topology defines novel autophagy-dependent exosome secretion from prostate cancer cells. Autophagy 2020, 17, 2200–2216. [Google Scholar] [CrossRef] [PubMed]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Maier, B.; Baillie, G.J.; Ferguson, C.; Parton, R.G.; Wolvetang, E.J.; Roost, M.S.; De Sousa, S.M.C.L.; et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 2015, 526, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Jaqaman, K.; Loerke, D.; Mettlen, M.; Kuwata, H.; Grinstein, S.; Schmid, S.; Danuser, G. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 2008, 5, 695–702. [Google Scholar] [CrossRef] [Green Version]
- Gan, P.P.; Pasquier, E.; Kavallaris, M. Class III β-Tubulin Mediates Sensitivity to Chemotherapeutic Drugs in Non–Small Cell Lung Cancer. Cancer Res. 2007, 67, 9356–9363. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, T.; Karnezis, A.N.; Murphy, S.P.; Hoang, T.; Freeman, B.C.; Phillips, B.; Morimoto, R.I. Cloning and Subcellular Localization of Human Mitochondrial hsp70. J. Biol. Chem. 1995, 270, 1705–1710. [Google Scholar] [CrossRef] [Green Version]
- Ouellet, M.; Guillebaud, G.; Gervais, V.; St-Pierre, D.L.; Germain, M. A novel algorithm identifies stress-induced alterations in mitochondrial connectivity and inner membrane structure from confocal images. PLOS Comput. Biol. 2017, 13, e1005612. [Google Scholar] [CrossRef] [Green Version]
- Lemasters, J.J.; Qian, T.; He, L.; Kim, J.-S.; Elmore, S.P.; Cascio, W.E.; Brenner, D. Role of Mitochondrial Inner Membrane Permeabilization in Necrotic Cell Death, Apoptosis, and Autophagy. Antioxidants Redox Signal. 2002, 4, 769–781. [Google Scholar] [CrossRef]
- Rossignol, R.; Gilkerson, R.; Aggeler, R.; Yamagata, K.; Remington, S.J.; Capaldi, R.A. Energy Substrate Modulates Mitochondrial Structure and Oxidative Capacity in Cancer Cells. Cancer Res. 2004, 64, 985–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, L.C.; Di Benedetto, G.; Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011, 13, 589–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambold, A.S.; Kostelecky, B.; Elia, N.; Lippincott-Schwartz, J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA 2011, 108, 10190–10195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Ma, X.; Fujioka, H.; Liu, J.; Chen, S.; Zhu, X. DJ-1 regulates the integrity and function of ER-mitochondria association through interaction with IP3R3-Grp75-VDAC1. Proc. Natl. Acad. Sci. USA 2019, 116, 25322–25328. [Google Scholar] [CrossRef]
- Canet-Avilés, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Baptista, M.J.; Ringe, D.; Petsko, G.A.; Cookson, M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarroll, J.A.; Sharbeen, G.; Liu, J.; Youkhana, J.; Goldstein, D.; McCarthy, N.; Limbri, L.F.; Dischl, D.; Ceyhan, G.O.; Erkan, M.; et al. beta III-Tubulin: A novel mediator of chemoresistance and metastases in pan-creatic cancer. Oncotarget 2015, 6, 2235–2249. [Google Scholar] [CrossRef] [Green Version]
- Al-Mehdi, A.-B.; Pastukh, V.M.; Swiger, B.M.; Reed, D.J.; Patel, M.R.; Bardwell, G.C.; Pastukh, V.V.; Alexeyev, M.F.; Gillespie, M.N. Perinuclear Mitochondrial Clustering Creates an Oxidant-Rich Nuclear Domain Required for Hypoxia-Induced Transcription. Sci. Signal. 2012, 5, ra47. [Google Scholar] [CrossRef] [Green Version]
- Strohecker, A.M.; Guo, J.Y.; Karsli-Uzunbas, G.; Price, S.M.; Chen, G.J.; Mathew, R.; McMahon, M.; White, E. Autophagy Sustains Mitochondrial Glutamine Metabolism and Growth of BrafV600E–Driven Lung Tumors. Cancer Discov. 2013, 3, 1272–1285. [Google Scholar] [CrossRef] [Green Version]
- Karsli-Uzunbas, G.; Guo, J.Y.; Price, S.; Teng, X.; Laddha, S.V.; Khor, S.; Kalaany, N.Y.; Jacks, T.; Chan, C.S.; Rabinowitz, J.D.; et al. Autophagy Is Required for Glucose Homeostasis and Lung Tumor Maintenance. Cancer Discov. 2014, 4, 914–927. [Google Scholar] [CrossRef] [Green Version]
- Caino, M.C.; Altieri, D.C. Cancer cells exploit adaptive mitochondrial dynamics to increase tumor cell invasion. Cell Cycle 2015, 14, 3242–3247. [Google Scholar] [CrossRef] [Green Version]
- Zala, D.; Hinckelmann, M.-V.; Yu, H.; Lyra Da Cunha, M.M.; Liot, G.; Cordelières, F.P.; Marco, S.; Saudou, F. Vesicular Glycolysis Provides On-Board Energy for Fast Axonal Transport. Cell 2013, 152, 479–491. [Google Scholar] [CrossRef] [Green Version]
- Parrado-Fernández, C.; Schreiner, B.; Ankarcrona, M.; Conti, M.M.; Cookson, M.R.; Kivipelto, M.; Cedazo-Minguez, A.; Sandebring-Matton, A. Reduction of PINK1 or DJ-1 impair mitochondrial motility in neurites and alter ER-mitochondria contacts. J. Cell. Mol. Med. 2018, 22, 5439–5449. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Heng, X.; Zhang, G.; Xiong, R.; Li, H.; Zhang, S.; Chen, S. DJ-1 deficiency perturbs microtubule dynamics and impairs striatal neurite outgrowth. Neurobiol. Aging 2013, 34, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, E.N.; Sheldon, K.L.; DeHart, D.N.; Patnaik, J.; Manevich, Y.; Townsend, D.M.; Bezrukov, S.M.; Rostovtseva, T.K.; Lemasters, J.J. Voltage-dependent Anion Channels Modulate Mitochondrial Metabolism in Cancer Cells. J. Biol. Chem. 2013, 288, 11920–11929. [Google Scholar] [CrossRef] [Green Version]
- Guzun, R.; Karu-Varikmaa, M.; Gonzalez-Granillo, M.; Kuznetsov, A.V.; Michel, L.; Cottet-Rousselle, C.; Saaremäe, M.; Kaambre, T.; Metsis, M.; Grimm, M.; et al. Mitochondria–cytoskeleton interaction: Distribution of β-tubulins in cardiomyocytes and HL-1 cells. Biochim. Biophys. Acta 2011, 1807, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Granillo, M.; Grichine, A.; Guzun, R.; Usson, Y.; Tepp, K.; Chekulayev, V.; Shevchuk, I.; Karu-Varikmaa, M.; Kuznetsov, A.V.; Grimm, M.; et al. Studies of the role of tubulin beta II isotype in regulation of mitochondrial respiration in intracellular energetic units in cardiac cells. J. Mol. Cell. Cardiol. 2012, 52, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Varikmaa, M.; Bagur, R.; Kaambre, T.; Grichine, A.; Timohhina, N.; Tepp, K.; Shevchuk, I.; Chekulayev, V.; Metsis, M.; Boucher, F.; et al. Role of mitochondria–cytoskeleton interactions in respiration regulation and mitochondrial organization in striated muscles. Biochim. Biophys. Acta 2014, 1837, 232–245. [Google Scholar] [CrossRef] [Green Version]
- Vulinovic, F.; Krajka, V.; Hausrat, T.J.; Seibler, P.; Alvarez-Fischer, D.; Madoev, H.; Park, J.-S.; Kumar, K.R.; Sue, C.M.; Lohmann, K.; et al. Motor protein binding and mitochondrial transport are altered by pathogenic TUBB4A variants. Hum. Mutat. 2018, 39, 1901–1915. [Google Scholar] [CrossRef]
- Rezania, V.; Azarenko, O.; Jordan, M.A.; Bolterauer, H.; Ludueña, R.F.; Huzil, J.T.; Tuszynski, J.A. Microtubule Assembly of Isotypically Purified Tubulin and Its Mixtures. Biophys. J. 2008, 95, 1993–2008. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parker, A.L.; Teo, W.S.; Brayford, S.; Moorthi, U.K.; Arumugam, S.; Ferguson, C.; Parton, R.G.; McCarroll, J.A.; Kavallaris, M. βIII-Tubulin Structural Domains Regulate Mitochondrial Network Architecture in an Isotype-Specific Manner. Cells 2022, 11, 776. https://doi.org/10.3390/cells11050776
Parker AL, Teo WS, Brayford S, Moorthi UK, Arumugam S, Ferguson C, Parton RG, McCarroll JA, Kavallaris M. βIII-Tubulin Structural Domains Regulate Mitochondrial Network Architecture in an Isotype-Specific Manner. Cells. 2022; 11(5):776. https://doi.org/10.3390/cells11050776
Chicago/Turabian StyleParker, Amelia L., Wee Siang Teo, Simon Brayford, Ullhas K. Moorthi, Senthil Arumugam, Charles Ferguson, Robert G. Parton, Joshua A. McCarroll, and Maria Kavallaris. 2022. "βIII-Tubulin Structural Domains Regulate Mitochondrial Network Architecture in an Isotype-Specific Manner" Cells 11, no. 5: 776. https://doi.org/10.3390/cells11050776





