Enhanced Wild-Type MET Receptor Levels in Mouse Hepatocytes Attenuates Insulin-Mediated Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animal Experimentation
2.3. Transcriptome Analysis by RNA-Seq
2.4. Analysis of Publicly Available RNA-Seq Data
2.5. Analysis of Plasma Insulin Levels
2.6. Glucose, Insulin, and Pyruvate Tolerance Tests (GTT, ITT, PTT)
2.7. Indirect Calorimetry
2.8. Homogenization and Preparation of Tissue Extracts
2.9. Isolation and Culture of Primary Mouse Hepatocytes
2.10. Western Blot Analysis
2.11. Co-Immunoprecipitation Assays
2.12. Data Analysis
3. Results
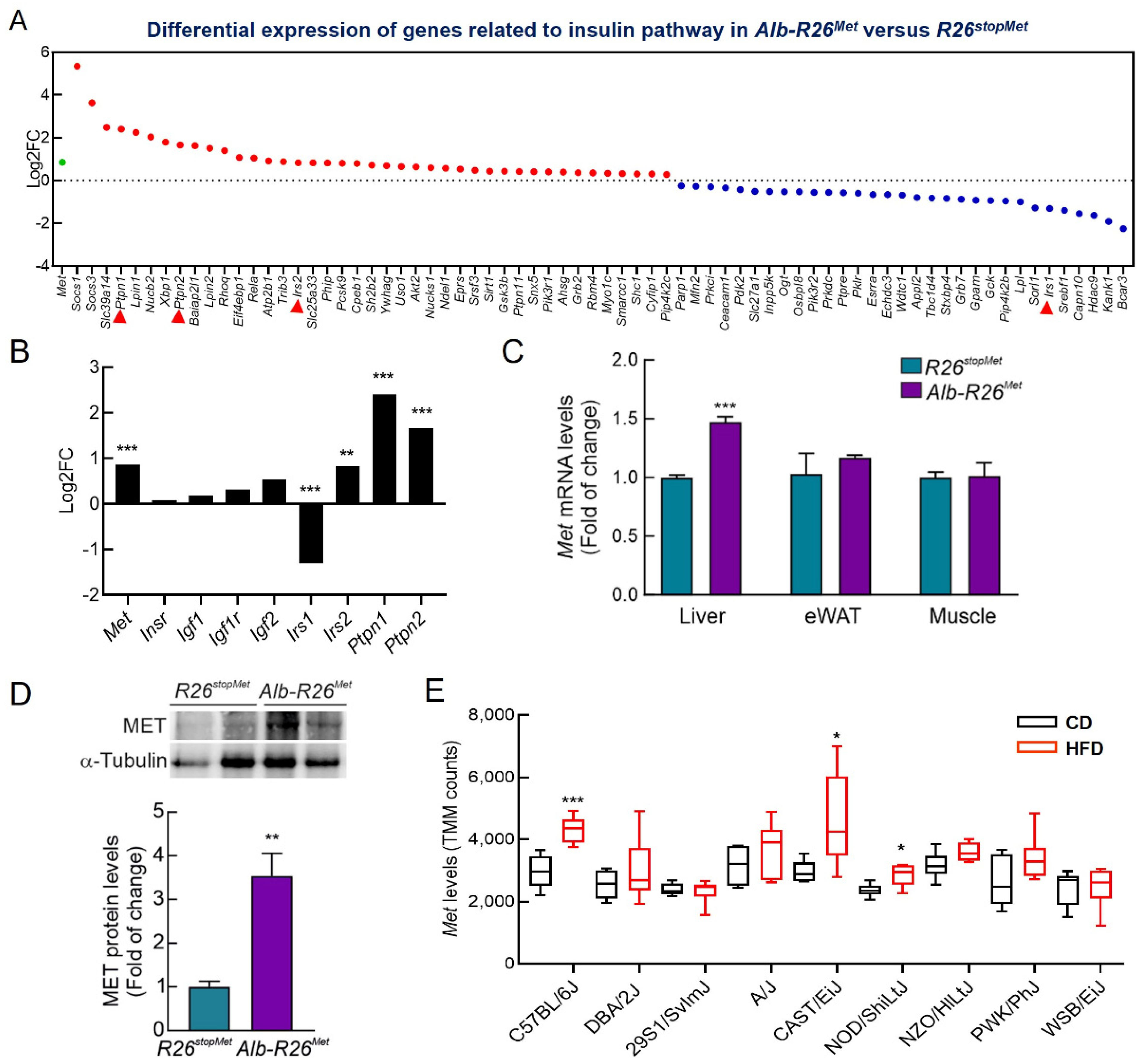
3.1. A Slight Increase in MET Levels Perturbs the Expression of a Set of Genes Related to Insulin Signaling
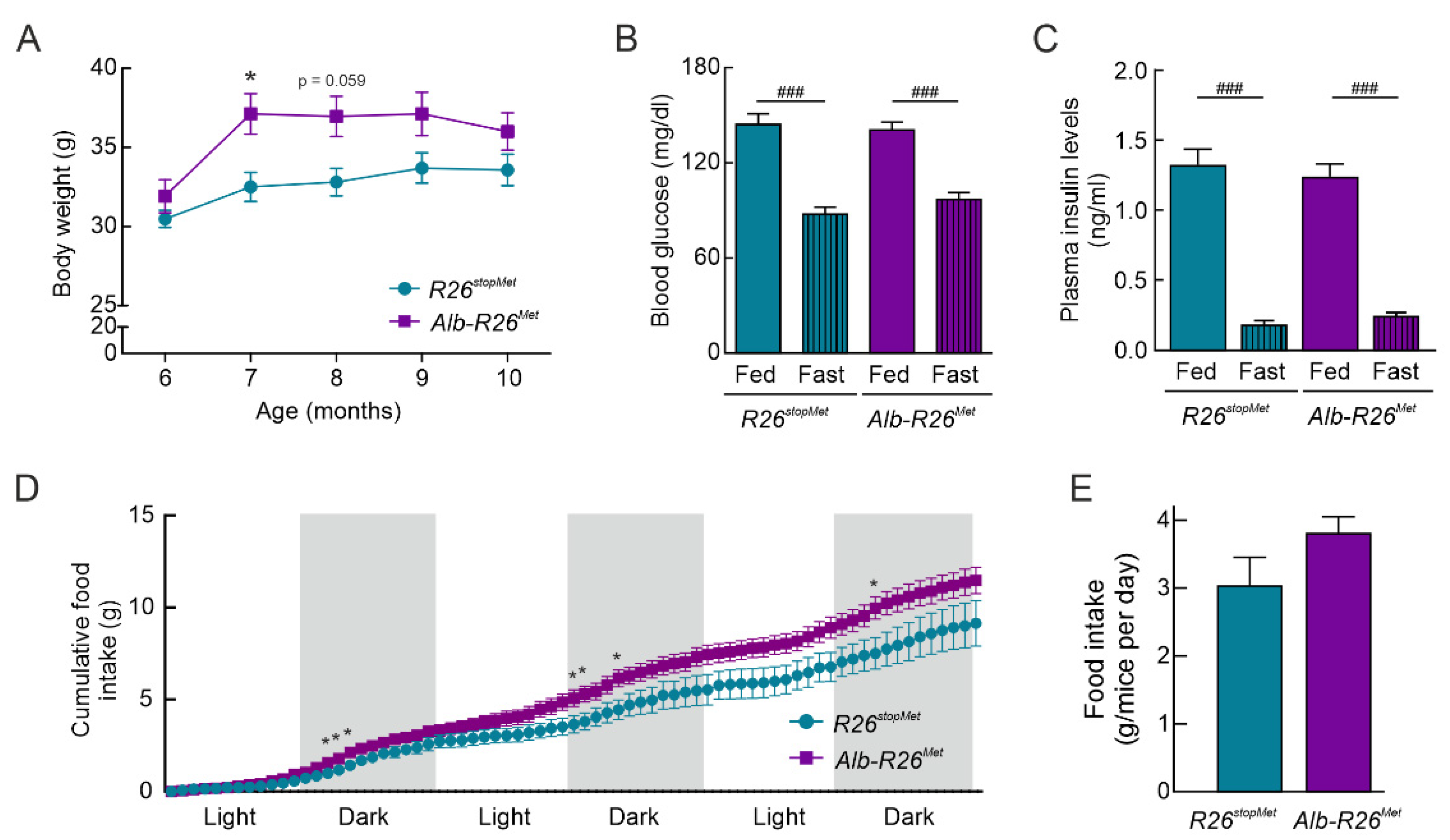
3.2. A Slight Enhancement of MET Levels in the Liver Results in Increased in Body Weight and Food Intake
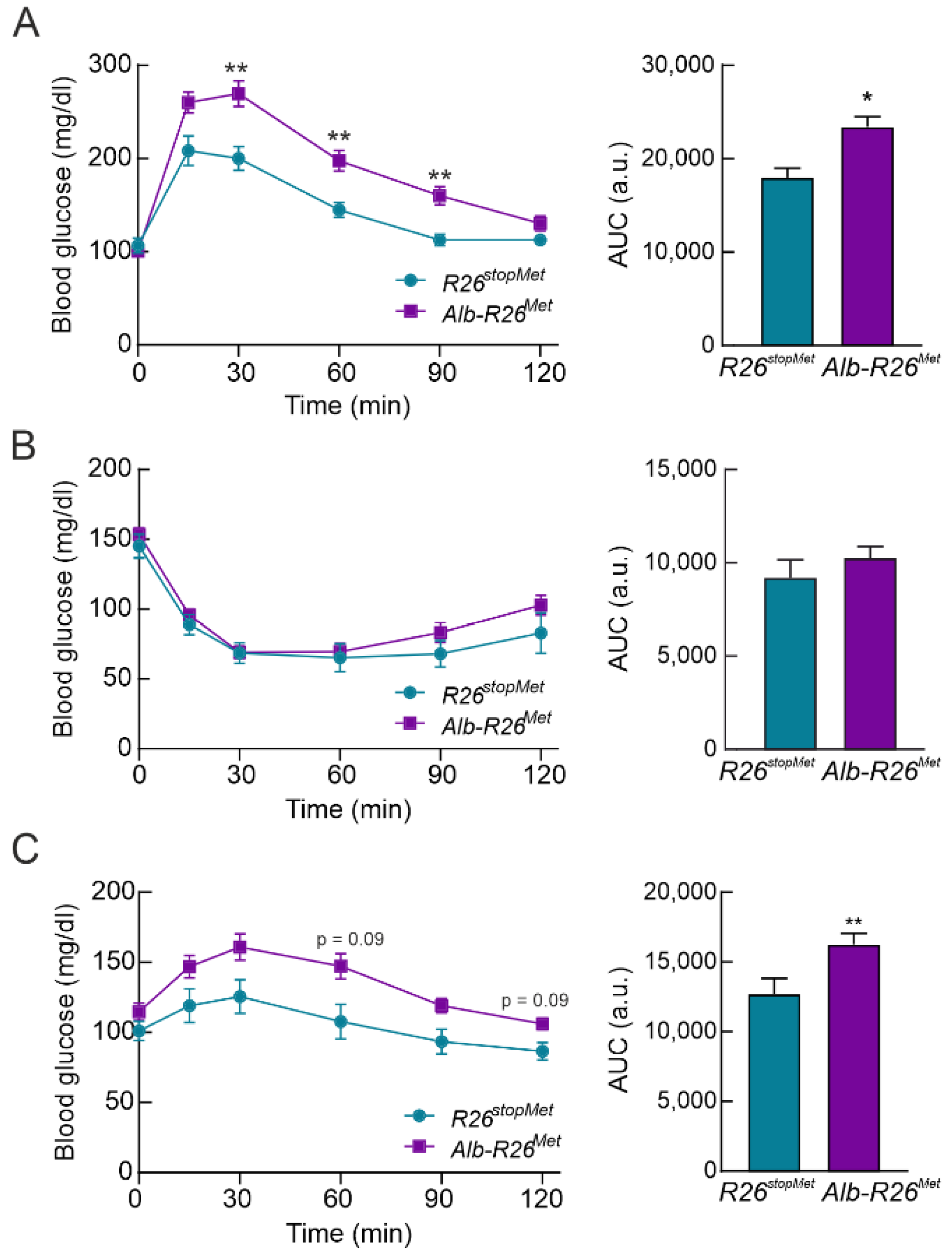
3.3. Hepatocyte-Specific Enhanced MET Levels Impair Glucose Homeostasis In Vivo
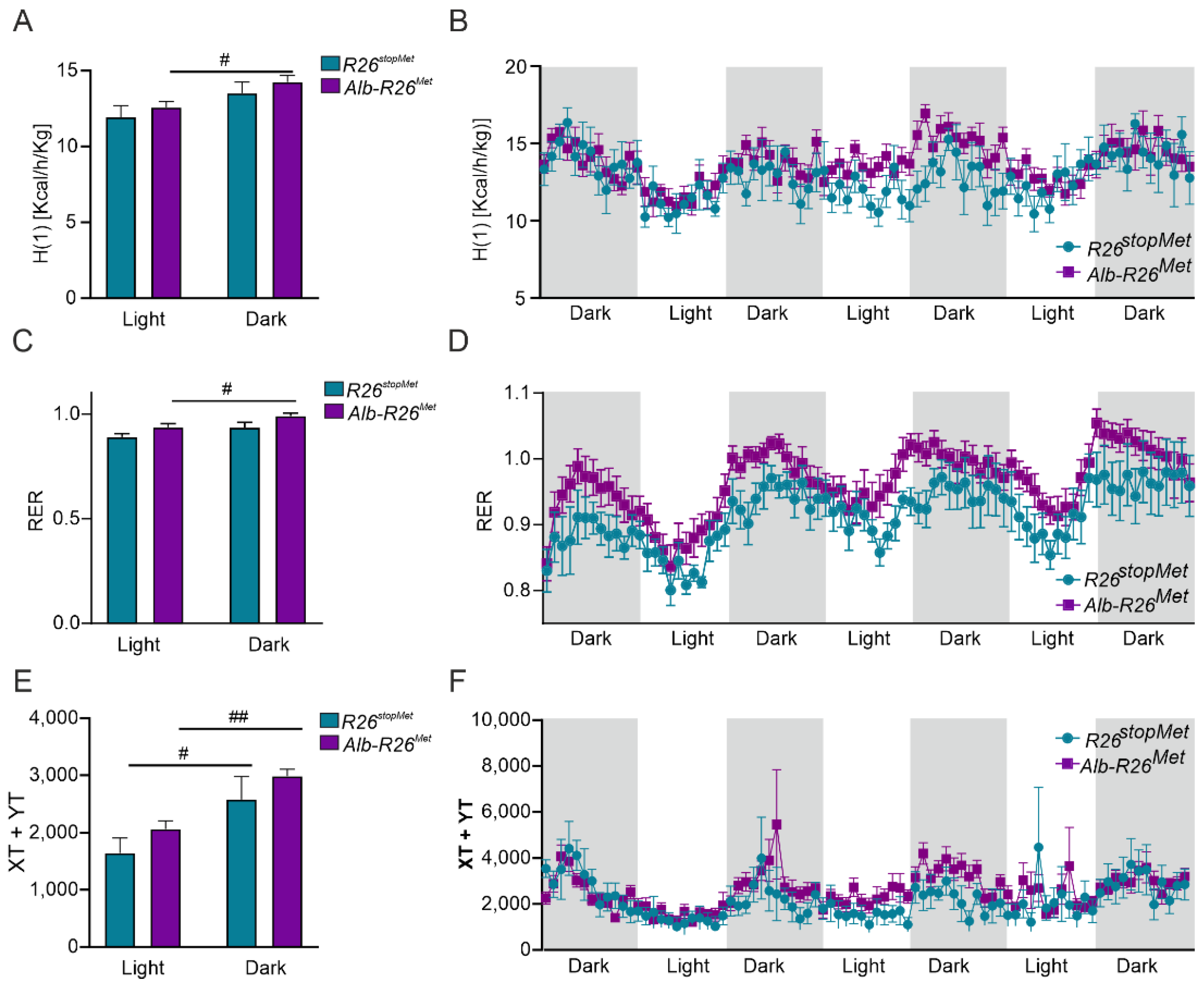
3.4. Enhanced MET Levels in Hepatocytes Do Not Severely Alter Whole-Body Energy Metabolism
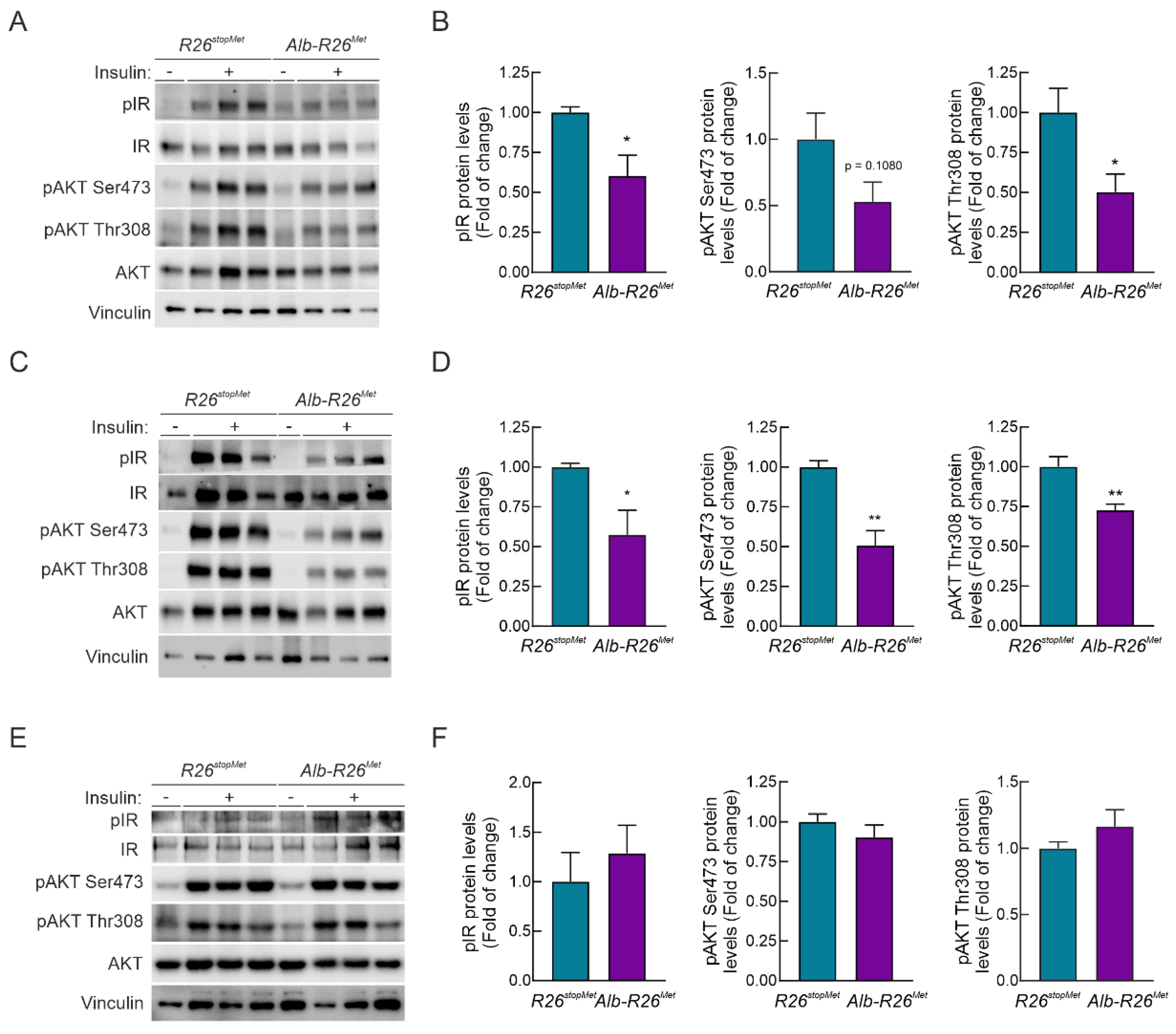
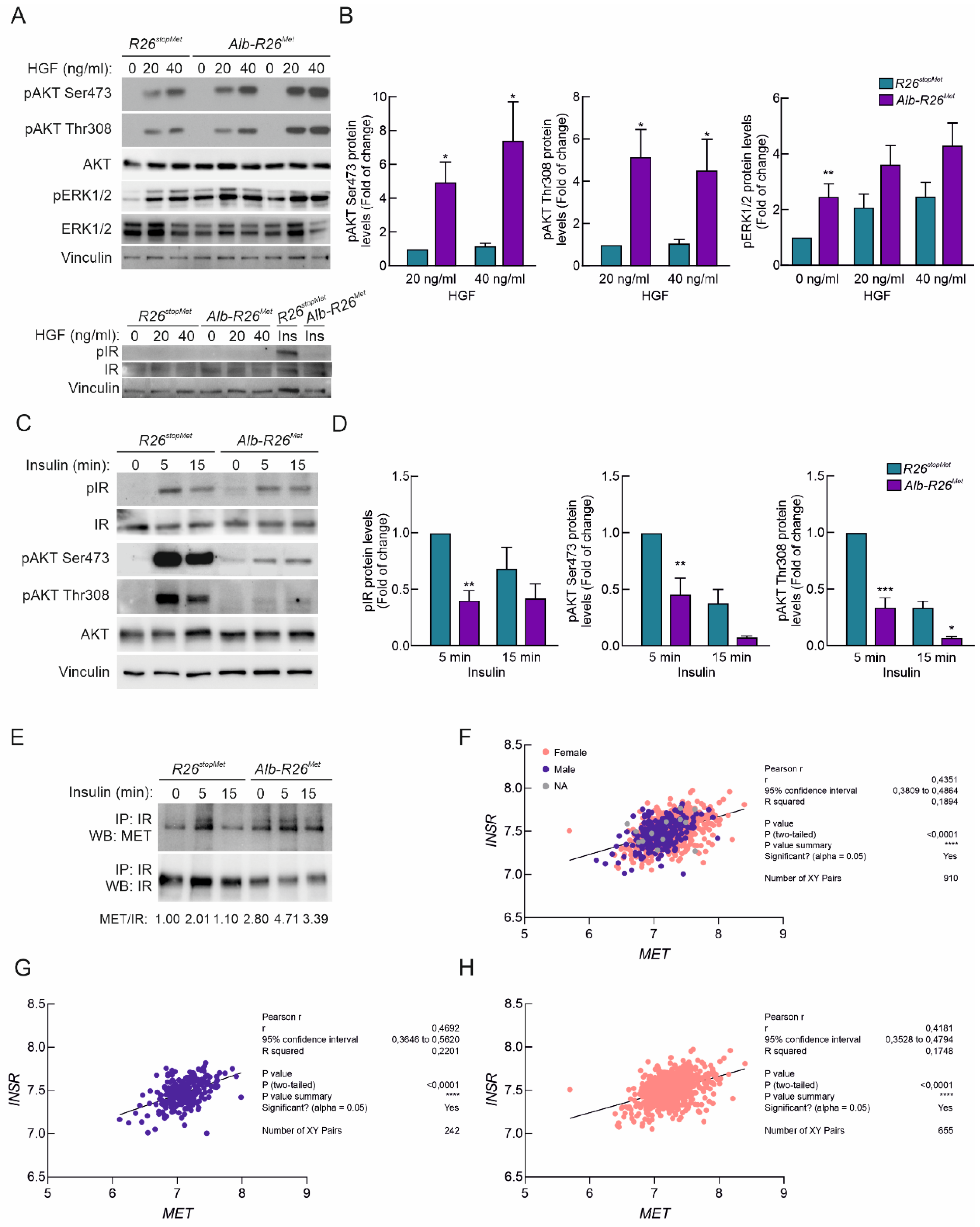
3.5. Enhanced MET Levels Restrain Insulin Signaling In Vivo
3.6. Enhanced MET Levels Attenuate Insulin Sensitivity in Primary Adult Hepatocytes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, M.; Jung, Y.; Jiang, Z.; Svensson, K.J. Regulation of Energy Metabolism by Receptor Tyrosine Kinase Ligands. Front. Physiol. 2020, 11, 354. [Google Scholar] [CrossRef]
- Schlessinger, J. Receptor tyrosine kinases: Legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 2014, 6, a008912. [Google Scholar] [CrossRef]
- Stuart, K.A.; Riordan, S.M.; Lidder, S.; Crostella, L.; Williams, R.; Skouteris, G.G. Hepatocyte growth factor/scatter factor-induced intracellular signalling. Int. J. Exp. Pathol. 2000, 81, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Comoglio, P.M.; Giordano, S.; Trusolino, L. Drug development of MET inhibitors: Targeting oncogene addiction and expedience. Nat. Rev. Drug Discov. 2008, 7, 504–516. [Google Scholar] [CrossRef]
- Oliveira, A.G.; Araujo, T.G.; Carvalho, B.M.; Rocha, G.Z.; Santos, A.; Saad, M.J.A. The Role of Hepatocyte Growth Factor (HGF) in Insulin Resistance and Diabetes. Front. Endocrinol. 2018, 9, 503. [Google Scholar] [CrossRef]
- Kosone, T.; Takagi, H.; Horiguchi, N.; Ariyama, Y.; Otsuka, T.; Sohara, N.; Kakizaki, S.; Sato, K.; Mori, M. HGF ameliorates a high-fat diet-induced fatty liver. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G204–G210. [Google Scholar] [CrossRef]
- Sanchez-Encinales, V.; Cozar-Castellano, I.; Garcia-Ocana, A.; Perdomo, G. Targeted delivery of HGF to the skeletal muscle improves glucose homeostasis in diet-induced obese mice. J. Physiol. Biochem. 2015, 71, 795–805. [Google Scholar] [CrossRef]
- Sakata, H.; Takayama, H.; Sharp, R.; Rubin, J.S.; Merlino, G.; LaRochelle, W.J. Hepatocyte growth factor/scatter factor overexpression induces growth, abnormal development, and tumor formation in transgenic mouse livers. Cell Growth Differ. 1996, 7, 1513–1523. [Google Scholar]
- Takayama, H.; LaRochelle, W.J.; Sabnis, S.G.; Otsuka, T.; Merlino, G. Renal tubular hyperplasia, polycystic disease, and glomerulosclerosis in transgenic mice overexpressing hepatocyte growth factor/scatter factor. Lab. Investig. 1997, 77, 131–138. [Google Scholar]
- Takayama, H.; LaRochelle, W.J.; Sharp, R.; Otsuka, T.; Kriebel, P.; Anver, M.; Aaronson, S.A.; Merlino, G. Diverse tumorigenesis associated with aberrant development in mice overexpressing hepatocyte growth factor/scatter factor. Proc. Natl. Acad. Sci. USA 1997, 94, 701–706. [Google Scholar] [CrossRef]
- Takayama, H.; Takagi, H.; Larochelle, W.J.; Kapur, R.P.; Merlino, G. Ulcerative proctitis, rectal prolapse, and intestinal pseudo-obstruction in transgenic mice overexpressing hepatocyte growth factor/scatter factor. Lab. Investig. 2001, 81, 297–305. [Google Scholar] [CrossRef][Green Version]
- Fan, Y.; Arechederra, M.; Richelme, S.; Daian, F.; Novello, C.; Calderaro, J.; Di Tommaso, L.; Morcrette, G.; Rebouissou, S.; Donadon, M.; et al. A phosphokinome-based screen uncovers new drug synergies for cancer driven by liver-specific gain of nononcogenic receptor tyrosine kinases. Hepatology 2017, 66, 1644–1661. [Google Scholar] [CrossRef]
- Arechederra, M.; Daian, F.; Yim, A.; Bazai, S.K.; Richelme, S.; Dono, R.; Saurin, A.J.; Habermann, B.H.; Maina, F. Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nat. Commun. 2018, 9, 3164. [Google Scholar] [CrossRef]
- Fan, Y.; Bazai, S.K.; Daian, F.; Arechederra, M.; Richelme, S.; Temiz, N.A.; Yim, A.; Habermann, B.H.; Dono, R.; Largaespada, D.A.; et al. Evaluating the landscape of gene cooperativity with receptor tyrosine kinases in liver tumorigenesis using transposon-mediated mutagenesis. J. Hepatol. 2019, 70, 470–482. [Google Scholar] [CrossRef]
- Liu, R.; Tang, W.; Han, X.; Geng, R.; Wang, C.; Zhang, Z. Hepatocyte growth factor-induced mesenchymal-epithelial transition factor activation leads to insulin-like growth factor 1 receptor inhibitor unresponsiveness in gastric cancer cells. Oncol. Lett. 2018, 16, 5983–5991. [Google Scholar] [CrossRef]
- Gordus, A.; Krall, J.A.; Beyer, E.M.; Kaushansky, A.; Wolf-Yadlin, A.; Sevecka, M.; Chang, B.H.; Rush, J.; MacBeath, G. Linear combinations of docking affinities explain quantitative differences in RTK signaling. Mol. Syst. Biol. 2009, 5, 235. [Google Scholar] [CrossRef]
- Jo, M.; Stolz, D.B.; Esplen, J.E.; Dorko, K.; Michalopoulos, G.K.; Strom, S.C. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J. Biol. Chem. 2000, 275, 8806–8811. [Google Scholar] [CrossRef]
- Brevet, M.; Shimizu, S.; Bott, M.J.; Shukla, N.; Zhou, Q.; Olshen, A.B.; Rusch, V.; Ladanyi, M. Coactivation of receptor tyrosine kinases in malignant mesothelioma as a rationale for combination targeted therapy. J. Thorac. Oncol. 2011, 6, 864–874. [Google Scholar] [CrossRef]
- Varkaris, A.; Gaur, S.; Parikh, N.U.; Song, J.H.; Dayyani, F.; Jin, J.K.; Logothetis, C.J.; Gallick, G.E. Ligand-independent activation of MET through IGF-1/IGF-1R signaling. Int. J. Cancer 2013, 133, 1536–1546. [Google Scholar] [CrossRef]
- Fafalios, A.; Ma, J.; Tan, X.; Stoops, J.; Luo, J.; Defrances, M.C.; Zarnegar, R. A hepatocyte growth factor receptor (Met)-insulin receptor hybrid governs hepatic glucose metabolism. Nat. Med. 2011, 17, 1577–1584. [Google Scholar] [CrossRef]
- Bertola, A.; Bonnafous, S.; Cormont, M.; Anty, R.; Tanti, J.F.; Tran, A.; Le Marchand-Brustel, Y.; Gual, P. Hepatocyte growth factor induces glucose uptake in 3T3-L1 adipocytes through A Gab1/phosphatidylinositol 3-kinase/Glut4 pathway. J. Biol. Chem. 2007, 282, 10325–10332. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ocana, A.; Vasavada, R.C.; Cebrian, A.; Reddy, V.; Takane, K.K.; Lopez-Talavera, J.C.; Stewart, A.F. Transgenic overexpression of hepatocyte growth factor in the beta-cell markedly improves islet function and islet transplant outcomes in mice. Diabetes 2001, 50, 2752–2762. [Google Scholar] [CrossRef] [PubMed]
- Roccisana, J.; Reddy, V.; Vasavada, R.C.; Gonzalez-Pertusa, J.A.; Magnuson, M.A.; Garcia-Ocana, A. Targeted inactivation of hepatocyte growth factor receptor c-met in beta-cells leads to defective insulin secretion and GLUT-2 downregulation without alteration of beta-cell mass. Diabetes 2005, 54, 2090–2102. [Google Scholar] [CrossRef]
- Perdomo, G.; Martinez-Brocca, M.A.; Bhatt, B.A.; Brown, N.F.; O’Doherty, R.M.; Garcia-Ocana, A. Hepatocyte growth factor is a novel stimulator of glucose uptake and metabolism in skeletal muscle cells. J. Biol. Chem. 2008, 283, 13700–13706. [Google Scholar] [CrossRef]
- Kroy, D.C.; Schumacher, F.; Ramadori, P.; Hatting, M.; Bergheim, I.; Gassler, N.; Boekschoten, M.V.; Muller, M.; Streetz, K.L.; Trautwein, C. Hepatocyte specific deletion of c-Met leads to the development of severe non-alcoholic steatohepatitis in mice. J. Hepatol. 2014, 61, 883–890. [Google Scholar] [CrossRef]
- Bhushan, B.; Banerjee, S.; Paranjpe, S.; Koral, K.; Mars, W.M.; Stoops, J.W.; Orr, A.; Bowen, W.C.; Locker, J.; Michalopoulos, G.K. Pharmacologic Inhibition of Epidermal Growth Factor Receptor Suppresses Nonalcoholic Fatty Liver Disease in a Murine Fast-Food Diet Model. Hepatology 2019, 70, 1546–1563. [Google Scholar] [CrossRef]
- Fan, Y.; Richelme, S.; Avazeri, E.; Audebert, S.; Helmbacher, F.; Dono, R.; Maina, F. Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis. PLoS Genet. 2015, 11, e1005533. [Google Scholar] [CrossRef]
- Genestine, M.; Caricati, E.; Fico, A.; Richelme, S.; Hassani, H.; Sunyach, C.; Lamballe, F.; Panzica, G.C.; Pettmann, B.; Helmbacher, F.; et al. Enhanced neuronal Met signalling levels in ALS mice delay disease onset. Cell Death Dis. 2011, 2, e130. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Mosquera, A.; Muntane, J.; Ferrandiz, F.; Rodriguez-Manas, L.; de Pablo, F.; Gonzalez-Canudas, J.; Valverde, A.M. Differential effects of metformin glycinate and hydrochloride in glucose production, AMPK phosphorylation and insulin sensitivity in hepatocytes from non-diabetic and diabetic mice. Food Chem. Toxicol. 2019, 123, 470–480. [Google Scholar] [CrossRef]
- Elchebly, M.; Payette, P.; Michaliszyn, E.; Cromlish, W.; Collins, S.; Loy, A.L.; Normandin, D.; Cheng, A.; Himms-Hagen, J.; Chan, C.C.; et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999, 283, 1544–1548. [Google Scholar] [CrossRef]
- Santin, I.; Moore, F.; Colli, M.L.; Gurzov, E.N.; Marselli, L.; Marchetti, P.; Eizirik, D.L. PTPN2, a candidate gene for type 1 diabetes, modulates pancreatic beta-cell apoptosis via regulation of the BH3-only protein Bim. Diabetes 2011, 60, 3279–3288. [Google Scholar] [CrossRef]
- Maina, F.; Casagranda, F.; Audero, E.; Simeone, A.; Comoglio, P.M.; Klein, R.; Ponzetto, C. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell 1996, 87, 531–542. [Google Scholar] [CrossRef]
- Toth, G.; Murphy, R.F.; Lovas, S. Stabilization of local structures by pi-CH and aromatic-backbone amide interactions involving prolyl and aromatic residues. Protein. Eng. 2001, 14, 543–547. [Google Scholar] [CrossRef][Green Version]
- Moumen, A.; Ieraci, A.; Patane, S.; Sole, C.; Comella, J.X.; Dono, R.; Maina, F. Met signals hepatocyte survival by preventing Fas-triggered FLIP degradation in a PI3k-Akt-dependent manner. Hepatology 2007, 45, 1210–1217. [Google Scholar] [CrossRef]
- Moumen, A.; Patane, S.; Porras, A.; Dono, R.; Maina, F. Met acts on Mdm2 via mTOR to signal cell survival during development. Development 2007, 134, 1443–1451. [Google Scholar] [CrossRef]
- Furlan, A.; Lamballe, F.; Stagni, V.; Hussain, A.; Richelme, S.; Prodosmo, A.; Moumen, A.; Brun, C.; Del Barco Barrantes, I.; Arthur, J.S.; et al. Met acts through Abl to regulate p53 transcriptional outcomes and cell survival in the developing liver. J. Hepatol. 2012, 57, 1292–1298. [Google Scholar] [CrossRef]
- Schmidt, C.; Bladt, F.; Goedecke, S.; Brinkmann, V.; Zschiesche, W.; Sharpe, M.; Gherardi, E.; Birchmeier, C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature 1995, 373, 699–702. [Google Scholar] [CrossRef]
- Ishikawa, T.; Factor, V.M.; Marquardt, J.U.; Raggi, C.; Seo, D.; Kitade, M.; Conner, E.A.; Thorgeirsson, S.S. Hepatocyte growth factor/c-met signaling is required for stem-cell-mediated liver regeneration in mice. Hepatology 2012, 55, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Garratt, A.N.; Wustefeld, T.; Strehle, M.; Trautwein, C.; Birchmeier, C. Met provides essential signals for liver regeneration. Proc. Natl. Acad. Sci. USA 2004, 101, 10608–10613. [Google Scholar] [CrossRef]
- Giordano, S.; Columbano, A. Met as a therapeutic target in HCC: Facts and hopes. J. Hepatol. 2014, 60, 442–452. [Google Scholar] [CrossRef]
- Karamouzis, M.V.; Konstantinopoulos, P.A.; Papavassiliou, A.G. Targeting MET as a strategy to overcome crosstalk-related resistance to EGFR inhibitors. Lancet Oncol. 2009, 10, 709–717. [Google Scholar] [CrossRef]
- Xu, A.M.; Huang, P.H. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010, 70, 3857–3860. [Google Scholar] [CrossRef] [PubMed]
- Maina, F. Strategies to overcome drug resistance of receptor tyrosine kinaseaddicted cancer cells. Curr. Med. Chem. 2014, 21, 1607–1617. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Du, Y.; Li, H.; Liu, C.; Wei, Y.; Chen, M.K.; Zhao, X.; Chu, Y.Y.; Qiu, Y.; Qin, L.; et al. EGFR and c-MET Cooperate to Enhance Resistance to PARP Inhibitors in Hepatocellular Carcinoma. Cancer Res. 2019, 79, 819–829. [Google Scholar] [CrossRef]
- Du, Y.; Yamaguchi, H.; Wei, Y.; Hsu, J.L.; Wang, H.L.; Hsu, Y.H.; Lin, W.C.; Yu, W.H.; Leonard, P.G.; Lee, G.R.t.; et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat. Med. 2016, 22, 194–201. [Google Scholar] [CrossRef]
- Watts, W.D.; Wright, L.S. The relationship of alcohol, tobacco, marijuana, and other illegal drug use to delinquency among Mexican-American, black, and white adolescent males. Adolescence 1990, 25, 171–181. [Google Scholar]
- Iroz, A.; Couty, J.P.; Postic, C. Hepatokines: Unlocking the multi-organ network in metabolic diseases. Diabetologia 2015, 58, 1699–1703. [Google Scholar] [CrossRef]
- Hiratsuka, A.; Adachi, H.; Fujiura, Y.; Yamagishi, S.; Hirai, Y.; Enomoto, M.; Satoh, A.; Hino, A.; Furuki, K.; Imaizumi, T. Strong association between serum hepatocyte growth factor and metabolic syndrome. J. Clin. Endocrinol. Metab. 2005, 90, 2927–2931. [Google Scholar] [CrossRef]
- Tsukagawa, E.; Adachi, H.; Hirai, Y.; Enomoto, M.; Fukami, A.; Ogata, K.; Kasahara, A.; Yokoi, K.; Imaizumi, T. Independent association of elevated serum hepatocyte growth factor levels with development of insulin resistance in a 10-year prospective study. Clin. Endocrinol. 2013, 79, 43–48. [Google Scholar] [CrossRef]
- Bancks, M.P.; Bielinski, S.J.; Decker, P.A.; Hanson, N.Q.; Larson, N.B.; Sicotte, H.; Wassel, C.L.; Pankow, J.S. Circulating level of hepatocyte growth factor predicts incidence of type 2 diabetes mellitus: The Multi-Ethnic Study of Atherosclerosis (MESA). Metabolism 2016, 65, 64–72. [Google Scholar] [CrossRef]
- Wang, X.; DeFrances, M.C.; Dai, Y.; Pediaditakis, P.; Johnson, C.; Bell, A.; Michalopoulos, G.K.; Zarnegar, R. A mechanism of cell survival: Sequestration of Fas by the HGF receptor Met. Mol. Cell 2002, 9, 411–421. [Google Scholar] [CrossRef]
- Huh, C.G.; Factor, V.M.; Sanchez, A.; Uchida, K.; Conner, E.A.; Thorgeirsson, S.S. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl. Acad. Sci. USA 2004, 101, 4477–4482. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, M.F.; Jiang, S.; Wu, J.; Liu, J.; Yuan, X.W.; Shen, D.; Zhang, J.Z.; Zhou, N.; He, J.; et al. Liver governs adipose remodelling via extracellular vesicles in response to lipid overload. Nat. Commun. 2020, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Bourebaba, L.; Marycz, K. Pathophysiological Implication of Fetuin-A Glycoprotein in the Development of Metabolic Disorders: A Concise Review. J. Clin. Med. 2019, 8, 2033. [Google Scholar] [CrossRef]
| Antibody | Commercial Brand | Reference | Dilution |
|---|---|---|---|
| Rabbit mAb phospho-AKT Ser473 | Cell Signaling Technologies | 4058 | 1:2000 |
| Rabbit mAb phospho-AKT Thr308 | Cell Signaling Technologies | 4056 | 1:1000 |
| Rabbit mAb AKT | Cell Signaling Technologies | 4691 | 1:20,000 |
| Rabbit mAb phospho-IRβ | Cell Signaling Technologies | 3024 | 1:1000 |
| Rabbit mAb IRβ | Cell Signaling Technologies | 3025 | 1:2000 |
| Rabbit mAb phospho-ERK1/2 | Cell Signaling Technologies | 4370 | 1:4000 |
| Rabbit pAb ERK1/2 | Cell Signaling Technologies | 9102 | 1:4000 |
| Mouse mAb MET | Santa Cruz Biotechnologies | sc-8057 | 1:1000 |
| Mouse mAb MET | Cell Signaling Technologies | 3127 | 1:1000 |
| Mouse mAb α-Tubulin | Sigma-Aldrich/Merck | T5168 | 1:5000 |
| Mouse mAb Vinculin | Santa Cruz Biotechnologies | sc-73614 | 1:7500 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rada, P.; Lamballe, F.; Carceller-López, E.; Hitos, A.B.; Sequera, C.; Maina, F.; Valverde, Á.M. Enhanced Wild-Type MET Receptor Levels in Mouse Hepatocytes Attenuates Insulin-Mediated Signaling. Cells 2022, 11, 793. https://doi.org/10.3390/cells11050793
Rada P, Lamballe F, Carceller-López E, Hitos AB, Sequera C, Maina F, Valverde ÁM. Enhanced Wild-Type MET Receptor Levels in Mouse Hepatocytes Attenuates Insulin-Mediated Signaling. Cells. 2022; 11(5):793. https://doi.org/10.3390/cells11050793
Chicago/Turabian StyleRada, Patricia, Fabienne Lamballe, Elena Carceller-López, Ana B. Hitos, Celia Sequera, Flavio Maina, and Ángela M. Valverde. 2022. "Enhanced Wild-Type MET Receptor Levels in Mouse Hepatocytes Attenuates Insulin-Mediated Signaling" Cells 11, no. 5: 793. https://doi.org/10.3390/cells11050793
APA StyleRada, P., Lamballe, F., Carceller-López, E., Hitos, A. B., Sequera, C., Maina, F., & Valverde, Á. M. (2022). Enhanced Wild-Type MET Receptor Levels in Mouse Hepatocytes Attenuates Insulin-Mediated Signaling. Cells, 11(5), 793. https://doi.org/10.3390/cells11050793







