A Novel LHX6 Reporter Cell Line for Tracking Human iPSC-Derived Cortical Interneurons
Abstract
:1. Introduction
2. Materials and Methods
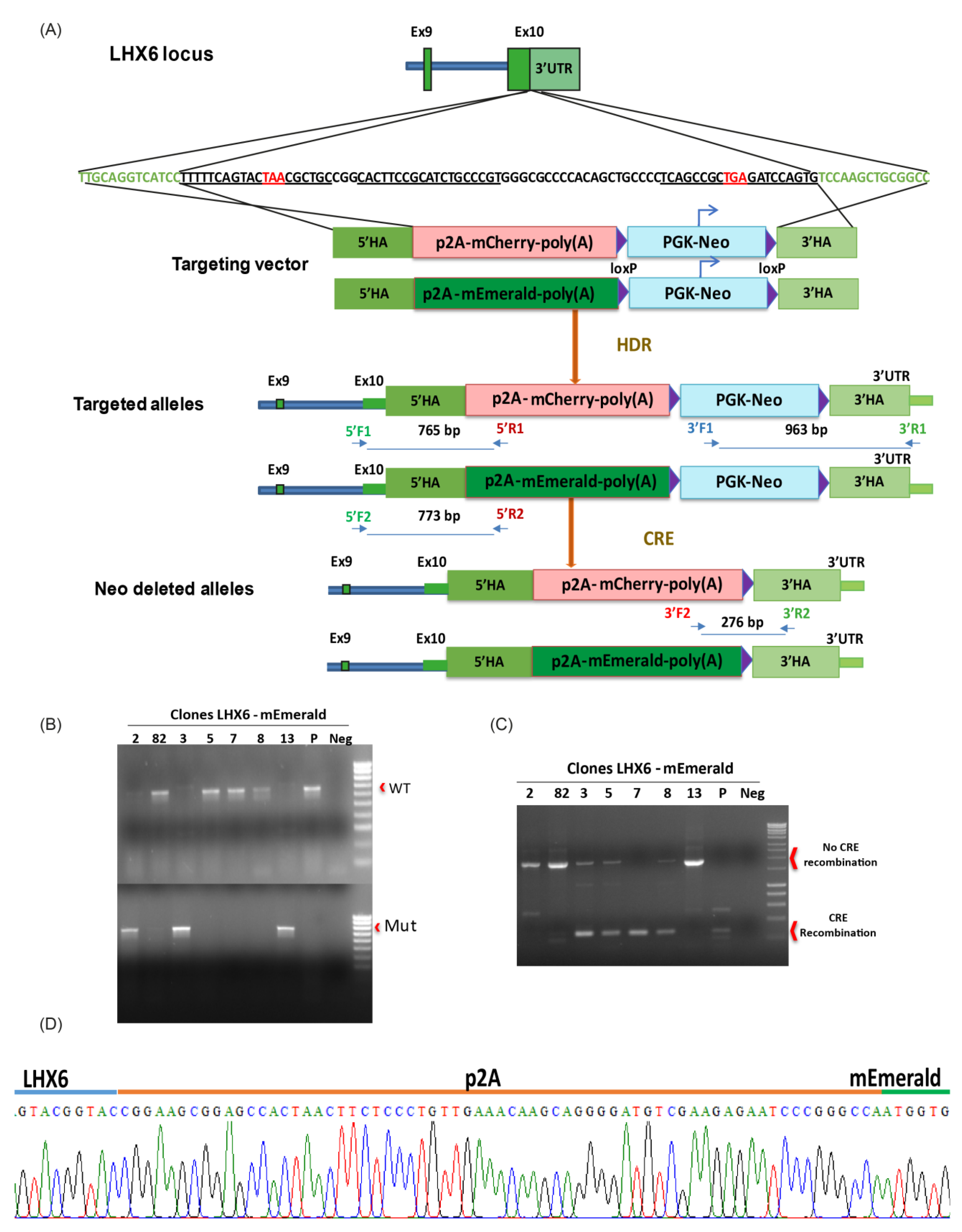
2.1. CRISPR/Cas9-Assisted Gene Targeting
2.2. HESC Culture and Neuronal Differentiation
2.3. Immunohistochemistry
2.4. Flow Cytometry
2.5. Statistical Analyses
3. Results
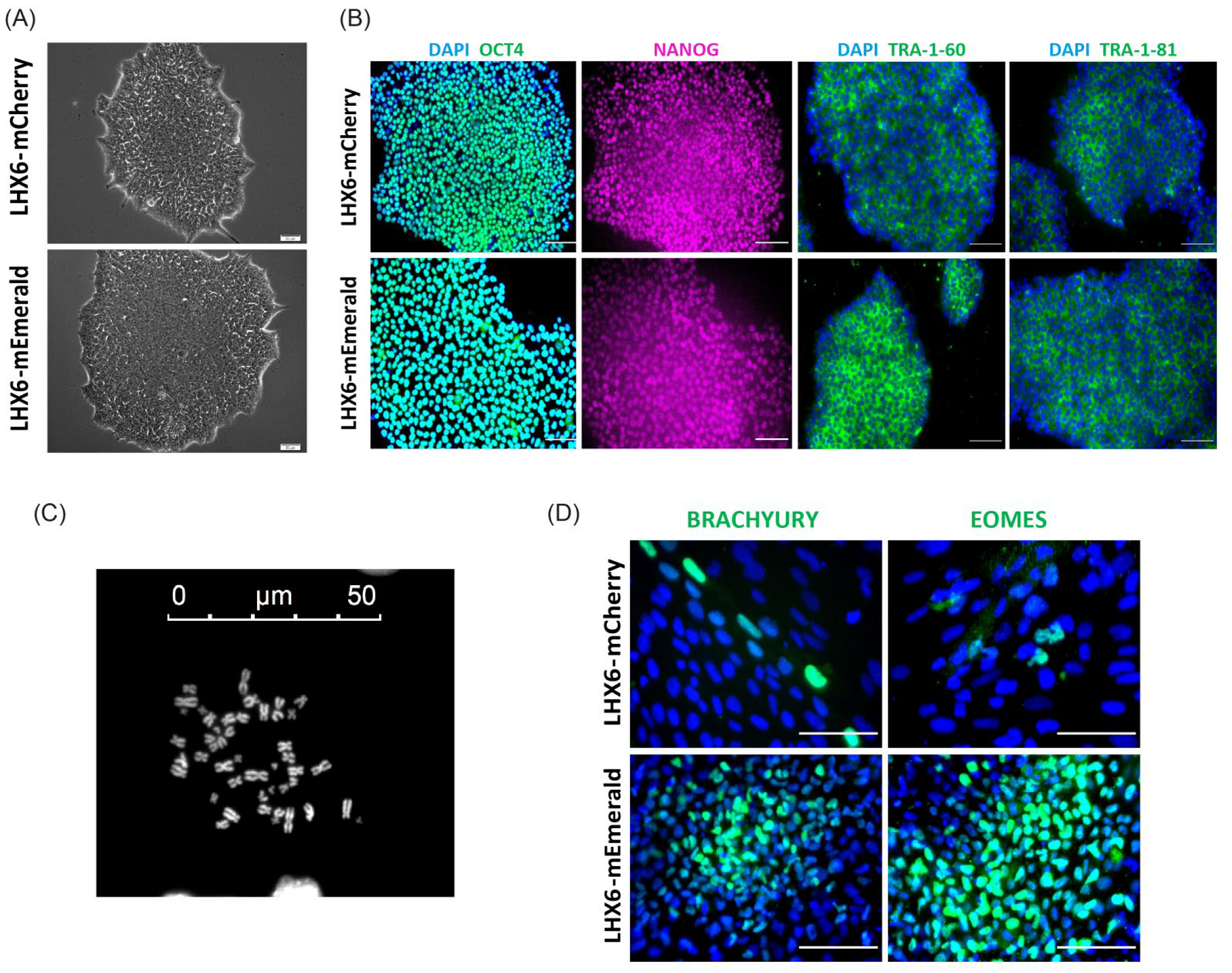
3.1. Targeting mCherry and mEmerald into the LHX6 Locus of hPSCs
3.2. mCherry and mEmerald Expression Faithfully Mirror That of LHX6 during hPSC Interneuron Differentiation
3.3. Production of mEmerald+ and mCherry+ Cells during PSC Differentiation Respond to External Cues
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buzsaki, G.; Geisler, C.; Henze, D.A.; Wang, X.J. Interneuron diversity series: Circuit complexity and axon wiring economy of cortical interneurons. Trends Neurosci. 2004, 27, 186–193. [Google Scholar] [CrossRef]
- Yizhar, O.; Fenno, L.E.; Prigge, M.; Schneider, F.; Davidson, T.J.; O’Shea, D.J.; Sohal, V.S.; Goshen, I.; Finkelstein, J.; Paz, J.T.; et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 2011, 477, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Marin, O. Interneuron dysfunction in psychiatric disorders. Nat. Reviews Neurosci. 2012, 13, 107–120. [Google Scholar] [CrossRef]
- Tremblay, R.; Lee, S.; Rudy, B. Gabaergic interneurons in the neocortex: From cellular properties to circuits. Neuron 2016, 91, 260–292. [Google Scholar] [CrossRef] [Green Version]
- Haider, B.; Duque, A.; Hasenstaub, A.R.; McCormick, D.A. Neocortical network activity in vivo is generated through a dynamic balance of excitation and inhibition. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 4535–4545. [Google Scholar] [CrossRef] [Green Version]
- Klausberger, T.; Somogyi, P. Neuronal diversity and temporal dynamics: The unity of hippocampal circuit operations. Science 2008, 321, 53–57. [Google Scholar] [CrossRef] [Green Version]
- DeFelipe, J. Chandelier cells and epilepsy. Brain 1999, 122 Pt 10, 1807–1822. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.M.; Campbell, D.B.; Stanwood, G.D.; Davis, C.; Noebels, J.L.; Levitt, P. Genetic disruption of cortical interneuron development causes region- and gaba cell type-specific deficits, epilepsy, and behavioral dysfunction. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 622–631. [Google Scholar] [CrossRef] [Green Version]
- Akbarian, S.; Huang, H.S. Molecular and cellular mechanisms of altered gad1/gad67 expression in schizophrenia and related disorders. Brain Res. Rev. 2006, 52, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.A. Cortical circuit dysfunction and cognitive deficits in schizophrenia--implications for preemptive interventions. Eur. J. Neurosci. 2012, 35, 1871–1878. [Google Scholar] [CrossRef] [Green Version]
- Zikopoulos, B.; Barbas, H. Altered neural connectivity in excitatory and inhibitory cortical circuits in autism. Front. Hum. Neurosci. 2013, 7, 609. [Google Scholar] [CrossRef] [Green Version]
- Schmid, L.C.; Mittag, M.; Poll, S.; Steffen, J.; Wagner, J.; Geis, H.R.; Schwarz, I.; Schmidt, B.; Schwarz, M.K.; Remy, S.; et al. Dysfunction of somatostatin-positive interneurons associated with memory deficits in an alzheimer’s disease model. Neuron 2016, 92, 114–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whittington, M.A.; Traub, R.D. Interneuron diversity series: Inhibitory interneurons and network oscillations in vitro. Trends Neurosci. 2003, 26, 676–682. [Google Scholar] [CrossRef]
- Lavdas, A.A.; Grigoriou, M.; Pachnis, V.; Parnavelas, J.G. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J. Neurosci. Off. J. Soc. Neurosci. 1999, 19, 7881–7888. [Google Scholar] [CrossRef] [Green Version]
- Laclef, C.; Metin, C. Conserved rules in embryonic development of cortical interneurons. Semin. Cell Dev. Biol. 2018, 76, 86–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudy, B.; Fishell, G.; Lee, S.; Hjerling-Leffler, J. Three groups of interneurons account for nearly 100% of neocortical gabaergic neurons. Dev. Neurobiol. 2011, 71, 45–61. [Google Scholar] [CrossRef] [Green Version]
- Sussel, L.; Marin, O.; Kimura, S.; Rubenstein, J.L. Loss of nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: Evidence for a transformation of the pallidum into the striatum. Development 1999, 126, 3359–3370. [Google Scholar] [CrossRef]
- Xu, Q.; Guo, L.; Moore, H.; Waclaw, R.R.; Campbell, K.; Anderson, S.A. Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron 2010, 65, 328–340. [Google Scholar] [CrossRef] [Green Version]
- Alifragis, P.; Liapi, A.; Parnavelas, J.G. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their gaba phenotype. J. Neurosci. Off. J. Soc. Neurosci. 2004, 24, 5643–5648. [Google Scholar] [CrossRef] [Green Version]
- Liodis, P.; Denaxa, M.; Grigoriou, M.; Akufo-Addo, C.; Yanagawa, Y.; Pachnis, V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 3078–3089. [Google Scholar] [CrossRef]
- Du, T.; Xu, Q.; Ocbina, P.J.; Anderson, S.A. Nkx2.1 specifies cortical interneuron fate by activating lhx6. Development 2008, 135, 1559–1567. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Wang, M.; Mi, D.; Lu, T.; Wang, B.; Dong, H.; Zhong, S.; Chen, Y.; Sun, L.; Zhou, X.; et al. Mouse and human share conserved transcriptional programs for interneuron development. Science 2021, 374, eabj6641. [Google Scholar] [CrossRef] [PubMed]
- Doetschman, T.C.; Eistetter, H.; Katz, M.; Schmidt, W.; Kemler, R. The in vitro development of blastocyst-derived embryonic stem cell lines: Formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 1985, 87, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Maroof, A.M.; Keros, S.; Tyson, J.A.; Ying, S.W.; Ganat, Y.M.; Merkle, F.T.; Liu, B.; Goulburn, A.; Stanley, E.G.; Elefanty, A.G.; et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 2013, 12, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Nicholas, C.R.; Chen, J.; Tang, Y.; Southwell, D.G.; Chalmers, N.; Vogt, D.; Arnold, C.M.; Chen, Y.J.; Stanley, E.G.; Elefanty, A.G.; et al. Functional maturation of hpsc-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 2013, 12, 573–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.G.; Yao, R.; Monnell, T.; Cho, J.H.; Vasudevan, A.; Koh, A.; Peeyush, K.T.; Moon, M.; Datta, D.; Bolshakov, V.Y.; et al. Efficient specification of interneurons from human pluripotent stem cells by dorsoventral and rostrocaudal modulation. Stem Cells 2014, 32, 1789–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyson, J.A.; Goldberg, E.M.; Maroof, A.M.; Xu, Q.; Petros, T.J.; Anderson, S.A. Duration of culture and sonic hedgehog signaling differentially specify pv versus sst cortical interneuron fates from embryonic stem cells. Development 2015, 142, 1267–1278. [Google Scholar] [CrossRef] [Green Version]
- Sun, A.X.; Yuan, Q.; Tan, S.; Xiao, Y.; Wang, D.; Khoo, A.T.; Sani, L.; Tran, H.D.; Kim, P.; Chiew, Y.S.; et al. Direct induction and functional maturation of forebrain gabaergic neurons from human pluripotent stem cells. Cell Rep. 2016, 16, 1942–1953. [Google Scholar] [CrossRef] [Green Version]
- Noakes, Z.; Keefe, F.; Tamburini, C.; Kelly, C.M.; Cruz Santos, M.; Dunnett, S.B.; Errington, A.C.; Li, M. Human pluripotent stem cell-derived striatal interneurons: Differentiation and maturation in vitro and in the rat brain. Stem Cell Rep. 2019, 12, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Mayer, C.; Hafemeister, C.; Bandler, R.C.; Machold, R.; Batista Brito, R.; Jaglin, X.; Allaway, K.; Butler, A.; Fishell, G.; Satija, R. Developmental diversification of cortical inhibitory interneurons. Nature 2018, 555, 457–462. [Google Scholar] [CrossRef]
- Mi, D.; Li, Z.; Lim, L.; Li, M.; Moissidis, M.; Yang, Y.; Gao, T.; Hu, T.X.; Pratt, T.; Price, D.J.; et al. Early emergence of cortical interneuron diversity in the mouse embryo. Science 2018, 360, 81–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Pevny, L.; Lovell-Badge, R.; Smith, A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr. Biol. CB 1998, 8, 971–974. [Google Scholar] [CrossRef] [Green Version]
- Ying, Q.L.; Stavridis, M.; Griffiths, D.; Li, M.; Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003, 21, 183–186. [Google Scholar] [CrossRef] [PubMed]
- DeRosa, B.A.; Belle, K.C.; Thomas, B.J.; Cukier, H.N.; Pericak-Vance, M.A.; Vance, J.M.; Dykxhoorn, D.M. Hvgat-mcherry: A novel molecular tool for analysis of gabaergic neurons derived from human pluripotent stem cells. Mol. Cell. Neurosci. 2015, 68, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Goulburn, A.L.; Alden, D.; Davis, R.P.; Micallef, S.J.; Ng, E.S.; Yu, Q.C.; Lim, S.M.; Soh, C.L.; Elliott, D.A.; Hatzistavrou, T.; et al. A targeted nkx2.1 human embryonic stem cell reporter line enables identification of human basal forebrain derivatives. Stem Cells 2011, 29, 462–473. [Google Scholar] [CrossRef]
- Nobrega-Pereira, S.; Kessaris, N.; Du, T.; Kimura, S.; Anderson, S.A.; Marin, O. Postmitotic nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron 2008, 59, 733–745. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, T.; Nishikawa, A.; Kume, S.; Chayama, K.; Yamamoto, T. Multiplex genome engineering in human cells using all-in-one crispr/cas9 vector system. Sci. Rep. 2014, 4, 5400. [Google Scholar] [CrossRef] [Green Version]
- Pham, C.T.; MacIvor, D.M.; Hug, B.A.; Heusel, J.W.; Ley, T.J. Long-range disruption of gene expression by a selectable marker cassette. Proc. Natl. Acad. Sci. USA 1996, 93, 13090–13095. [Google Scholar] [CrossRef] [Green Version]
- Scarff, K.L.; Ung, K.S.; Sun, J.; Bird, P.I. A retained selection cassette increases reporter gene expression without affecting tissue distribution in spi3 knockout/gfp knock-in mice. Genesis 2003, 36, 149–157. [Google Scholar] [CrossRef]
- Zhu, Z.; Verma, N.; Gonzalez, F.; Shi, Z.D.; Huangfu, D. A crispr/cas-mediated selection-free knockin strategy in human embryonic stem cells. Stem Cell Rep. 2015, 4, 1103–1111. [Google Scholar] [CrossRef] [Green Version]
- Tosic, J.; Kim, G.J.; Pavlovic, M.; Schroder, C.M.; Mersiowsky, S.L.; Barg, M.; Hofherr, A.; Probst, S.; Kottgen, M.; Hein, L.; et al. Eomes and brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat. Cell Biol. 2019, 21, 1518–1531. [Google Scholar] [CrossRef]
- Grigoriou, M.; Tucker, A.S.; Sharpe, P.T.; Pachnis, V. Expression and regulation of lhx6 and lhx7, a novel subfamily of lim homeodomain encoding genes, suggests a role in mammalian head development. Development 1998, 125, 2063–2074. [Google Scholar] [CrossRef] [PubMed]
- Colasante, G.; Lignani, G.; Rubio, A.; Medrihan, L.; Yekhlef, L.; Sessa, A.; Massimino, L.; Giannelli, S.G.; Sacchetti, S.; Caiazzo, M.; et al. Rapid conversion of fibroblasts into functional forebrain gabaergic interneurons by direct genetic reprogramming. Cell Stem Cell 2015, 17, 719–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, A.; McGuire, T.; Peng, C.Y.; Kessler, J.A. Differential effects of bmp signaling on parvalbumin and somatostatin interneuron differentiation. Development 2009, 136, 2633–2642. [Google Scholar] [CrossRef] [Green Version]
- Arshad, A.; Vose, L.R.; Vinukonda, G.; Hu, F.; Yoshikawa, K.; Csiszar, A.; Brumberg, J.C.; Ballabh, P. Extended production of cortical interneurons into the third trimester of human gestation. Cereb. Cortex 2016, 26, 2242–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Santos, M.; Cardo, L.F.; Li, M. A Novel LHX6 Reporter Cell Line for Tracking Human iPSC-Derived Cortical Interneurons. Cells 2022, 11, 853. https://doi.org/10.3390/cells11050853
Cruz-Santos M, Cardo LF, Li M. A Novel LHX6 Reporter Cell Line for Tracking Human iPSC-Derived Cortical Interneurons. Cells. 2022; 11(5):853. https://doi.org/10.3390/cells11050853
Chicago/Turabian StyleCruz-Santos, Maria, Lucia Fernandez Cardo, and Meng Li. 2022. "A Novel LHX6 Reporter Cell Line for Tracking Human iPSC-Derived Cortical Interneurons" Cells 11, no. 5: 853. https://doi.org/10.3390/cells11050853
APA StyleCruz-Santos, M., Cardo, L. F., & Li, M. (2022). A Novel LHX6 Reporter Cell Line for Tracking Human iPSC-Derived Cortical Interneurons. Cells, 11(5), 853. https://doi.org/10.3390/cells11050853




