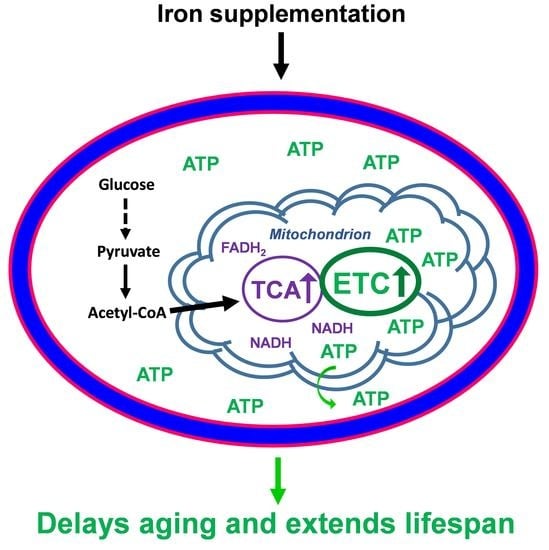
Iron Supplementation Delays Aging and Extends Cellular Lifespan through Potentiation of Mitochondrial Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Gene Deletion
2.2. Medium Composition and Chemicals
2.3. Yeast Growth Conditions
2.4. Chronological Aging Assay
2.5. Oxidative Resistance Assay
2.6. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
2.7. Mitochondrial Membrane Potential and Structure Analysis
2.8. ATP Analysis
2.9. Statistical Analysis
3. Results
3.1. Iron Supplementation Extends the Cellular Lifespan of Yeast
3.2. Iron Supplementation Increases Oxidative Stress Resistance
3.3. Iron Supplementation Potentiates Mitochondrial Functions
3.4. Iron Supplementation Increases the ATP Level Required for Extension of Cellular Lifespan
3.5. Iron Supplementation Prevents Accelerated Aging of AMPK Knockout Mutant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bloom, D.E.; Canning, D.; Lubet, A. Global Population Aging: Facts, Challenges, Solutions & Perspectives. Chall. Solut. Perspect. Daedalus 2015, 144, 80–92. [Google Scholar]
- United Nations Department of Economic and Social Affairs, Population Division. World Population Prospects 2019; Highlights (ST/ESA/SERA/423); UN: New York, NY, USA, 2019. [Google Scholar]
- Robine, J. Ageing Populations: We Are Living Longer Lives, but Are We Healthier? United Nations, Department of Economics and Social Affairs, Population Division: New York, NY, USA, 2021; UN DESA/POP/2021/TP/NO. [Google Scholar]
- Niccoli, T.; Partridge, L. Ageing as a risk factor for disease. Curr. Biol. 2012, 22, R741–R752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Harman, D. The aging process: Major risk factor for disease and death. Proc. Natl. Acad. Sci. USA 1991, 88, 5360–5363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, L.; Deelen, J.; Slagboom, P.E. Facing up to the global challenges of ageing. Nature 2018, 561, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Kennedy, B.K.; Longo, V.D.; Seals, D.; Melov, S. Medical research: Treat ageing. Nature 2014, 511, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span-from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [Green Version]
- Partridge, L.; Fuentealba, M.; Kennedy, B.K. The quest to slow ageing through drug discovery. Nat. Rev. Drug Discov. 2020, 19, 513–532. [Google Scholar] [CrossRef]
- Zimmermann, A.; Hofer, S.; Pendl, T.; Kainz, K.; Madeo, F.; Carmona-Gutierrez, D. Yeast as a tool to identify anti-aging compounds. FEMS Yeast Res. 2018, 18, foy020. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Burtner, C.R.; Kennedy, B.K. Recent developments in yeast aging. PLoS Genet. 2007, 3, e84. [Google Scholar] [CrossRef]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Longo, V.D.; Fabrizio, P. Chronological Aging in Saccharomyces cerevisiae. Aging Res. Yeast 2011, 57, 101–121. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, A.S.; Gubbi, S.; Barzilai, N. Benefits of Metformin in Attenuating the Hallmarks of Aging. Cell Metab. 2020, 32, 15–30. [Google Scholar] [CrossRef]
- Alfatah, M.; Wong, J.H.; Krishnan, V.G.; Lee, Y.C.; Sin, Q.F.; Goh, C.J.H.; Kong, K.W.; Lee, W.T.; Lewis, J.; Hoon, S.; et al. TORC1 regulates the transcriptional response to glucose and developmental cycle via the Tap42-Sit4-Rrd1/2 pathway in Saccharomyces cerevisiae. BMC Biol. 2021, 19, 1–22. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Hall, M.N.; Lin, S.C.; Hardie, D.G. AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab. 2020, 31, 472–492. [Google Scholar] [CrossRef]
- Lin, S.C.; Hardie, D.G. AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab. 2018, 27, 299–313. [Google Scholar] [CrossRef] [Green Version]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Blagosklonny, M.V. Rapamycin for longevity: Opinion article. Aging 2019, 11, 8048–8067. [Google Scholar] [CrossRef]
- Yiannikourides, A.; Latunde-Dada, G.O. A Short Review of Iron Metabolism and Pathophysiology of Iron Disorders. Medicines 2019, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Lynch, S.; Pfeiffer, C.M.; Georgieff, M.K.; Brittenham, G.; Fairweather-Tait, S.; Hurrell, R.F.; McArdle, H.J.; Raiten, D.J. Biomarkers of Nutrition for Development (BOND)-Iron review. J. Nutr. 2018, 148, 1001S–1067S. [Google Scholar] [CrossRef] [Green Version]
- Abbaspour, N.; Hurrell, R.; Kelishadi, R. Review on iron and its importance for human health. J. Res. Med. Sci. 2014, 19, 164. [Google Scholar]
- Beard, J.L. Iron biology in immune function, muscle metabolism and neuronal functioning. J. Nutr. 2001, 131, 568S–580S. [Google Scholar] [CrossRef] [PubMed]
- Van Dijken, J.P.; Bauer, J.; Brambilla, L.; Duboc, P.; Francois, J.M.; Gancedo, C.; Giuseppin, M.L.F.; Heijnen, J.J.; Hoare, M.; Lange, H.C. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzym. Microb. Technol. 2000, 26, 706–714. [Google Scholar] [CrossRef]
- Mülleder, M.; Capuano, F.; Pir, P.; Christen, S.; Sauer, U.; Oliver, S.G.; Ralser, M. A prototrophic deletion mutant collection for yeast metabolomics and systems biology. Nat. Biotechnol. 2012, 30, 1176–1178. [Google Scholar] [CrossRef] [Green Version]
- Longtine, M.S.; Mckenzie, A., III; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Powers, R.W.; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef] [Green Version]
- Murakami, C.J.; Burtner, C.R.; Kennedy, B.; Kaeberlein, M. A Method for High-Throughput Quantitative Analysis of Yeast Chronological Life Span. J. Gerontol. Ser. A 2008, 63, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hassett, R.F.; Romeo, A.M.; Kosman, D.J. Regulation of High Affinity Iron Uptake in the YeastSaccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 7628–7636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, D.M.; Cloonan, S.M. Mitochondrial Iron in Human Health and Disease. Annu. Rev. Physiol. 2019, 81, 453–482. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.T.; Manz, D.H.; Torti, F.M.; Torti, S.V. Mitochondria and Iron: Current questions. Expert Rev. Hematol. 2016, 10, 65–79. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Zhou, Q.; Wu, D.; Chen, L. Mitochondrial iron metabolism and its role in diseases. Clin. Chim. Acta 2021, 513, 6–12. [Google Scholar] [CrossRef]
- Levi, S.; Rovida, E. The role of iron in mitochondrial function. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 629–636. [Google Scholar] [CrossRef]
- Sun, N.; Youle, R.J.; Finkel, T. The Mitochondrial Basis of Aging. Mol. Cell 2016, 61, 654–666. [Google Scholar] [CrossRef] [Green Version]
- Kauppila, T.E.; Kauppila, J.H.; Larsson, N.G. Mammalian Mitochondria and Aging: An Update. Cell Metab. 2017, 25, 57–71. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.Y.; Blum, A.; Liu, J.; Finkel, T. The role of mitochondria in aging. J. Clin. Investig. 2018, 128, 3662–3670. [Google Scholar] [CrossRef] [Green Version]
- Sreedhar, A.; Aguilera-Aguirre, L.; Singh, K.K. Mitochondria in skin health, aging, and disease. Cell Death Dis. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Roca-Portoles, A.; Tait, S.W. Mitochondrial quality control: From molecule to organelle. Cell. Mol. Life Sci. 2021, 78, 3853–3866. [Google Scholar] [CrossRef]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef]
- Galy, B.; Ferring-Appel, D.; Sauer, S.W.; Kaden, S.; Lyoumi, S.; Puy, H.; Kölker, S.; Gröne, H.-J.; Hentze, M.W. Iron Regulatory Proteins Secure Mitochondrial Iron Sufficiency and Function. Cell Metab. 2010, 12, 194–201. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef] [Green Version]
- Cappel, D.; Deja, S.; Duarte, J.A.G.; Kucejova, B.; Inigo, M.M.; Fletcher, J.; Fu, X.; Berglund, E.D.; Liu, T.; Elmquist, J.K.; et al. Pyruvate-Carboxylase-Mediated Anaplerosis Promotes Antioxidant Capacity by Sustaining TCA Cycle and Redox Metabolism in Liver. Cell Metab. 2019, 29, 1291–1305.e8. [Google Scholar] [CrossRef]
- Inigo, M.; Deja, S.; Burgess, S.C. Ins and Outs of the TCA Cycle: The Central Role of Anaplerosis. Annu. Rev. Nutr. 2021, 41, 19–47. [Google Scholar] [CrossRef]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef]
- Ocampo, A.; Liu, J.; Schroeder, E.A.; Shadel, G.S.; Barrientos, A. Mitochondrial Respiratory Thresholds Regulate Yeast Chronological Life Span and its Extension by Caloric Restriction. Cell Metab. 2012, 16, 55–67. [Google Scholar] [CrossRef] [Green Version]
- Hedbacker, K.; Carlson, M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008, 13, 2408. [Google Scholar] [CrossRef] [Green Version]
- Reznick, R.M.; Zong, H.; Li, J.; Morino, K.; Moore, I.K.; Yu, H.J.; Liu, Z.-X.; Dong, J.; Mustard, K.J.; Hawley, S.A.; et al. Aging-Associated Reductions in AMP-Activated Protein Kinase Activity and Mitochondrial Biogenesis. Cell Metab. 2007, 5, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Burkewitz, K.; Zhang, Y.; Mair, W.B. AMPK at the nexus of energetics and aging. Cell Metab. 2014, 20, 10–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.X.; Xiong, Y.; Guan, K.L. Nutrient Sensing, Metabolism, and Cell Growth Control. Mol. Cell 2013, 49, 379–387. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.P.; Lei, Q.Y. Metabolite sensing and signaling in cell metabolism. Signal Transduct. Target. Ther. 2018, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cappellini, M.D.; Musallam, K.M.; Taher, A.T. Iron deficiency anaemia revisited. J. Intern. Med. 2020, 287, 153–170. [Google Scholar] [CrossRef]
- Neidlein, S.; Wirth, R.; Pourhassan, M. Iron deficiency, fatigue and muscle strength and function in older hospitalized patients. Eur. J. Clin. Nutr. 2020, 75, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Pasricha, S.R.; Tye-Din, J.; Muckenthaler, M.U.; Swinkels, D.W. Iron deficiency. Lancet 2021, 397, 233–248. [Google Scholar] [CrossRef]
- Busti, F.; Campostrini, N.; Martinelli, N.; Girelli, D. Iron deficiency in the elderly population, revisited in the hepcidin era. Front. Pharmacol. 2014, 5, 83. [Google Scholar] [CrossRef]
- Fairweather-Tait, S.J.; Wawer, A.; Gillings, R.; Jennings, A.; Myint, P.K. Iron status in the elderly. Mech. Ageing Dev. 2013, 136–137, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Wawer, A.A.; Jennings, A.; Fairweather-Tait, S.J. Iron status in the elderly: A review of recent evidence. Mech. Ageing Dev. 2018, 175, 55–73. [Google Scholar] [CrossRef] [Green Version]
- Shahmirzadi, A.A.; Edgar, D.; Liao, C.-Y.; Hsu, Y.-M.; Lucanic, M.; Shahmirzadi, A.A.; Wiley, C.D.; Gan, G.; Kim, D.E.; Kasler, H.G.; et al. Alpha-Ketoglutarate, an Endogenous Metabolite, Extends Lifespan and Compresses Morbidity in Aging Mice. Cell Metab. 2020, 32, 447–456.e6. [Google Scholar] [CrossRef]
- Kwon, Y.-Y.; Choi, K.-M.; Cho, C.; Lee, A.C.-K. Mitochondrial Efficiency-Dependent Viability of Saccharomyces cerevisiae Mutants Carrying Individual Electron Transport Chain Component Deletions. Mol. Cells 2015, 38, 1054–1063. [Google Scholar] [CrossRef] [Green Version]
- Kaya, A.; Ma, S.; Wasko, B.; Lee, M.; Kaeberlein, M.; Gladyshev, V.N. Defining molecular basis for longevity traits in natural yeast isolates. NPJ Aging Mech. Dis. 2015, 1, 15001. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.E.; Coody, T.; Jeong, M.-Y.; Berg, J.; Winge, D.R.; Hughes, A.L. Cysteine Toxicity Drives Age-Related Mitochondrial Decline by Altering Iron Homeostasis. Cell 2020, 180, 296–310.e8. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.L.; Ven, T.N.; Crane, M.M.; Brunner, M.L.C.; Pun, A.K.; Helget, K.L.; Brower, K.; Chen, D.E.; Doan, H.; Dillard-Telm, J.D.; et al. Loss of vacuolar acidity results in iron-sulfur cluster defects and divergent homeostatic responses during aging in Saccharomyces cerevisiae. GeroScience 2020, 42, 749–764. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jing, J.L.; Ning, T.C.Y.; Natali, F.; Eisenhaber, F.; Alfatah, M. Iron Supplementation Delays Aging and Extends Cellular Lifespan through Potentiation of Mitochondrial Function. Cells 2022, 11, 862. https://doi.org/10.3390/cells11050862
Jing JL, Ning TCY, Natali F, Eisenhaber F, Alfatah M. Iron Supplementation Delays Aging and Extends Cellular Lifespan through Potentiation of Mitochondrial Function. Cells. 2022; 11(5):862. https://doi.org/10.3390/cells11050862
Chicago/Turabian StyleJing, Jovian Lin, Trishia Cheng Yi Ning, Federica Natali, Frank Eisenhaber, and Mohammad Alfatah. 2022. "Iron Supplementation Delays Aging and Extends Cellular Lifespan through Potentiation of Mitochondrial Function" Cells 11, no. 5: 862. https://doi.org/10.3390/cells11050862
APA StyleJing, J. L., Ning, T. C. Y., Natali, F., Eisenhaber, F., & Alfatah, M. (2022). Iron Supplementation Delays Aging and Extends Cellular Lifespan through Potentiation of Mitochondrial Function. Cells, 11(5), 862. https://doi.org/10.3390/cells11050862