MicroRNA Roles in Cell Reprogramming Mechanisms
Abstract
:1. Introduction
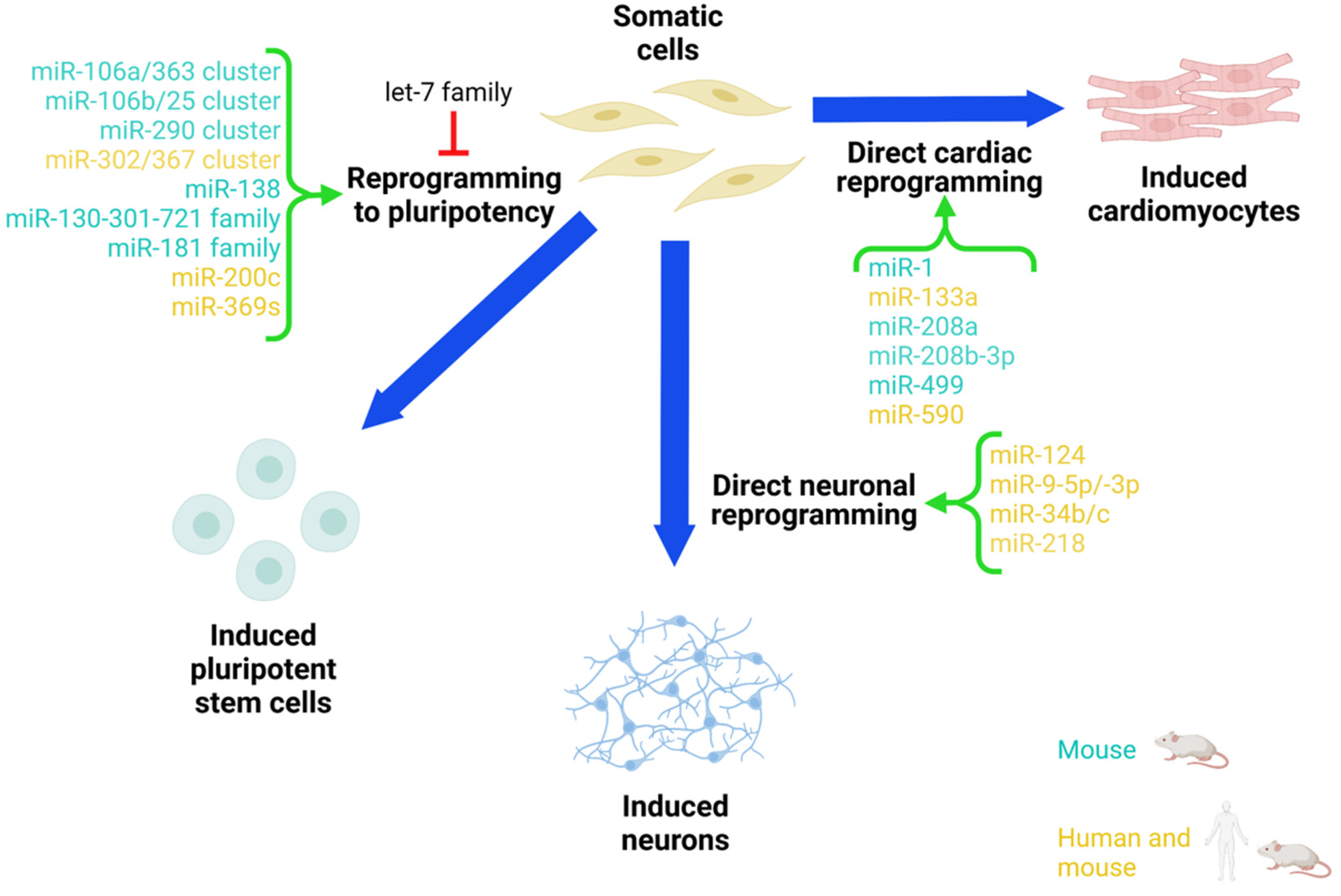
2. iPSC Reprogramming Approach
miRNA in iPSC Reprogramming
3. Neural Direct Reprogramming Approach
3.1. miRNAs in Direct Neuronal Reprogramming In Vitro
3.2. miRNAs in Direct Neuronal Reprogramming In Vivo
3.3. Challenges and Opportunities of miRNA-Mediated Neuronal Reprogramming as a Therapeutic Strategy to Treat Neurodegenerative Diseases
4. Direct Cardiac Reprogramming Approach
4.1. miRNA-Mediated Direct Cardiac Reprogramming In Vitro
4.2. miRNA-Mediated Direct Cardiac Reprogramming In Vivo
4.3. Challenges and Opportunities of miRNA-Mediated Cardiac Reprogramming as a Therapeutic Strategy to Treat Heart Failure Patients
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zeng, X. The role of induced pluripotent stem cells in regenerative medicine: Neurodegenerative diseases. Stem Cell Res. Ther. 2011, 2, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunaseeli, I.; Doss, M.X.; Antzelevitch, C.; Hescheler, J.; Sachinidis, A. Induced pluripotent stem cells as a model for accelerated patient- and disease-specific drug discovery. Curr. Med. Chem. 2010, 17, 759–766. [Google Scholar] [CrossRef] [Green Version]
- Lassar, A.B.; Paterson, B.M.; Weintraub, H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 1986, 47, 649–656. [Google Scholar] [CrossRef]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Graf, T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell 2011, 9, 504–516. [Google Scholar] [CrossRef] [Green Version]
- Ladewig, J.; Koch, P.; Brustle, O. Leveling Waddington: The emergence of direct programming and the loss of cell fate hierarchies. Nat. Rev. Mol. Cell Biol. 2013, 14, 225–236. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Sudhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.P.; Yang, N.; Vierbuchen, T.; Ostermeier, A.; Fuentes, D.R.; Yang, T.Q.; Citri, A.; Sebastiano, V.; Marro, S.; Sudhof, T.C.; et al. Induction of human neuronal cells by defined transcription factors. Nature 2011, 476, 220–223. [Google Scholar] [CrossRef]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011, 476, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, U.; Kirkeby, A.; Torper, O.; Wood, J.; Nelander, J.; Dufour, A.; Bjorklund, A.; Lindvall, O.; Jakobsson, J.; Parmar, M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 10343–10348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victor, M.B.; Richner, M.; Hermanstyne, T.O.; Ransdell, J.L.; Sobieski, C.; Deng, P.Y.; Klyachko, V.A.; Nerbonne, J.M.; Yoo, A.S. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 2014, 84, 311–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011, 476, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.L.; Zang, T.; Zou, Y.; Chang, J.C.; Gibson, J.R.; Huber, K.M.; Zhang, C.L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013, 4, 2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, E.Y.; Ichida, J.K.; Wainger, B.J.; Toma, J.S.; Rafuse, V.F.; Woolf, C.J.; Eggan, K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 2011, 9, 205–218. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Shi, Y.; Yao, X.; Wang, X.; Shen, L.; Rao, Z.; Yuan, J.; Liu, Y.; Zhou, Z.; Zhang, Z.; et al. Conversion of Astrocytes and Fibroblasts into Functional Noradrenergic Neurons. Cell Rep. 2019, 28, 682–697.e7. [Google Scholar] [CrossRef]
- Blanchard, J.W.; Eade, K.T.; Szucs, A.; Lo Sardo, V.; Tsunemoto, R.K.; Williams, D.; Sanna, P.P.; Baldwin, K.K. Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 2015, 18, 25–35. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, L.; Wang, J.; Zhang, Y.; Kou, Z.; Gao, S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell 2009, 5, 135–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Nashun, B.; Hill, P.W.; Hajkova, P. Reprogramming of cell fate: Epigenetic memory and the erasure of memories past. EMBO J. 2015, 34, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Yamanaka, S. Induced pluripotent stem cells: Opportunities and challenges. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 2198–2207. [Google Scholar] [CrossRef] [Green Version]
- Bao, X.; Zhu, X.; Liao, B.; Benda, C.; Zhuang, Q.; Pei, D.; Qin, B.; Esteban, M.A. MicroRNAs in somatic cell reprogramming. Curr. Opin. Cell Biol. 2013, 25, 208–214. [Google Scholar] [CrossRef]
- Moradi, S.; Asgari, S.; Baharvand, H. Concise review: Harmonies played by microRNAs in cell fate reprogramming. Stem Cells 2014, 32, 3–15. [Google Scholar] [CrossRef]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef]
- Suh, M.R.; Lee, Y.; Kim, J.Y.; Kim, S.K.; Moon, S.H.; Lee, J.Y.; Cha, K.Y.; Chung, H.M.; Yoon, H.S.; Moon, S.Y.; et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004, 270, 488–498. [Google Scholar] [CrossRef] [Green Version]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [Green Version]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divisato, G.; Passaro, F.; Russo, T.; Parisi, S. The Key Role of MicroRNAs in Self-Renewal and Differentiation of Embryonic Stem Cells. Int. J. Mol. Sci. 2020, 21, 6285. [Google Scholar] [CrossRef] [PubMed]
- Judson, R.L.; Babiarz, J.E.; Venere, M.; Blelloch, R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009, 27, 459–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F.; et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar] [CrossRef] [Green Version]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A.; et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar] [CrossRef] [Green Version]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef]
- Lipchina, I.; Elkabetz, Y.; Hafner, M.; Sheridan, R.; Mihailovic, A.; Tuschl, T.; Sander, C.; Studer, L.; Betel, D. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011, 25, 2173–2186. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Guo, X.; Hong, W.; Liu, Q.; Wei, T.; Lu, C.; Gao, L.; Ye, D.; Zhou, Y.; Chen, J.; et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc. Natl. Acad. Sci. USA 2013, 110, 2858–2863. [Google Scholar] [CrossRef] [Green Version]
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009, 460, 1132–1135. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Suzuki, J.; Wang, Y.V.; Menendez, S.; Morera, L.B.; Raya, A.; Wahl, G.M.; Izpisua Belmonte, J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009, 460, 1140–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marion, R.M.; Strati, K.; Li, H.; Murga, M.; Blanco, R.; Ortega, S.; Fernandez-Capetillo, O.; Serrano, M.; Blasco, M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009, 460, 1149–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, D.; Wang, G.; Liu, Y.; Huang, W.; Wu, M.; Zhu, S.; Jia, W.; Deng, A.M.; Liu, H.; Kang, J. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells 2012, 30, 1645–1654. [Google Scholar] [CrossRef]
- Li, Z.; Yang, C.S.; Nakashima, K.; Rana, T.M. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011, 30, 823–834. [Google Scholar] [CrossRef] [Green Version]
- Pfaff, N.; Fiedler, J.; Holzmann, A.; Schambach, A.; Moritz, T.; Cantz, T.; Thum, T. miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep. 2011, 12, 1153–1159. [Google Scholar] [CrossRef]
- Judson, R.L.; Greve, T.S.; Parchem, R.J.; Blelloch, R. MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1227–1235. [Google Scholar] [CrossRef] [Green Version]
- Masserdotti, G.; Gascon, S.; Gotz, M. Direct neuronal reprogramming: Learning from and for development. Development 2016, 143, 2494–2510. [Google Scholar] [CrossRef] [Green Version]
- Colasante, G.; Lignani, G.; Rubio, A.; Medrihan, L.; Yekhlef, L.; Sessa, A.; Massimino, L.; Giannelli, S.G.; Sacchetti, S.; Caiazzo, M.; et al. Rapid Conversion of Fibroblasts into Functional Forebrain GABAergic Interneurons by Direct Genetic Reprogramming. Cell Stem Cell 2015, 17, 719–734. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Qiu, B.; Guan, W.; Wang, Q.; Wang, M.; Li, W.; Gao, L.; Shen, L.; Huang, Y.; Xie, G.; et al. Direct Conversion of Normal and Alzheimer’s Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell 2015, 17, 204–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, A.F.; Grigsby, C.L.; Kulangara, K.; Wang, H.; Yasuda, R.; Leong, K.W. Nonviral direct conversion of primary mouse embryonic fibroblasts to neuronal cells. Mol. Ther. Nucleic Acids 2012, 1, e32. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.; Fraser, I.; McKee, E.; Scheck, K.; Chang, L.; Willerth, S.M. Transdifferentiating Astrocytes Into Neurons Using ASCL1 Functionalized With a Novel Intracellular Protein Delivery Technology. Front. Bioeng. Biotechnol. 2018, 6, 173. [Google Scholar] [CrossRef]
- Lau, S.; Rylander Ottosson, D.; Jakobsson, J.; Parmar, M. Direct neural conversion from human fibroblasts using self-regulating and nonintegrating viral vectors. Cell Rep. 2014, 9, 1673–1680. [Google Scholar] [CrossRef] [Green Version]
- Mertens, J.; Reid, D.; Lau, S.; Kim, Y.; Gage, F.H. Aging in a Dish: iPSC-Derived and Directly Induced Neurons for Studying Brain Aging and Age-Related Neurodegenerative Diseases. Annu. Rev. Genet. 2018, 52, 271–293. [Google Scholar] [CrossRef]
- Huh, C.J.; Zhang, B.; Victor, M.B.; Dahiya, S.; Batista, L.F.; Horvath, S.; Yoo, A.S. Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. Elife 2016, 5, e18648. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.L.; Zang, T.; Zhang, C.L. Direct Lineage Reprogramming Reveals Disease-Specific Phenotypes of Motor Neurons from Human ALS Patients. Cell Rep. 2016, 14, 115–128. [Google Scholar] [CrossRef] [Green Version]
- Wapinski, O.L.; Vierbuchen, T.; Qu, K.; Lee, Q.Y.; Chanda, S.; Fuentes, D.R.; Giresi, P.G.; Ng, Y.H.; Marro, S.; Neff, N.F.; et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 2013, 155, 621–635. [Google Scholar] [CrossRef] [Green Version]
- Treutlein, B.; Lee, Q.Y.; Camp, J.G.; Mall, M.; Koh, W.; Shariati, S.A.; Sim, S.; Neff, N.F.; Skotheim, J.M.; Wernig, M.; et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 2016, 534, 391–395. [Google Scholar] [CrossRef] [Green Version]
- Wapinski, O.L.; Lee, Q.Y.; Chen, A.C.; Li, R.; Corces, M.R.; Ang, C.E.; Treutlein, B.; Xiang, C.; Baubet, V.; Suchy, F.P.; et al. Rapid Chromatin Switch in the Direct Reprogramming of Fibroblasts to Neurons. Cell Rep. 2017, 20, 3236–3247. [Google Scholar] [CrossRef] [Green Version]
- De Gregorio, R.; Pulcrano, S.; De Sanctis, C.; Volpicelli, F.; Guatteo, E.; von Oerthel, L.; Latagliata, E.C.; Esposito, R.; Piscitelli, R.M.; Perrone-Capano, C.; et al. miR-34b/c Regulates Wnt1 and Enhances Mesencephalic Dopaminergic Neuron Differentiation. Stem Cell Rep. 2018, 10, 1237–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Della Valle, F.; Thimma, M.P.; Caiazzo, M.; Pulcrano, S.; Celii, M.; Adroub, S.A.; Liu, P.; Alanis-Lobato, G.; Broccoli, V.; Orlando, V. Transdifferentiation of Mouse Embryonic Fibroblasts into Dopaminergic Neurons Reactivates LINE-1 Repetitive Elements. Stem Cell Rep. 2020, 14, 60–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abernathy, D.G.; Kim, W.K.; McCoy, M.J.; Lake, A.M.; Ouwenga, R.; Lee, S.W.; Xing, X.; Li, D.; Lee, H.J.; Heuckeroth, R.O.; et al. MicroRNAs Induce a Permissive Chromatin Environment that Enables Neuronal Subtype-Specific Reprogramming of Adult Human Fibroblasts. Cell Stem Cell 2017, 21, 332–348.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.W.; Oh, Y.M.; Lu, Y.L.; Kim, W.K.; Yoo, A.S. MicroRNAs Overcome Cell Fate Barrier by Reducing EZH2-Controlled REST Stability during Neuronal Conversion of Human Adult Fibroblasts. Dev. Cell 2018, 46, 73–84.e77. [Google Scholar] [CrossRef] [Green Version]
- Cates, K.; McCoy, M.J.; Kwon, J.S.; Liu, Y.; Abernathy, D.G.; Zhang, B.; Liu, S.; Gontarz, P.; Kim, W.K.; Chen, S.; et al. Deconstructing Stepwise Fate Conversion of Human Fibroblasts to Neurons by MicroRNAs. Cell Stem Cell 2021, 28, 127–140.e129. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [Green Version]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008, 10, 987–993. [Google Scholar] [CrossRef]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Ouyang, K.; Huang, J.; Zhou, Y.; Ouyang, H.; Li, H.; Wang, G.; Wu, Q.; Wei, C.; Bi, Y.; et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013, 152, 82–96. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Qian, H.; Hu, J.; Zhou, B.; Zhou, Y.; Hu, X.; Karakhanyan, A.; Pang, Z.; Fu, X.D. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 2016, 19, 807–815. [Google Scholar] [CrossRef] [Green Version]
- Rivetti di Val Cervo, P.; Romanov, R.A.; Spigolon, G.; Masini, D.; Martin-Montanez, E.; Toledo, E.M.; La Manno, G.; Feyder, M.; Pifl, C.; Ng, Y.H.; et al. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 2017, 35, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Pascale, E.; Divisato, G.; Palladino, R.; Auriemma, M.; Ngalya, E.F.; Caiazzo, M. Noncoding RNAs and Midbrain DA Neurons: Novel Molecular Mechanisms and Therapeutic Targets in Health and Disease. Biomolecules 2020, 10, 1269. [Google Scholar] [CrossRef] [PubMed]
- Soreq, H. Novel roles of non-coding brain RNAs in health and disease. Front. Mol. Neurosci. 2014, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, J.H.K.; Groenink, L.; Caiazzo, M. Cell reprogramming approaches in gene- and cell-based therapies for Parkinson’s disease. J. Control. Release 2018, 286, 114–124. [Google Scholar] [CrossRef]
- Bocchi, R.; Masserdotti, G.; Gotz, M. Direct neuronal reprogramming: Fast forward from new concepts toward therapeutic approaches. Neuron 2021, 110, 366–393. [Google Scholar] [CrossRef]
- Qian, H.; Kang, X.; Hu, J.; Zhang, D.; Liang, Z.; Meng, F.; Zhang, X.; Xue, Y.; Maimon, R.; Dowdy, S.F.; et al. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 2020, 582, 550–556. [Google Scholar] [CrossRef]
- Zhou, H.; Su, J.; Hu, X.; Zhou, C.; Li, H.; Chen, Z.; Xiao, Q.; Wang, B.; Wu, W.; Sun, Y.; et al. Glia-to-Neuron Conversion by CRISPR-CasRx Alleviates Symptoms of Neurological Disease in Mice. Cell 2020, 181, 590–603.e516. [Google Scholar] [CrossRef]
- Wang, L.L.; Serrano, C.; Zhong, X.; Ma, S.; Zou, Y.; Zhang, C.L. Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell 2021, 184, 5465–5481.e16. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Cardiac fibrosis. Cardiovasc. Res. 2021, 117, 1450–1488. [Google Scholar] [CrossRef]
- Passaro, F.; Tocchetti, C.G.; Spinetti, G.; Paudice, F.; Ambrosone, L.; Costagliola, C.; Cacciatore, F.; Abete, P.; Testa, G. Targeting fibrosis in the failing heart with nanoparticles. Adv. Drug Deliv. Rev. 2021, 174, 461–481. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, J.; Qian, L. Direct cardiac reprogramming comes of age: Recent advance and remaining challenges. Semin. Cell Dev. Biol. 2021, 122, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Testa, G.; Di Benedetto, G.; Passaro, F. Advanced Technologies to Target Cardiac Cell Fate Plasticity for Heart Regeneration. Int. J. Mol. Sci. 2021, 22, 9517. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.R.; Ilinykh, A.; Ivey, M.J.; Kuwabara, J.T.; D’Antoni, M.L.; Debuque, R.; Chandran, A.; Wang, L.; Arora, K.; Rosenthal, N.A.; et al. Revisiting Cardiac Cellular Composition. Circ. Res. 2016, 118, 400–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passaro, F.; Testa, G.; Ambrosone, L.; Costagliola, C.; Tocchetti, C.G.; di Nezza, F.; Russo, M.; Pirozzi, F.; Abete, P.; Russo, T.; et al. Nanotechnology-Based Cardiac Targeting and Direct Cardiac Reprogramming: The Betrothed. Stem Cells Int. 2017, 2017, 4940397. [Google Scholar] [CrossRef] [Green Version]
- Soufi, A.; Garcia, M.F.; Jaroszewicz, A.; Osman, N.; Pellegrini, M.; Zaret, K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161, 555–568. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.D.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Delgado-Olguin, P.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef] [Green Version]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef] [Green Version]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, H.; Wang, Z.; Garry, G.A.; Malladi, V.S.; Botten, G.A.; Ye, W.; Zhou, H.; Osterwalder, M.; Dickel, D.E.; Visel, A.; et al. Cardiac Reprogramming Factors Synergistically Activate Genome-wide Cardiogenic Stage-Specific Enhancers. Cell Stem Cell 2019, 25, 69–86.e5. [Google Scholar] [CrossRef]
- Addis, R.C.; Ifkovits, J.L.; Pinto, F.; Kellam, L.D.; Esteso, P.; Rentschler, S.; Christoforou, N.; Epstein, J.A.; Gearhart, J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell Cardiol. 2013, 60, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Dickson, M.E.; Kim, M.S.; Bassel-Duby, R.; Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 11864–11869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.D.; Wang, G.G.; Liu, J.; et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Testa, G.; Russo, M.; Di Benedetto, G.; Barbato, M.; Parisi, S.; Pirozzi, F.; Tocchetti, C.G.; Abete, P.; Bonaduce, D.; Russo, T.; et al. Bmi1 inhibitor PTC-209 promotes Chemically-induced Direct Cardiac Reprogramming of cardiac fibroblasts into cardiomyocytes. Sci. Rep. 2020, 10, 7129. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhao, L.; Yu, X.; Zhang, J.; Qian, L.; Jin, J.; Lu, R.; Zhou, Y. Inhibition of EZH2 primes the cardiac gene activation via removal of epigenetic repression during human direct cardiac reprogramming. Stem Cell Res. 2021, 53, 102365. [Google Scholar] [CrossRef] [PubMed]
- Garry, G.A.; Bezprozvannaya, S.; Chen, K.; Zhou, H.; Hashimoto, H.; Morales, M.G.; Liu, N.; Bassel-Duby, R.; Olson, E.N. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat. Cell Biol. 2021, 23, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.A.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, T.M.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation 2017, 135, 978–995. [Google Scholar] [CrossRef]
- Abad, M.; Hashimoto, H.; Zhou, H.; Morales, M.G.; Chen, B.; Bassel-Duby, R.; Olson, E.N. Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity. Stem Cell Rep. 2017, 8, 548–560. [Google Scholar] [CrossRef]
- Passaro, F.; Testa, G. Implications of Cellular Aging in Cardiac Reprogramming. Front. Cardiovasc. Med. 2018, 5, 43. [Google Scholar] [CrossRef] [Green Version]
- Muraoka, N.; Nara, K.; Tamura, F.; Kojima, H.; Yamakawa, H.; Sadahiro, T.; Miyamoto, K.; Isomi, M.; Haginiwa, S.; Tani, H.; et al. Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac reprogramming. Nat. Commun. 2019, 10, 674. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, N.; Huang, Y.; Spencer, C.I.; Fu, J.D.; Yu, C.; Liu, K.; Nie, B.; Xu, T.; Li, K.; et al. Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts. Cell Stem Cell 2016, 18, 368–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgkinson, C.P.; Kang, M.H.; Dal-Pra, S.; Mirotsou, M.; Dzau, V.J. MicroRNAs and Cardiac Regeneration. Circ. Res. 2015, 116, 1700–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dal-Pra, S.; Hodgkinson, C.P.; Mirotsou, M.; Kirste, I.; Dzau, V.J. Demethylation of H3K27 Is Essential for the Induction of Direct Cardiac Reprogramming by miR Combo. Circ. Res. 2017, 120, 1403–1413. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.; Kim, S.; Lee, H.; Rah, W.; Cho, H.C.; Kim, N.K.; Bae, S.; Shin, D.H.; Lee, M.G.; Park, I.H.; et al. Regeneration of infarcted mouse hearts by cardiovascular tissue formed via the direct reprogramming of mouse fibroblasts. Nat. Biomed. Eng. 2021, 5, 880–896. [Google Scholar] [CrossRef]
- Paoletti, C.; Divieto, C.; Tarricone, G.; Di Meglio, F.; Nurzynska, D.; Chiono, V. MicroRNA-Mediated Direct Reprogramming of Human Adult Fibroblasts Toward Cardiac Phenotype. Front. Bioeng. Biotechnol. 2020, 8, 529. [Google Scholar] [CrossRef]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [Green Version]
- Riching, A.S.; Zhao, Y.; Cao, Y.; Londono, P.; Xu, H.; Song, K. Suppression of Pro-fibrotic Signaling Potentiates Factor-mediated Reprogramming of Mouse Embryonic Fibroblasts into Induced Cardiomyocytes. J. Vis. Exp. 2018, 136, e57687. [Google Scholar] [CrossRef]
- Nam, Y.J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; DiMaio, J.M.; Baker, L.A.; Bassel-Duby, R.; et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 2013, 110, 5588–5593. [Google Scholar] [CrossRef] [Green Version]
- Christoforou, N.; Chakraborty, S.; Kirkton, R.D.; Adler, A.F.; Addis, R.C.; Leong, K.W. Core Transcription Factors, MicroRNAs, and Small Molecules Drive Transdifferentiation of Human Fibroblasts Towards The Cardiac Cell Lineage. Sci. Rep. 2017, 7, 40285. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.P.; Mathison, M.; Patel, V.; Sanagasetti, D.; Gibson, B.W.; Yang, J.; Rosengart, T.K. MiR-590 Promotes Transdifferentiation of Porcine and Human Fibroblasts Toward a Cardiomyocyte-Like Fate by Directly Repressing Specificity Protein 1. J. Am. Heart Assoc. 2016, 5, e003922. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.P.; Pinnamaneni, J.P.; Pugazenthi, A.; Sanagasetti, D.; Mathison, M.; Wang, K.; Yang, J.; Rosengart, T.K. Enhanced Generation of Induced Cardiomyocytes Using a Small-Molecule Cocktail to Overcome Barriers to Cardiac Cellular Reprogramming. J. Am. Heart Assoc. 2020, 9, e015686. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, M.H.; Hu, J.; Pratt, R.E.; Hodgkinson, C.P.; Asokan, A.; Dzau, V.J. Optimizing delivery for efficient cardiac reprogramming. Biochem. Biophys. Res. Commun. 2020, 533, 9–16. [Google Scholar] [CrossRef]
- Jayawardena, T.M.; Finch, E.A.; Zhang, L.; Zhang, H.; Hodgkinson, C.P.; Pratt, R.E.; Rosenberg, P.B.; Mirotsou, M.; Dzau, V.J. MicroRNA induced cardiac reprogramming in vivo: Evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015, 116, 418–424. [Google Scholar] [CrossRef]
- Escors, D.; Breckpot, K. Lentiviral vectors in gene therapy: Their current status and future potential. Arch. Immunol. Ther. Exp. 2010, 58, 107–119. [Google Scholar] [CrossRef] [Green Version]
- Fausther-Bovendo, H.; Kobinger, G.P. Pre-existing immunity against Ad vectors: Humoral, cellular, and innate response, what’s important? Hum. Vaccines Immunother. 2014, 10, 2875–2884. [Google Scholar] [CrossRef] [Green Version]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef] [Green Version]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L., 3rd; Strohl, W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 2017, 31, 317–334. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Xue, S.; Du, M.; Lian, F. Highly Efficient MicroRNA Delivery Using Functionalized Carbon Dots for Enhanced Conversion of Fibroblasts to Cardiomyocytes. Int. J. Nanomed. 2021, 16, 3741–3754. [Google Scholar] [CrossRef]
- Wang, Q.; Song, Y.; Chen, J.; Li, Q.; Gao, J.; Tan, H.; Zhu, Y.; Wang, Z.; Li, M.; Yang, H.; et al. Direct in vivo reprogramming with non-viral sequential targeting nanoparticles promotes cardiac regeneration. Biomaterials 2021, 276, 121028. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control. Release 2019, 313, 80–95. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [Green Version]
| miRNAs | Functions in iPSC Reprogramming | Direct Targets | References |
|---|---|---|---|
| miR-290 cluster (mmu-miR-291a-3p, mmu-miR-294, and mmu-miR-295) | Significantly increases the number of mouse iPSC colonies | MEK pathway -directly repressing Akt1 | [33] |
| miR-let7 family | Its inhibition via LIN28 facilitates induced pluripotency. | c-Myc, Lin28b, and Hmga2 | [35] |
| miR-302/367 cluster | Induces pluripotency of mouse and human fibroblasts without exogenous expression of other transcription factors | TGFβ receptor 2 NR2F2 | [37] |
| hsa/mmu-miR-200c, hsa/mmu -miR-302s, and hsa/mmu miR-369s | Used to induce pluripotency of mouse and human somatic cells without integration of any viral-based vectors | Aof1 and Zeb1/2 | [36] |
| miR-130/ 301/721 family | Enhances the efficiency of iPSC generation by repressing the homeobox transcription factor Meox2. | Meox2 | [46] |
| mmu-miR-138 | Targets the 3’-UTR of the p53 mRNA and significantly increases reprogramming efficiency | p53 signaling pathway | [44] |
| miR-106b-25/ miR-106a-363 clusters | Overexpression of members of these two clusters significantly enhances iPSC generation. | Tgfbr2 and p21 | [45] |
| miR-181 family | Is transiently induced during the initial phase of iPSC reprogramming | Cpsf6, Nr2c2, Marcks, Bptf, Igf2bp2, Nol8, Bclaf1, and Lin7c | [47] |
| miRNAs | Functions in Neuronal Cell Reprogramming | Direct Targets | References |
|---|---|---|---|
| hsa-miR-9-5p/-3p hsa-miR-124 | Neuronal fate induction | PTPB1, REST, CoREST, SCP1, and BAF53a | [11] |
| hsa-miR-124/hsa-miR-9-5p/3p + CTIP2/DLX1/DLX2/MYT1L | Promotes the differentiation from human adult fibroblasts to GABA medium spiny neurons | PTPB1, REST, CoREST, SCP1, and BAF53a | [13] |
| hsa-miR-124/hsa-miR-9-5p/3p + ISL1/LHX3 | Promotes the differentiation from human adult fibroblasts to motor neurons | PTPB1, REST, CoREST, SCP1, and BAF53a | [63] |
| miR-218 + ASCL1/NEUROD1/ LMX1A | Promotes in vivo astrocyte-to-neuron conversion | Onecut2 | [71] |
| miR-34b-5p/miR-34c-5p + ASCL1/NURR1 | Increases the generation of iDANs | Wnt1 | [61] |
| miRNAs | Functions in Cardiac Cell Reprogramming | Direct Targets | References |
|---|---|---|---|
| mmu-miR-1/mmu-miR-133a/mmu-miR-208a/mmu-miR-499 | This combination (miRcombo) induces transdifferentiation to iCMs. | Twf1, Col16a1, and Ezh2 | [102] |
| mmu-miR-208b-3p + ascorbic acid/BMP4 | This combination (MAB) induces transdifferentiation to iCMs. | Gata4 | [105] |
| mmu-miR-133a + GMT/GMTMM cocktails | Increases the efficiency of iCM generation | Snai1 | [107] |
| mmu-miR-1/mmu-mmu-miR-133a + GHMT cocktail | Increases the efficiency of iCM generation | Twf1 and Snai1 | [109] |
| hsa-miR-590 + GMT cocktail | Increases the maturation of iCMs | Sp1 | [111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pascale, E.; Caiazza, C.; Paladino, M.; Parisi, S.; Passaro, F.; Caiazzo, M. MicroRNA Roles in Cell Reprogramming Mechanisms. Cells 2022, 11, 940. https://doi.org/10.3390/cells11060940
Pascale E, Caiazza C, Paladino M, Parisi S, Passaro F, Caiazzo M. MicroRNA Roles in Cell Reprogramming Mechanisms. Cells. 2022; 11(6):940. https://doi.org/10.3390/cells11060940
Chicago/Turabian StylePascale, Emilia, Carmen Caiazza, Martina Paladino, Silvia Parisi, Fabiana Passaro, and Massimiliano Caiazzo. 2022. "MicroRNA Roles in Cell Reprogramming Mechanisms" Cells 11, no. 6: 940. https://doi.org/10.3390/cells11060940
APA StylePascale, E., Caiazza, C., Paladino, M., Parisi, S., Passaro, F., & Caiazzo, M. (2022). MicroRNA Roles in Cell Reprogramming Mechanisms. Cells, 11(6), 940. https://doi.org/10.3390/cells11060940







