Molecular Characterization of Portuguese Patients with Hereditary Cerebellar Ataxia
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Study
2.2. Genetic Analysis
2.3. Minigene Assays
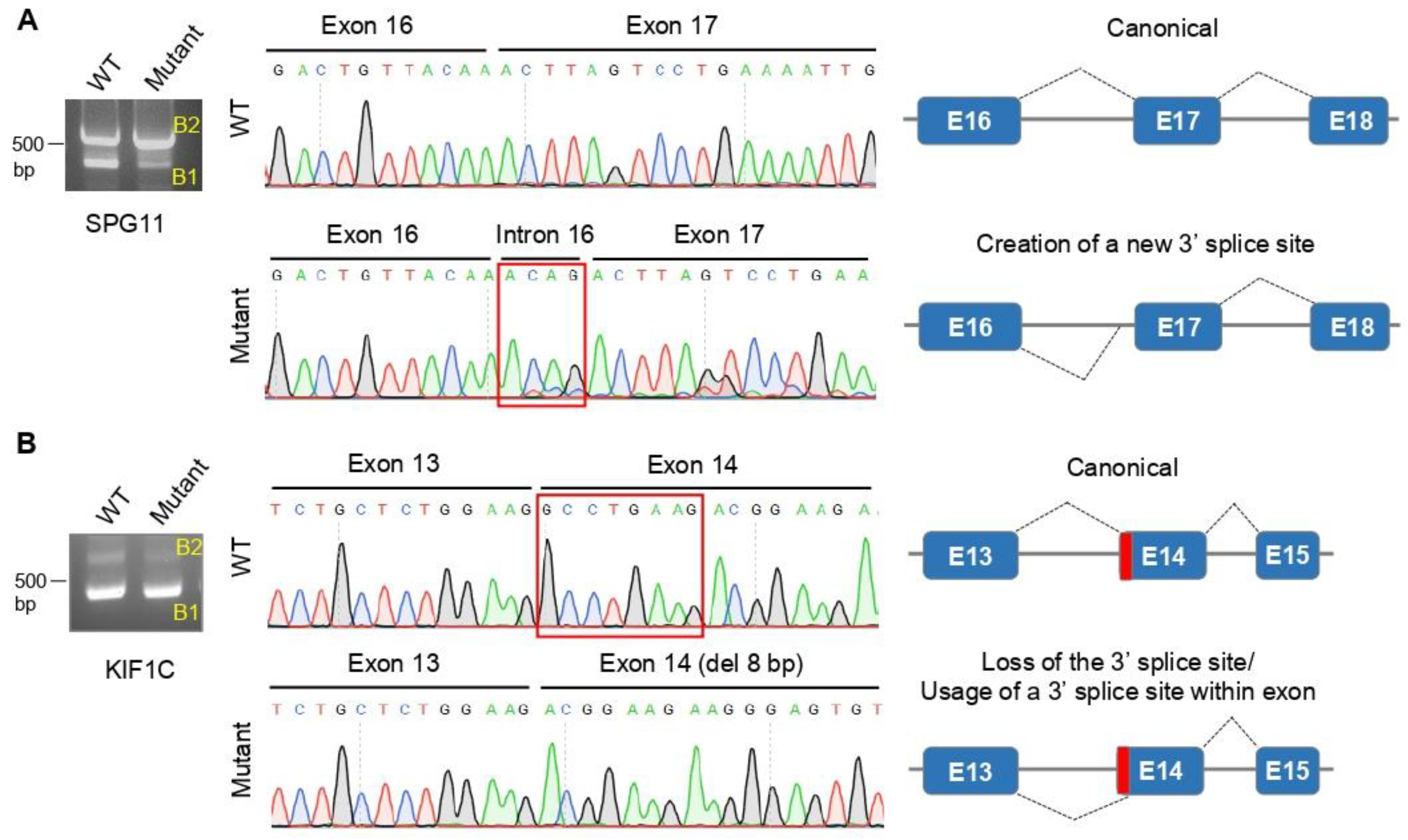
3. Results
3.1. Genetic and Molecular Characterization
3.2. Clinical Characterization
3.2.1. Spastic Ataxia
3.2.2. Ataxia and Neuropathy
3.2.3. Ataxia and Oculomotor Apraxia
3.2.4. Ataxia and Dystonia/Ataxia and Cognitive Impairment
4. Discussion
4.1. Heterozygous De Novo Variants
4.2. Molecular Mechanisms
4.2.1. Spastic Ataxia
4.2.2. Ataxia and Neuropathy
4.2.3. Ataxia and Oculomotor Apraxia
4.2.4. Ataxia and Cognitive Impairment or Dystonia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anheim, M.; Tranchant, C.; Koenig, M. The autosomal recessive cerebellar ataxias. N. Engl. J. Med. 2012, 366, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Ruano, L.; Melo, C.; Silva, M.C.; Coutinho, P. The global epidemiology of hereditary ataxia and spastic paraplegia: A systematic review of prevalence studies. Neuroepidemiology 2014, 42, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Sequeiros, J.; Martins, S.; Silveira, I. Epidemiology and population genetics of degenerative ataxias. Handb. Clin. Neurol. 2012, 103, 227–251. [Google Scholar] [CrossRef] [PubMed]
- Fogel, B.L.; Lee, H.; Deignan, J.L.; Strom, S.P.; Kantarci, S.; Wang, X.; Quintero-Rivera, F.; Vilain, E.; Grody, W.W.; Perlman, S.; et al. Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol. 2014, 71, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- van de Warrenburg, B.P.; Schouten, M.I.; de Bot, S.T.; Vermeer, S.; Meijer, R.; Pennings, M.; Gilissen, C.; Willemsen, M.A.; Scheffer, H.; Kamsteeg, E.J. Clinical exome sequencing for cerebellar ataxia and spastic paraplegia uncovers novel gene-disease associations and unanticipated rare disorders. Eur. J. Hum. Genet. 2016, 24, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.J.; Watchon, M.; Laird, A.S. Aberrant Cerebellar Circuitry in the Spinocerebellar Ataxias. Front. Neurosci. 2020, 14, 707. [Google Scholar] [CrossRef] [PubMed]
- Eidhof, I.; van de Warrenburg, B.P.; Schenck, A. Integrative network and brain expression analysis reveals mechanistic modules in ataxia. J. Med. Genet. 2019, 56, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Duenas, A.M.; Goold, R.; Giunti, P. Molecular pathogenesis of spinocerebellar ataxias. Brain 2006, 129, 1357–1370. [Google Scholar] [CrossRef] [PubMed]
- Fogel, B.L. Autosomal-recessive cerebellar ataxias. Handb. Clin. Neurol. 2018, 147, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, P.; Ruano, L.; Loureiro, J.L.; Cruz, V.T.; Barros, J.; Tuna, A.; Barbot, C.; Guimarães, J.; Alonso, I.; Silveira, I.; et al. Hereditary Ataxia and Spastic Paraplegia in Portugal. JAMA Neurol. 2013, 70, 746. [Google Scholar] [CrossRef]
- Bras, J.; Alonso, I.; Barbot, C.; Costa, M.M.; Darwent, L.; Orme, T.; Sequeiros, J.; Hardy, J.; Coutinho, P.; Guerreiro, R. Mutations in PNKP cause recessive ataxia with oculomotor apraxia type 4. Am. J. Hum. Genet 2015, 96, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Damasio, J.; Kun-Rodrigues, C.; Barbot, C.; Sequeiros, J.; Bras, J.; Alonso, I.; Guerreiro, R. Novel MAG Variant Causes Cerebellar Ataxia with Oculomotor Apraxia: Molecular Basis and Expanded Clinical Phenotype. J. Clin. Med. 2020, 9, 1212. [Google Scholar] [CrossRef] [PubMed]
- Cortese, A.; Simone, R.; Sullivan, R.; Vandrovcova, J.; Tariq, H.; Yau, W.Y.; Humphrey, J.; Jaunmuktane, Z.; Sivakumar, P.; Polke, J.; et al. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat. Genet. 2019, 51, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.11–11.10.33. [Google Scholar] [CrossRef]
- Faust, G.G.; Hall, I.M. SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 2014, 30, 2503–2505. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jian, X.; Boerwinkle, E. dbNSFP v2.0: A database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013, 34, E2393–E2402. [Google Scholar] [CrossRef] [PubMed]
- Smedley, D.; Jacobsen, J.O.; Jager, M.; Kohler, S.; Holtgrewe, M.; Schubach, M.; Siragusa, E.; Zemojtel, T.; Buske, O.J.; Washington, N.L.; et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc. 2015, 10, 2004–2015. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef] [PubMed]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.; Edwards, K.J.; Day, I.N.; Gaunt, T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Salgado, D.; Desvignes, J.P.; Rai, G.; Blanchard, A.; Miltgen, M.; Pinard, A.; Levy, N.; Collod-Beroud, G.; Beroud, C. UMD-Predictor: A High-Throughput Sequencing Compliant System for Pathogenicity Prediction of any Human cDNA Substitution. Hum. Mutat. 2016, 37, 439–446. [Google Scholar] [CrossRef]
- Shapiro, M.B.; Senapathy, P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987, 15, 7155–7174. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Lin, X.; Salzberg, S.L. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 2001, 29, 1185–1190. [Google Scholar] [CrossRef]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Beroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- da Gloria, V.G.; Martins de Araujo, M.; Mafalda Santos, A.; Leal, R.; de Almeida, S.F.; Carmo, A.M.; Moreira, A. T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1. J. Immunol. 2014, 193, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Rosenfeld, J.A.; Yamamoto, S.; Harel, T.; Zuo, Z.; Hall, M.; Wierenga, K.J.; Pastore, M.T.; Bartholomew, D.; Delgado, M.R.; et al. Clinically severe CACNA1A alleles affect synaptic function and neurodegeneration differentially. PLoS Genet. 2017, 13, e1006905. [Google Scholar] [CrossRef] [PubMed]
- Seixas, A.I.; Loureiro, J.R.; Costa, C.; Ordonez-Ugalde, A.; Marcelino, H.; Oliveira, C.L.; Loureiro, J.L.; Dhingra, A.; Brandao, E.; Cruz, V.T.; et al. A Pentanucleotide ATTTC Repeat Insertion in the Non-coding Region of DAB1, Mapping to SCA37, Causes Spinocerebellar Ataxia. Am. J. Hum. Genet. 2017, 101, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Coutelier, M.; Hammer, M.B.; Stevanin, G.; Monin, M.L.; Davoine, C.S.; Mochel, F.; Labauge, P.; Ewenczyk, C.; Ding, J.; Gibbs, J.R.; et al. Efficacy of Exome-Targeted Capture Sequencing to Detect Mutations in Known Cerebellar Ataxia Genes. JAMA Neurol. 2018, 75, 591–599. [Google Scholar] [CrossRef]
- Pedroso, J.L.; Rocha, C.R.; Macedo-Souza, L.I.; De Mario, V.; Marques, W., Jr.; Barsottini, O.G.; Bulle Oliveira, A.S.; Menck, C.F.; Kok, F. Mutation in PNKP presenting initially as axonal Charcot-Marie-Tooth disease. Neurol. Genet. 2015, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Van Goethem, G.; Martin, J.J.; Dermaut, B.; Löfgren, A.; Wibail, A.; Ververken, D.; Tack, P.; Dehaene, I.; Van Zandijcke, M.; Moonen, M.; et al. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul. Disord. 2003, 13, 133–142. [Google Scholar] [CrossRef]
- Synofzik, M.; Smets, K.; Mallaret, M.; Di Bella, D.; Gallenmuller, C.; Baets, J.; Schulze, M.; Magri, S.; Sarto, E.; Mustafa, M.; et al. SYNE1 ataxia is a common recessive ataxia with major non-cerebellar features: A large multi-centre study. Brain 2016, 139, 1378–1393. [Google Scholar] [CrossRef] [PubMed]
- Dor, T.; Cinnamon, Y.; Raymond, L.; Shaag, A.; Bouslam, N.; Bouhouche, A.; Gaussen, M.; Meyer, V.; Durr, A.; Brice, A.; et al. KIF1C mutations in two families with hereditary spastic paraparesis and cerebellar dysfunction. J. Med. Genet. 2014, 51, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Anheim, M.; Monga, B.; Fleury, M.; Charles, P.; Barbot, C.; Salih, M.; Delaunoy, J.P.; Fritsch, M.; Arning, L.; Synofzik, M.; et al. Ataxia with oculomotor apraxia type 2: Clinical, biological and genotype/phenotype correlation study of a cohort of 90 patients. Brain 2009, 132, 2688–2698. [Google Scholar] [CrossRef] [PubMed]
- Ohba, C.; Haginoya, K.; Osaka, H.; Kubota, K.; Ishiyama, A.; Hiraide, T.; Komaki, H.; Sasaki, M.; Miyatake, S.; Nakashima, M.; et al. De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J. Hum. Genet. 2015, 60, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Morais, S.; Raymond, L.; Mairey, M.; Coutinho, P.; Brandao, E.; Ribeiro, P.; Loureiro, J.L.; Sequeiros, J.; Brice, A.; Alonso, I.; et al. Massive sequencing of 70 genes reveals a myriad of missing genes or mechanisms to be uncovered in hereditary spastic paraplegias. Eur. J. Hum. Genet. 2017, 25, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.; Srour, M.; Kim, D.; Hamdan, F.F.; Lim, S.H.; Brunel-Guitton, C.; Decarie, J.C.; Rossignol, E.; Mitchell, G.A.; Schreiber, A.; et al. De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum. Mutat. 2015, 36, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Nicita, F.; Ginevrino, M.; Travaglini, L.; D’Arrigo, S.; Zorzi, G.; Borgatti, R.; Terrone, G.; Catteruccia, M.; Vasco, G.; Brankovic, V.; et al. Heterozygous KIF1A variants underlie a wide spectrum of neurodevelopmental and neurodegenerative disorders. J. Med. Genet. 2021, 58, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Toru, S.; Murakoshi, T.; Ishikawa, K.; Saegusa, H.; Fujigasaki, H.; Uchihara, T.; Nagayama, S.; Osanai, M.; Mizusawa, H.; Tanabe, T. Spinocerebellar ataxia type 6 mutation alters P-type calcium channel function. J. Biol. Chem. 2000, 275, 10893–10898. [Google Scholar] [CrossRef] [PubMed]
- Wappl, E.; Koschak, A.; Poteser, M.; Sinnegger, M.J.; Walter, D.; Eberhart, A.; Groschner, K.; Glossmann, H.; Kraus, R.L.; Grabner, M.; et al. Functional consequences of P/Q-type Ca2+ channel Cav2.1 missense mutations associated with episodic ataxia type 2 and progressive ataxia. J. Biol. Chem. 2002, 277, 6960–6966. [Google Scholar] [CrossRef] [PubMed]
- Heinzen, E.L.; Swoboda, K.J.; Hitomi, Y.; Gurrieri, F.; Nicole, S.; de Vries, B.; Tiziano, F.D.; Fontaine, B.; Walley, N.M.; Heavin, S.; et al. De novo mutations in ATP1A3 cause alternating hemiplegia of childhood. Nat. Genet. 2012, 44, 1030–1034. [Google Scholar] [CrossRef]
- Demos, M.K.; van Karnebeek, C.D.; Ross, C.J.; Adam, S.; Shen, Y.; Zhan, S.H.; Shyr, C.; Horvath, G.; Suri, M.; Fryer, A.; et al. A novel recurrent mutation in ATP1A3 causes CAPOS syndrome. Orphanet J. Rare Dis. 2014, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Klockgether, T.; Mariotti, C.; Paulson, H.L. Spinocerebellar ataxia. Nat. Rev. Dis. Primers 2019, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Gur-Hartman, T.; Berkowitz, O.; Yosovich, K.; Roubertie, A.; Zanni, G.; Macaya, A.; Heimer, G.; Duenas, B.P.; Sival, D.A.; Pode-Shakked, B.; et al. Clinical phenotypes of infantile onset CACNA1A-related disorder. Eur. J. Paediatr. Neurol. EJPN Off. J. Eur. Paediatr. Neurol. Soc. 2021, 30, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Salles, P.A.; Mata, I.F.; Brunger, T.; Lal, D.; Fernandez, H.H. ATP1A3-Related Disorders: An Ever-Expanding Clinical Spectrum. Front. Neurol. 2021, 12, 637890. [Google Scholar] [CrossRef] [PubMed]
- Roubergue, A.; Roze, E.; Vuillaumier-Barrot, S.; Fontenille, M.J.; Meneret, A.; Vidailhet, M.; Fontaine, B.; Doummar, D.; Philibert, B.; Riant, F.; et al. The multiple faces of the ATP1A3-related dystonic movement disorder. Mov. Disord. Off. J. Mov. Disord. Soc. 2013, 28, 1457–1459. [Google Scholar] [CrossRef] [PubMed]
- Bereznyakova, O.; Dupre, N. Spastic ataxias. Handb. Clin. Neurol. 2018, 155, 191–203. [Google Scholar] [CrossRef] [PubMed]
- de Bot, S.T.; Willemsen, M.A.; Vermeer, S.; Kremer, H.P.; van de Warrenburg, B.P. Reviewing the genetic causes of spastic-ataxias. Neurology 2012, 79, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Parfitt, D.A.; Michael, G.J.; Vermeulen, E.G.; Prodromou, N.V.; Webb, T.R.; Gallo, J.M.; Cheetham, M.E.; Nicoll, W.S.; Blatch, G.L.; Chapple, J.P. The ataxia protein sacsin is a functional co-chaperone that protects against polyglutamine-expanded ataxin-1. Hum. Mol. Genet. 2009, 18, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
- Girard, M.; Lariviere, R.; Parfitt, D.A.; Deane, E.C.; Gaudet, R.; Nossova, N.; Blondeau, F.; Prenosil, G.; Vermeulen, E.G.; Duchen, M.R.; et al. Mitochondrial dysfunction and Purkinje cell loss in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS). Proc. Natl. Acad. Sci. USA 2012, 109, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Lariviere, R.; Gaudet, R.; Gentil, B.J.; Girard, M.; Conte, T.C.; Minotti, S.; Leclerc-Desaulniers, K.; Gehring, K.; McKinney, R.A.; Shoubridge, E.A.; et al. Sacs knockout mice present pathophysiological defects underlying autosomal recessive spastic ataxia of Charlevoix-Saguenay. Hum. Mol. Genet. 2015, 24, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Lariviere, R.; Sgarioto, N.; Marquez, B.T.; Gaudet, R.; Choquet, K.; McKinney, R.A.; Watt, A.J.; Brais, B. Sacs R272C missense homozygous mice develop an ataxia phenotype. Mol. Brain 2019, 12, 19. [Google Scholar] [CrossRef]
- Longo, F.; De Ritis, D.; Miluzio, A.; Fraticelli, D.; Baets, J.; Scarlato, M.; Santorelli, F.M.; Biffo, S.; Maltecca, F. Assessment of Sacsin Turnover in Patients With ARSACS: Implications for Molecular Diagnosis and Pathogenesis. Neurology 2021, 97, e2315–e2327. [Google Scholar] [CrossRef] [PubMed]
- Bouhlal, Y.; Amouri, R.; El Euch-Fayeche, G.; Hentati, F. Autosomal recessive spastic ataxia of Charlevoix-Saguenay: An overview. Parkinsonism Relat. Disord. 2011, 17, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Gabrych, D.R.; Lau, V.Z.; Niwa, S.; Silverman, M.A. Going Too Far Is the Same as Falling Short(dagger): Kinesin-3 Family Members in Hereditary Spastic Paraplegia. Front. Cell Neurosci. 2019, 13, 419. [Google Scholar] [CrossRef] [PubMed]
- Caballero Oteyza, A.; Battaloglu, E.; Ocek, L.; Lindig, T.; Reichbauer, J.; Rebelo, A.P.; Gonzalez, M.A.; Zorlu, Y.; Ozes, B.; Timmann, D.; et al. Motor protein mutations cause a new form of hereditary spastic paraplegia. Neurology 2014, 82, 2007–2016. [Google Scholar] [CrossRef] [PubMed]
- Manole, A.; Chelban, V.; Haridy, N.A.; Hamed, S.A.; Berardo, A.; Reilly, M.M.; Houlden, H. Severe axonal neuropathy is a late manifestation of SPG11. J. Neurol. 2016, 263, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Renvoise, B.; Chang, J.; Singh, R.; Yonekawa, S.; FitzGibbon, E.J.; Mankodi, A.; Vanderver, A.; Schindler, A.; Toro, C.; Gahl, W.A.; et al. Lysosomal abnormalities in hereditary spastic paraplegia types SPG15 and SPG11. Ann. Clin. Transl. Neurol. 2014, 1, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Perez-Branguli, F.; Mishra, H.K.; Prots, I.; Havlicek, S.; Kohl, Z.; Saul, D.; Rummel, C.; Dorca-Arevalo, J.; Regensburger, M.; Graef, D.; et al. Dysfunction of spatacsin leads to axonal pathology in SPG11-linked hereditary spastic paraplegia. Hum. Mol. Genet. 2014, 23, 4859–4874. [Google Scholar] [CrossRef]
- Apel, E.D.; Lewis, R.M.; Grady, R.M.; Sanes, J.R. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 2000, 275, 31986–31995. [Google Scholar] [CrossRef] [PubMed]
- Gros-Louis, F.; Dupre, N.; Dion, P.; Fox, M.A.; Laurent, S.; Verreault, S.; Sanes, J.R.; Bouchard, J.P.; Rouleau, G.A. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat. Genet. 2007, 39, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, T.; Nakamura, K.; Ishikawa, M.; Yamaguchi, T.; Takano, K.; Wakui, K.; Kosho, T.; Yoshida, K.; Fukushima, Y.; Sekijima, Y. A novel frameshift mutation of SYNE1 in a Japanese family with autosomal recessive cerebellar ataxia type 8. Hum. Genome Var. 2017, 4, 17052. [Google Scholar] [CrossRef]
- Razafsky, D.; Hodzic, D. A variant of Nesprin1 giant devoid of KASH domain underlies the molecular etiology of autosomal recessive cerebellar ataxia type I. Neurobiol. Dis. 2015, 78, 57–67. [Google Scholar] [CrossRef]
- Vermeer, S.; Hoischen, A.; Meijer, R.P.; Gilissen, C.; Neveling, K.; Wieskamp, N.; de Brouwer, A.; Koenig, M.; Anheim, M.; Assoum, M.; et al. Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am. J. Hum. Genet. 2010, 87, 813–819. [Google Scholar] [CrossRef]
- Balreira, A.; Boczonadi, V.; Barca, E.; Pyle, A.; Bansagi, B.; Appleton, M.; Graham, C.; Hargreaves, I.P.; Rasic, V.M.; Lochmuller, H.; et al. ANO10 mutations cause ataxia and coenzyme Q(1)(0) deficiency. J. Neurol. 2014, 261, 2192–2198. [Google Scholar] [CrossRef] [PubMed]
- Lestienne, P. Evidence for a direct role of the DNA polymerase gamma in the replication of the human mitochondrial DNA in vitro. Biochem. Biophys. Res. Commun. 1987, 146, 1146–1153. [Google Scholar] [CrossRef]
- Luoma, P.T.; Luo, N.; Loscher, W.N.; Farr, C.L.; Horvath, R.; Wanschitz, J.; Kiechl, S.; Kaguni, L.S.; Suomalainen, A. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum. Mol. Genet. 2005, 14, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Hahn, D.; Jackson, C.B.; Kern, I.; Chardot, C.; Belli, D.C.; Gallati, S.; Nuoffer, J.M. Molecular and biochemical characterisation of a novel mutation in POLG associated with Alpers syndrome. BMC Neurol. 2011, 11, 4. [Google Scholar] [CrossRef]
- Van Goethem, G.; Luoma, P.; Rantamaki, M.; Al Memar, A.; Kaakkola, S.; Hackman, P.; Krahe, R.; Lofgren, A.; Martin, J.J.; De Jonghe, P.; et al. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology 2004, 63, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Moreira, M.C.; Klur, S.; Watanabe, M.; Nemeth, A.H.; Le Ber, I.; Moniz, J.C.; Tranchant, C.; Aubourg, P.; Tazir, M.; Schols, L.; et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004, 36, 225–227. [Google Scholar] [CrossRef]
- Airoldi, G.; Guidarelli, A.; Cantoni, O.; Panzeri, C.; Vantaggiato, C.; Bonato, S.; Grazia D’Angelo, M.; Falcone, S.; De Palma, C.; Tonelli, A.; et al. Characterization of two novel SETX mutations in AOA2 patients reveals aspects of the pathophysiological role of senataxin. Neurogenetics 2010, 11, 91–100. [Google Scholar] [CrossRef]
- Bernard, V.; Minnerop, M.; Burk, K.; Kreuz, F.; Gillessen-Kaesbach, G.; Zuhlke, C. Exon deletions and intragenic insertions are not rare in ataxia with oculomotor apraxia 2. BMC Med. Genet. 2009, 10, 87. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Bennett, C.L.; Huynh, H.M.; Blair, I.P.; Puls, I.; Irobi, J.; Dierick, I.; Abel, A.; Kennerson, M.L.; Rabin, B.A.; et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 2004, 74, 1128–1135. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Hashemi, S.H.; Anderson, S.K.; Huang, Y.; Moreira, M.C.; Lynch, D.R.; Glass, I.A.; Chance, P.F.; Bennett, C.L. Senataxin, the yeast Sen1p orthologue: Characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol. Dis. 2006, 23, 97–108. [Google Scholar] [CrossRef]
- Fogel, B.L.; Perlman, S. Novel mutations in the senataxin DNA/RNA helicase domain in ataxia with oculomotor apraxia 2. Neurology 2006, 67, 2083–2084. [Google Scholar] [CrossRef] [PubMed]
- Roda, R.H.; Rinaldi, C.; Singh, R.; Schindler, A.B.; Blackstone, C. Ataxia with oculomotor apraxia type 2 fibroblasts exhibit increased susceptibility to oxidative DNA damage. J. Clin. Neurosci. 2014, 21, 1627–1631. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sawyer, S.L.; Schwartzentruber, J.; Beaulieu, C.L.; Dyment, D.; Smith, A.; Warman Chardon, J.; Yoon, G.; Rouleau, G.A.; Suchowersky, O.; Siu, V.; et al. Exome sequencing as a diagnostic tool for pediatric-onset ataxia. Hum. Mutat. 2014, 35, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Gilmore, E.C.; Marshall, C.A.; Haddadin, M.; Reynolds, J.J.; Eyaid, W.; Bodell, A.; Barry, B.; Gleason, D.; Allen, K.; et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 2010, 42, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.J.; Walker, A.K.; Gilmore, E.C.; Walsh, C.A.; Caldecott, K.W. Impact of PNKP mutations associated with microcephaly, seizures and developmental delay on enzyme activity and DNA strand break repair. Nucleic Acids Res. 2012, 40, 6608–6619. [Google Scholar] [CrossRef] [PubMed]
- Kalasova, I.; Hailstone, R.; Bublitz, J.; Bogantes, J.; Hofmann, W.; Leal, A.; Hanzlikova, H.; Caldecott, K.W. Pathological mutations in PNKP trigger defects in DNA single-strand break repair but not DNA double-strand break repair. Nucleic Acids Res. 2020, 48, 6672–6684. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Magri, S.; Nanetti, L.; Sarto, E.; Di Bella, D.; Salsano, E.; Pantaleoni, C.; Mariotti, C.; Taroni, F. From congenital microcephaly to adult onset cerebellar ataxia: Distinct and overlapping phenotypes in patients with PNKP gene mutations. Am J. Med. Genet. A 2019, 179, 2277–2283. [Google Scholar] [CrossRef] [PubMed]
- Bolhuis, P.A.; Ponne, N.J.; Bikker, H.; Baas, F.; Vianney de Jong, J.M. Molecular basis of an adult form of Sandhoff disease: Substitution of glutamine for arginine at position 505 of the beta-chain of beta-hexosaminidase results in a labile enzyme. Biochim. Biophys. Acta 1993, 1182, 142–146. [Google Scholar] [CrossRef]
- Banerjee, P.; Siciliano, L.; Oliveri, D.; McCabe, N.R.; Boyers, M.J.; Horwitz, A.L.; Li, S.C.; Dawson, G. Molecular basis of an adult form of beta-hexosaminidase B deficiency with motor neuron disease. Biochem. Biophys. Res. Commun. 1991, 181, 108–115. [Google Scholar] [CrossRef]
- Redonnet-Vernhet, I.; Mahuran, D.J.; Salvayre, R.; Dubas, F.; Levade, T. Significance of two point mutations present in each HEXB allele of patients with adult GM2 gangliosidosis (Sandhoff disease) homozygosity for the Ile207-->Val substitution is not associated with a clinical or biochemical phenotype. Biochim. Biophys. Acta 1996, 1317, 127–133. [Google Scholar] [CrossRef]
- Zampieri, S.; Filocamo, M.; Buratti, E.; Stroppiano, M.; Vlahovicek, K.; Rosso, N.; Bignulin, E.; Regis, S.; Carnevale, F.; Bembi, B.; et al. Molecular and functional analysis of the HEXB gene in Italian patients affected with Sandhoff disease: Identification of six novel alleles. Neurogenetics 2009, 10, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Modoni, A.; Sabatelli, M.; Madia, F.; Piemonte, F.; Tozzi, G.; Ricci, E.; Tonali, P.A.; Silvestri, G. Chronic GM2 gangliosidosis type Sandhoff associated with a novel missense HEXB gene mutation causing a double pathogenic effect. Mol. Genet. Metab. 2007, 91, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, S.; Cattarossi, S.; Oller Ramirez, A.M.; Rosano, C.; Lourenco, C.M.; Passon, N.; Moroni, I.; Uziel, G.; Pettinari, A.; Stanzial, F.; et al. Sequence and copy number analyses of HEXB gene in patients affected by Sandhoff disease: Functional characterization of 9 novel sequence variants. PLoS ONE 2012, 7, e41516. [Google Scholar] [CrossRef] [PubMed]
- Yasui, N.; Takaoka, Y.; Nishio, H.; Nurputra, D.K.; Sekiguchi, K.; Hamaguchi, H.; Kowa, H.; Maeda, E.; Sugano, A.; Miura, K.; et al. Molecular pathology of Sandhoff disease with p.Arg505Gln in HEXB: Application of simulation analysis. J. Hum. Genet. 2013, 58, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Edvardson, S.; Hama, H.; Shaag, A.; Gomori, J.M.; Berger, I.; Soffer, D.; Korman, S.H.; Taustein, I.; Saada, A.; Elpeleg, O. Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am. J. Hum. Genet. 2008, 83, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Dick, K.J.; Eckhardt, M.; Paisan-Ruiz, C.; Alshehhi, A.A.; Proukakis, C.; Sibtain, N.A.; Maier, H.; Sharifi, R.; Patton, M.A.; Bashir, W.; et al. Mutation of FA2H underlies a complicated form of hereditary spastic paraplegia (SPG35). Hum. Mutat. 2010, 31, E1251–E1260. [Google Scholar] [CrossRef] [PubMed]
- Kruer, M.C.; Paisan-Ruiz, C.; Boddaert, N.; Yoon, M.Y.; Hama, H.; Gregory, A.; Malandrini, A.; Woltjer, R.L.; Munnich, A.; Gobin, S.; et al. Defective FA2H leads to a novel form of neurodegeneration with brain iron accumulation (NBIA). Ann. Neurol. 2010, 68, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Engert, J.C.; Berube, P.; Mercier, J.; Dore, C.; Lepage, P.; Ge, B.; Bouchard, J.P.; Mathieu, J.; Melancon, S.B.; Schalling, M.; et al. ARSACS, a spastic ataxia common in northeastern Quebec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat. Genet. 2000, 24, 120–125. [Google Scholar] [CrossRef]
- Alfares, A.; Alfadhel, M.; Wani, T.; Alsahli, S.; Alluhaydan, I.; Al Mutairi, F.; Alothaim, A.; Albalwi, M.; Al Subaie, L.; Alturki, S.; et al. A multicenter clinical exome study in unselected cohorts from a consanguineous population of Saudi Arabia demonstrated a high diagnostic yield. Mol. Genet. Metab. 2017, 121, 91–95. [Google Scholar] [CrossRef]
- Stevanin, G.; Azzedine, H.; Denora, P.; Boukhris, A.; Tazir, M.; Lossos, A.; Rosa, A.L.; Lerer, I.; Hamri, A.; Alegria, P.; et al. Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain 2008, 131, 772–784. [Google Scholar] [CrossRef]
- Orlacchio, A.; Babalini, C.; Borreca, A.; Patrono, C.; Massa, R.; Basaran, S.; Munhoz, R.P.; Rogaeva, E.A.; St George-Hyslop, P.H.; Bernardi, G.; et al. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain 2010, 133, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Montecchiani, C.; Pedace, L.; Lo Giudice, T.; Casella, A.; Mearini, M.; Gaudiello, F.; Pedroso, J.L.; Terracciano, C.; Caltagirone, C.; Massa, R.; et al. ALS5/SPG11/KIAA1840 mutations cause autosomal recessive axonal Charcot-Marie-Tooth disease. Brain 2016, 139, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bethmann, C.; Worth, N.F.; Davies, J.D.; Wasner, C.; Feuer, A.; Ragnauth, C.D.; Yi, Q.; Mellad, J.A.; Warren, D.T.; et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007, 16, 2816–2833. [Google Scholar] [CrossRef] [PubMed]
- Van Goethem, G.; Dermaut, B.; Lofgren, A.; Martin, J.J.; Van Broeckhoven, C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet 2001, 28, 211–212. [Google Scholar] [CrossRef]
- Naviaux, R.K.; Nguyen, K.V. POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann. Neurol. 2004, 55, 706–712. [Google Scholar] [CrossRef]
- Winterthun, S.; Ferrari, G.; He, L.; Taylor, R.W.; Zeviani, M.; Turnbull, D.M.; Engelsen, B.A.; Moen, G.; Bindoff, L.A. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology 2005, 64, 1204–1208. [Google Scholar] [CrossRef]
- Vissing, J.; Ravn, K.; Danielsen, E.R.; Duno, M.; Wibrand, F.; Wevers, R.A.; Schwartz, M. Multiple mtDNA deletions with features of MNGIE. Neurology 2002, 59, 926–929. [Google Scholar] [CrossRef]
- Fogel, B.L.; Cho, E.; Wahnich, A.; Gao, F.; Becherel, O.J.; Wang, X.; Fike, F.; Chen, L.; Criscuolo, C.; De Michele, G.; et al. Mutation of senataxin alters disease-specific transcriptional networks in patients with ataxia with oculomotor apraxia type 2. Hum. Mol. Genet. 2014, 23, 4758–4769. [Google Scholar] [CrossRef]
- Bolhuis, P.A.; Oonk, J.G.; Kamp, P.E.; Ris, A.J.; Michalski, J.C.; Overdijk, B.; Reuser, A.J. Ganglioside storage, hexosaminidase lability, and urinary oligosaccharides in adult Sandhoff’s disease. Neurology 1987, 37, 75–81. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]

| Family | Ind. Code (Gender) | Tested | Gene | Disease Causing Variants | Consanguinity | Age | First Symptom | Ataxia | OMA | Diplopia | Pyramidal Signs | Peripheral Neuropathy | Dystonia | Cognitive Impairment | Other Symptoms/Signs | Imaging | Others | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Onset | Obs. | Wheelchair | Death | |||||||||||||||||
| Spastic ataxia | ||||||||||||||||||||
| AR53 | IV.1 (F) | Yes | SACS | c.8148del; c.8148del | Yes | 6 | 53 | 47 | NA | Ataxia | Yes | No | No | Yes | No | No | No | NAv | ||
| AR77 | II.3 (M) | Yes | SACS | c.1373C >T; c. 5440_5449del | No | <1 | 31 | NA | NA | Delayed motor milestones | Yes | No | No | Yes | Yes | No | No | Seizures 1-16y Nystagmus | MRI: cerebellar atrophy | |
| AR120 | II.1 (F) | Yes | SACS | c.3066del; c.2656_2666del | No | 15 | 66 | 29 | NA | Ataxia | Yes | No | No | Yes | Yes | No | No | Nystagmus | NAv | |
| AR252 | II.1 (M) | Yes | SACS | c.814C > T; c.814C > T | Not clear | <1 | 57 | 54 | NA | Delayed motor milestones | Yes | No | No | Yes | Yes | No | No | Kyphoscoliosis | NAv | |
| AR96 | IV.1 (M) | Yes | KIF1C | c.393_396del; c.393_396del | Yes | 9 | 52 | 41 | NA | UL tremor | Yes | No | No | Yes | Yes | No | Mild deterioration | Nystagmus | MRI: pallidum, putamen, thalamus hypointensities (T2/FLAIR) | |
| IV.2 (F) | Yes | 8 | 50 | 44 | NA | UL tremor | Yes | No | No | Yes | Yes | No | Mild deterioration | Nystagmus | MRI: vermis atrophy | |||||
| AR111 | III.3 (F) | Yes | KIF1C | c.1166-2A > G; c.1166-2A > G | Yes | <1 | 41 | NA | NA | Delayed motor milestones | Yes | No | No | Yes | Yes | No | No | Vertical gaze restriction Mild dysmorphism Pes cavus | CT: brainstem atrophy | |
| AR103 | III.3 (F) | Yes | ANO10 | c.132dup; c.132dup | Yes | 7 | 40 | NA | NA | Ataxia | Yes | No | Yes | Yes | No | No | No | Nystagmus Migraine | MRI: cerebellar atrophy | |
| AR108 | II.3 (F) | Yes | SPG11 | c.1951C > T; c.3039-5T > G | No | 7 | 43 | NA | NA | Cognitive regression | Yes | No | No | Yes | No | No | Yes | MRI: Thin corpus callosum | ||
| II.2 (M) | No | 8 | 47 | NA | NA | Cognitive regression | Yes | No | No | Yes | No | No | Yes | MRI: Chiari type1 | ||||||
| AR109 | III.2 (M) | Yes | SYNE1 | c.17270C > G; c.17270C > G | Not clear | NAv | 67 | NA | NA | Ataxia | Yes | No | No | Yes | No | No | No | NAv | ||
| AR267 | III.4 (M) | Yes | CACNA1A | c.4996C > G | No | 19 | 21 | NA | NA | Ataxia | Yes | No | No | Yes | No | No | Yes | MRI: cerebellar atrophy | ||
| Ataxia and neuropathy | ||||||||||||||||||||
| AR4 | VI.27 (M) | Yes | SETX | c.1514G > A; c.1514G > A | Yes | 14 | 37 | NA | NA | Ataxia | Yes | No * | Yes | No | Yes | No | Nystagmus | CT: cerebellar atrophy | ||
| VI.7 (F) | No | 13 | 49 | NA | NA | Ataxia | Yes | No * | Yes | No | Yes | No | Nystagmus | CT: cerebellar atrophy | ||||||
| VI.10 (F) | No | 11 | 43 | NA | NA | Ataxia | Yes | No * | Yes | No | Yes | No | Nystagmus | CT: cerebellar atrophy | ||||||
| VI.19 (F) | No | 12 | 43 | NA | NA | Ataxia | Yes | No * | Yes | No | Yes | No | Nystagmus Obesity | CT: cerebellar atrophy | ||||||
| VI.20 (F) | No | 13 | 42 | NA | NA | Ataxia | Yes | No * | Yes | No | Yes | No | Mild retardation | Nystagmus | CT: cerebellar atrophy | |||||
| VI.31 (F) | No | 11 | 32 | NA | NA | Ataxia | Yes | No * | Yes | No | Yes | No | Nystagmus | CT: cerebellar atrophy | ||||||
| AR49 | II.3 (F) | Yes | KIF1A | c.761G > A | No | 5 | 26 | NA | NA | Ataxia | Yes | No | NAv | Yes | Yes | No | Mild retardation | CT: cortical cerebellar atrophy | ||
| AR92 | II.1 (M) | Yes | PNKP | c.1123G > T; c.1123G > T | Not clear | 6 | 33 | 16 | 35 | Ataxia | Yes | No | No | No | Yes | No | No | NAv | ||
| II.4 (M) | No | 7 | 27 | 16 | NA | Ataxia | Yes | No | No | No | Yes | No | No | NAv | αFP: N | |||||
| AR126 | V.7 (F) | Yes | POLG | c.2243G > C; c.2243G > C | Yes | 25 | 41 | NA | NA | Ataxia | Yes | No | No | No | Yes | No | Yes | Optic atrophy Vertical gaze palsy Nystagmus Epilepsy | NAv | |
| Ataxia and oculomotor apraxia | ||||||||||||||||||||
| AR86 | II.2 (F) | Yes | PNKP | c.1123G > T; c.1253_1269dup | No | 8 | 17 | NA | NA | Ataxia | Yes | Yes | No | No | Yes | No | Yes | Nystagmus Obesity | MRI: cerebellar atrophy (++vermis) | |
| AR117 | II.1 (M) | Yes | PNKP | c.1221_1223del; c.1123G > T | Yes | 8 | 32 | 16 | 37 | Ataxia | Yes | Yes | NAv | No | Yes | Yes (UL) | No | Obesity | NAv | |
| II.2 (F) | No | 7 | 35 | 15 | 42 | Ataxia | Yes | Yes | NAv | No | Yes | Yes (UL) | No | Obesity | MRI: cerebellar atrophy | αFP: ↑ Proteins: ↓ Albumin: ↓ Cholesterol: ↑ | ||||
| II.3 (F) | No | 7 | 29 | 15 | 55 | Ataxia | Yes | Yes | NAv | No | Yes | Yes (UL) | No | Obesity | MRI: cerebellar atrophy | Proteins: ↓ Albumin: ↓ Cholesterol: ↑ | ||||
| Ataxia and dystonia | ||||||||||||||||||||
| AR2 | IV.1 (M) | Yes | HEXB | c.1514G > A; c.1514G > A | Yes | 24 | 51 | NA | NA | Ataxia | Yes | No | No | Yes | No | Yes (OM) | No | Nystagmus Muscle atrophy ↓ vibration hallux | MRI: cerebellar, atrophy | |
| AR278 | III.1 (M) | Yes | ATP1A3 | c.374T > A | No | 11 | 44 | NA | NA | Parox dystonia | Yes | No | No | No | No | Yes | Yes | Supranuclear vertical gaze palsy | MRI: cerebellar atrophy | |
| Ataxia and cognitive impairment | ||||||||||||||||||||
| AR16 | V.1 (F) | Yes | FA2H | c.619_620del; c.619_620del | Yes | <1 | 14 | NA | NA | Delayed motor/cognitive milestones | Yes | No | No | Yes | No | No | Yes | Seizures, Aggressive behavior | MRI: cerebellar atrophy, mild cortical atrophy | VEP: ↑ BEP, SSEP: N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, M.; Damásio, J.; Carmona, S.; Neto, J.L.; Dehghani, N.; Guedes, L.C.; Barbot, C.; Barros, J.; Brás, J.; Sequeiros, J.; et al. Molecular Characterization of Portuguese Patients with Hereditary Cerebellar Ataxia. Cells 2022, 11, 981. https://doi.org/10.3390/cells11060981
Santos M, Damásio J, Carmona S, Neto JL, Dehghani N, Guedes LC, Barbot C, Barros J, Brás J, Sequeiros J, et al. Molecular Characterization of Portuguese Patients with Hereditary Cerebellar Ataxia. Cells. 2022; 11(6):981. https://doi.org/10.3390/cells11060981
Chicago/Turabian StyleSantos, Mariana, Joana Damásio, Susana Carmona, João Luís Neto, Nadia Dehghani, Leonor Correia Guedes, Clara Barbot, José Barros, José Brás, Jorge Sequeiros, and et al. 2022. "Molecular Characterization of Portuguese Patients with Hereditary Cerebellar Ataxia" Cells 11, no. 6: 981. https://doi.org/10.3390/cells11060981
APA StyleSantos, M., Damásio, J., Carmona, S., Neto, J. L., Dehghani, N., Guedes, L. C., Barbot, C., Barros, J., Brás, J., Sequeiros, J., & Guerreiro, R. (2022). Molecular Characterization of Portuguese Patients with Hereditary Cerebellar Ataxia. Cells, 11(6), 981. https://doi.org/10.3390/cells11060981







