Evaluation of Antibiotic-Based Selection Methods for Camelina sativa Stable Transformants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Plasmid Cloning and Transformation
2.3. G8O and GES qRT-PCR
3. Results
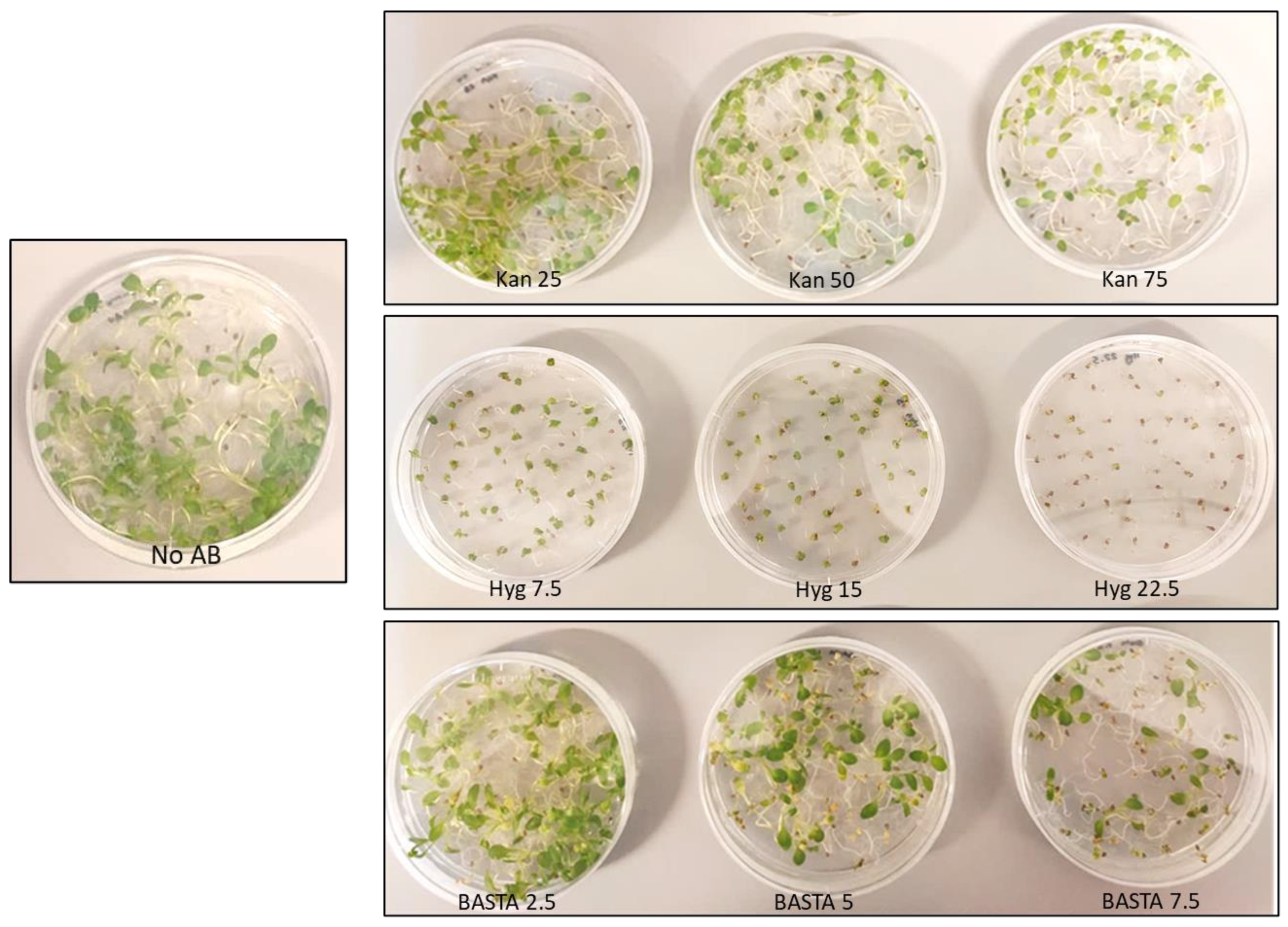
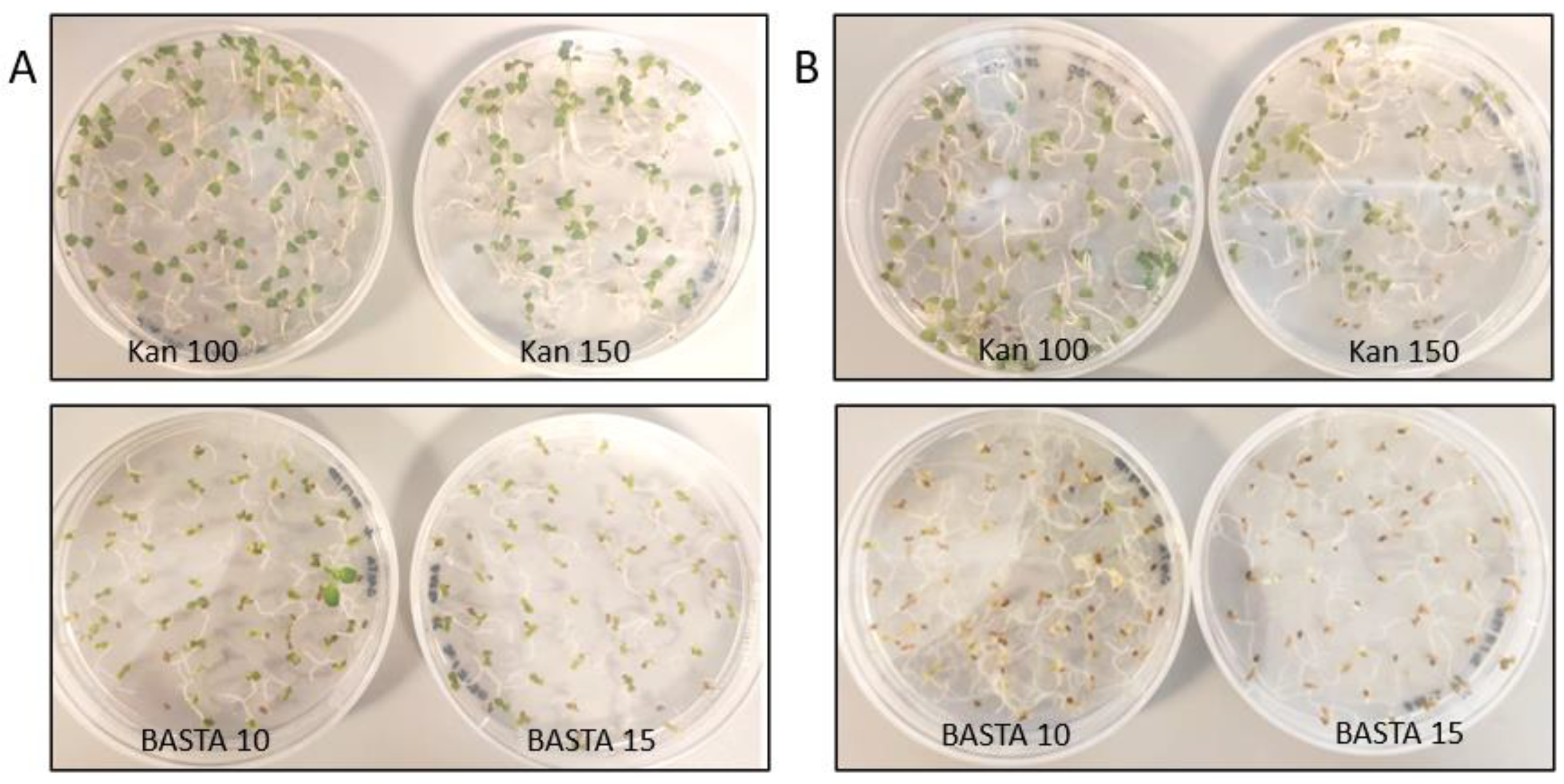
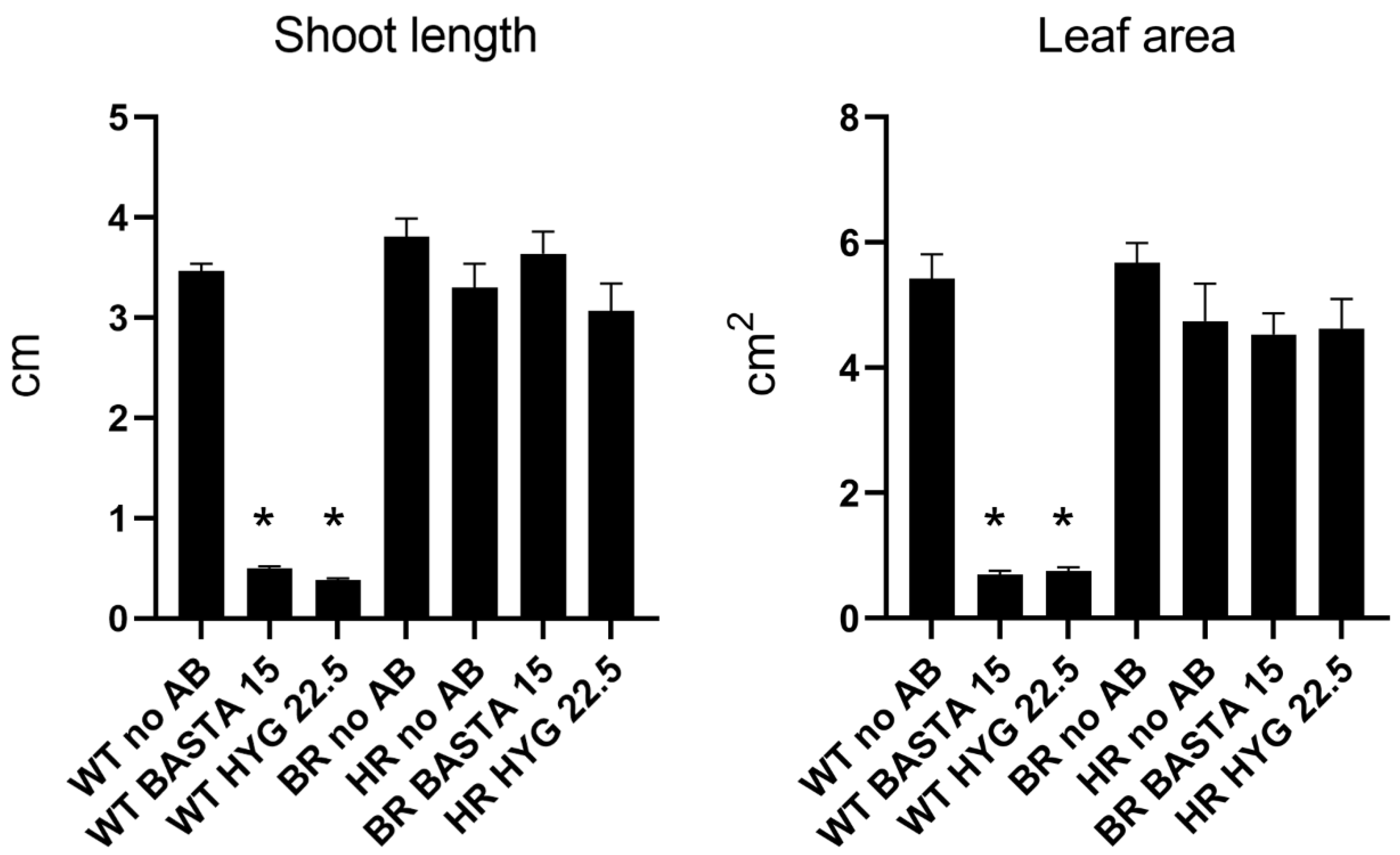
3.1. Antibiotic Concentration Evaluation on Wild Type Camelina
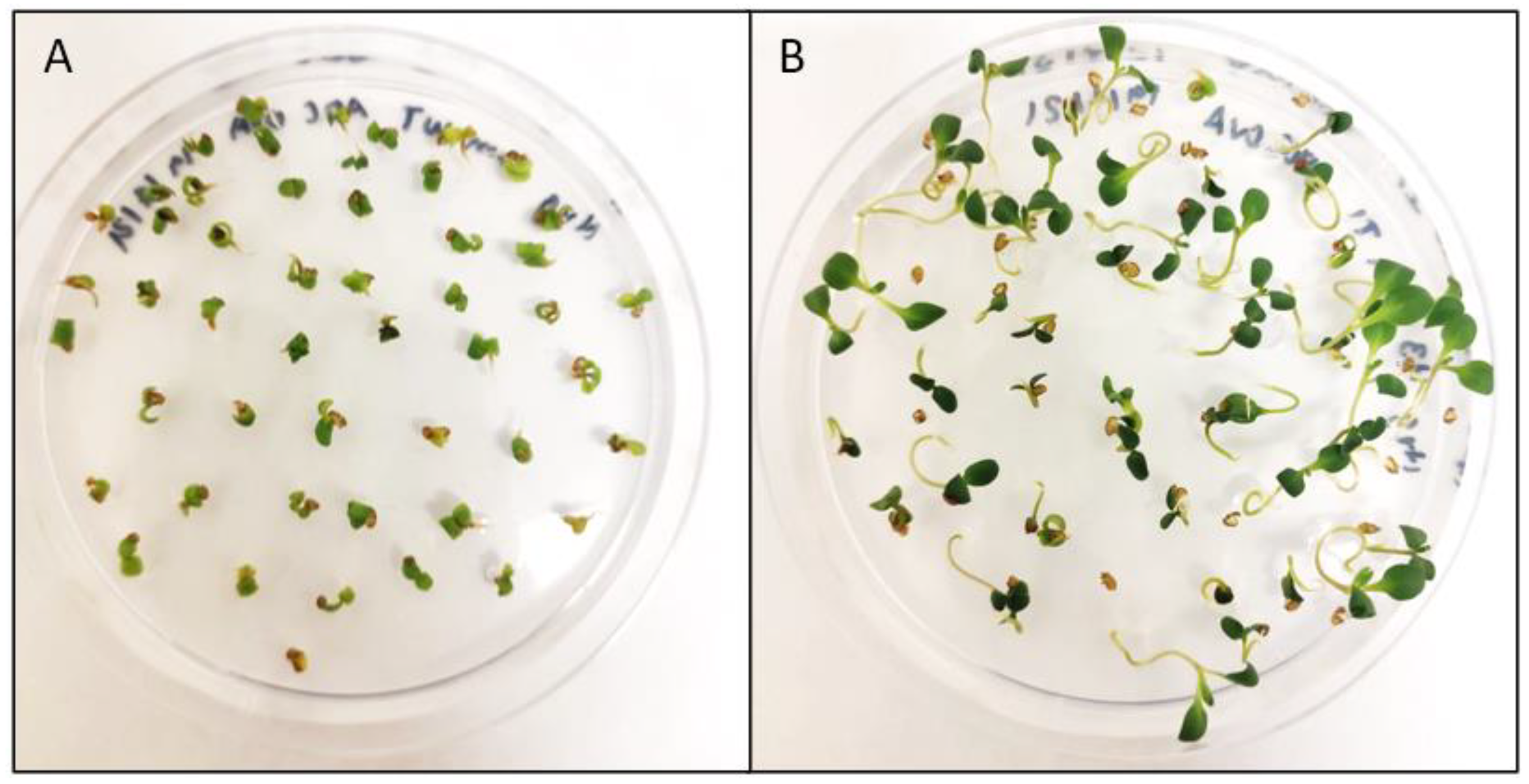
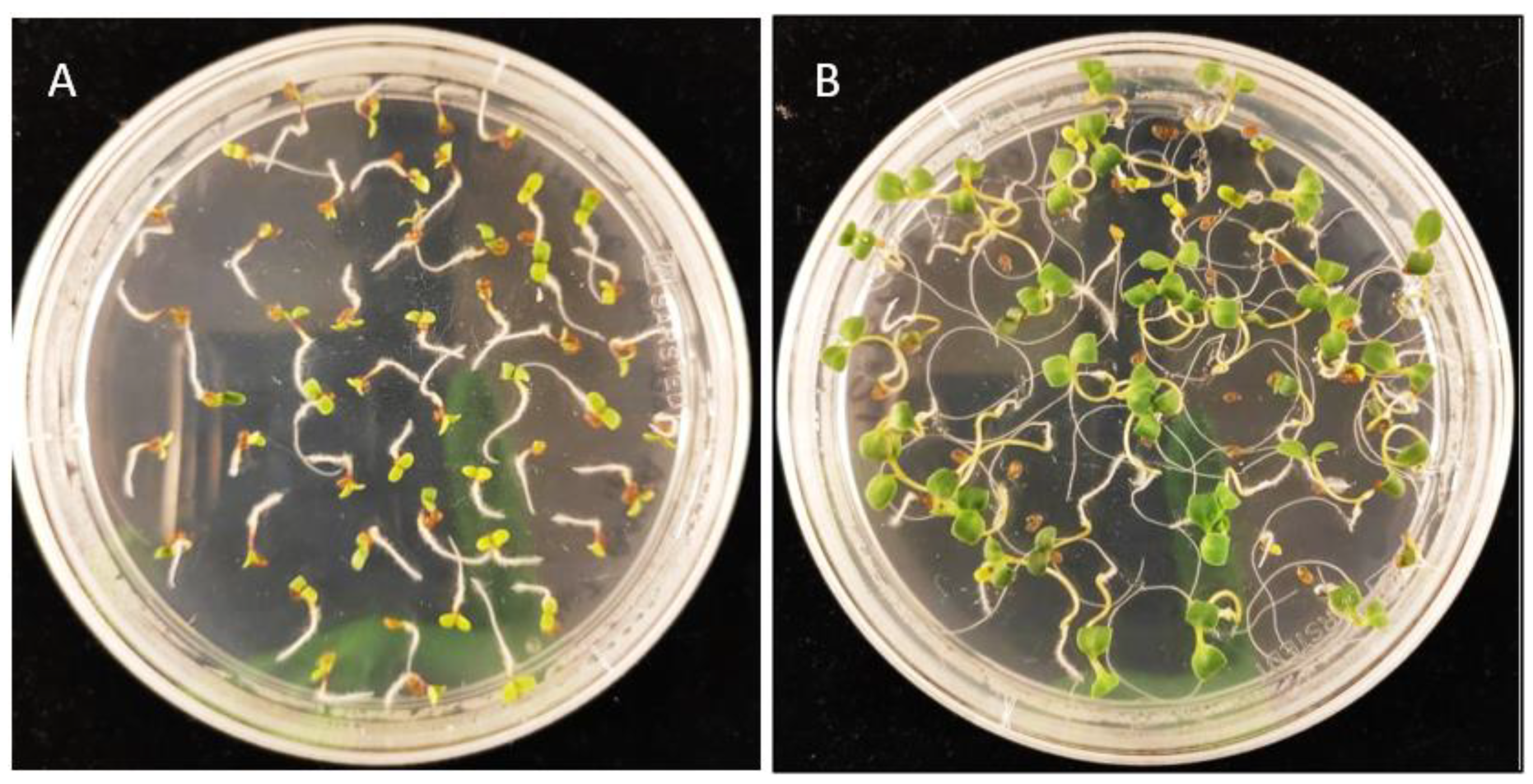
3.2. Camelina Transformant Selection
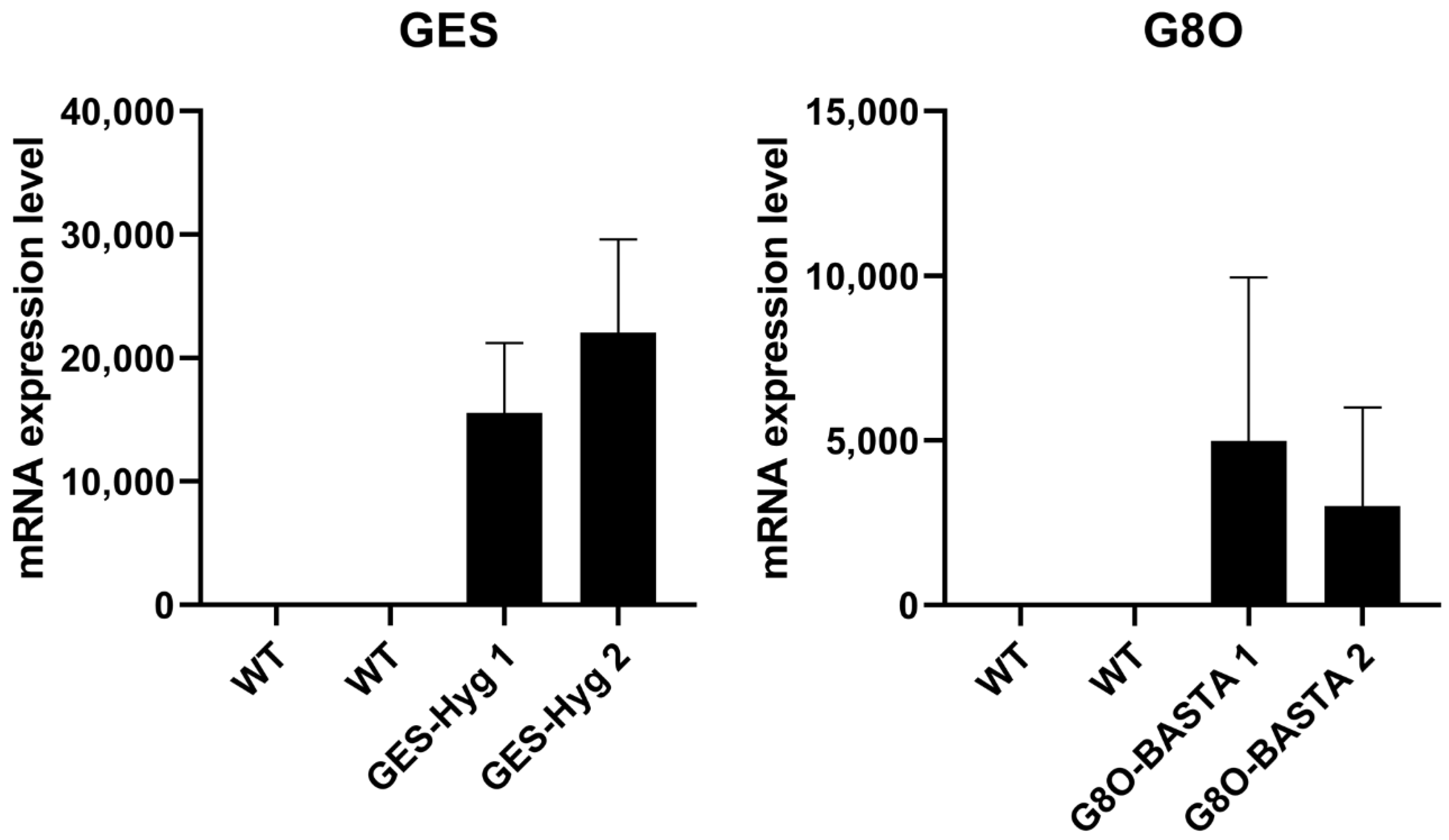
3.3. Selection of Transgenic Lines Overexpressing Seco-Iridoid Pathway Enzymes from Catharanthus roseus
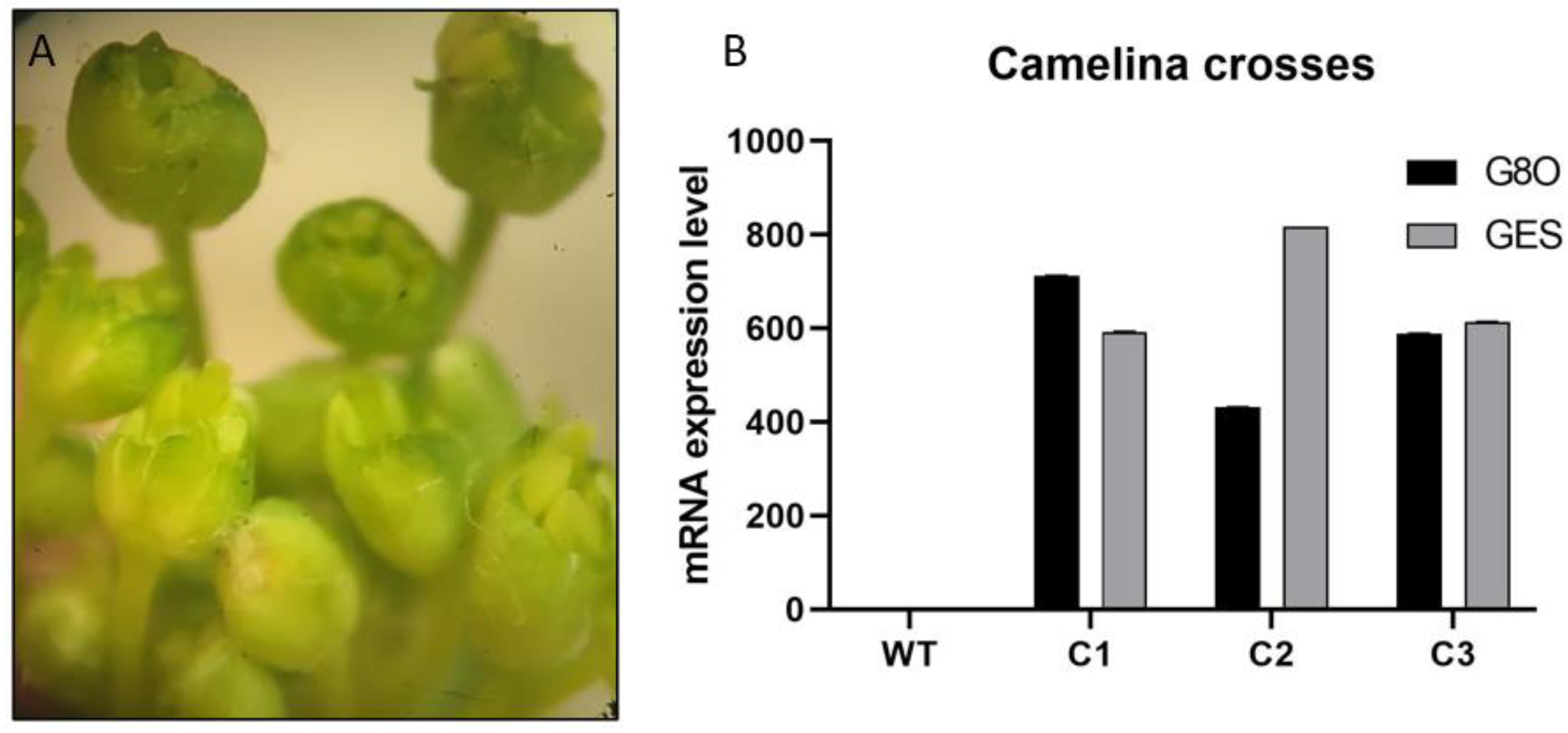
3.4. Selection of F1 Lines Overexpressing Both GES and G8O Seco-Iridoid Pathway Enzymes by Crossing of Camelina
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Camelina | Camelina sativa |
| Arabidopsis | Arabidopsis thaliana |
| DsRed | Discosoma sp. red protein |
| Kan | Kanamycin |
| Hyg | Hygromycin |
| BASTA | phosphinothricin herbicide |
| qRT-PCR | quantitative real time polymerase chain reaction |
| TAG | triacylglycerol |
| cDNA | complementary DNA |
References
- Zanetti, F.; Monti, A.; Berti, M.T. Challenges and opportunities for new industrial oilseed crops in EU-27: A review. Ind. Crop Prod. 2013, 50, 580–595. [Google Scholar] [CrossRef]
- Bakhshi, B.; Rostami-Ahmadvandi, H.; Fanaei, H.R. Camelina, an adaptable oilseed crop for the warm and dried regions of Iran. Cent. Asian J. Plant Sci. Innov. 2021, 1, 39–45. [Google Scholar]
- Liu, X.; Brost, J.; Hutcheon, C.; Guilfoil, R.; Wilson, A.K.; Leung, S.; Shewmaker, C.K.; Rooke, S.; Nguyen, T.; Kiser, J.; et al. Transformation of the oilseed crop Camelina sativa by Agrobacterium-mediated floral dip and simple large-scale screening of transformants. Vitr. Cell. Dev. Biol. Plant 2012, 48, 462–468. [Google Scholar] [CrossRef]
- Bansal, S.; Durrett, T.P. Camelina sativa: An ideal platform for the metabolic engineering and field production of industrial lipids. Biochimie 2016, 120, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Vollmann, J.; Eynck, C. Camelina as a sustainable oilseed crop: Contributions of plant breeding and genetic engineering. Biotechnol. J. 2015, 10, 525–535. [Google Scholar] [CrossRef]
- Fröhlich, A.; Rice, B. Evaluation of Camelina sativa oil as a feedstock for biodiesel production. Ind. Crop Prod. 2005, 21, 25–31. [Google Scholar] [CrossRef]
- Kagale, S.; Koh, C.; Nixon, J.; Bollina, V.; Clarke, W.E.; Tuteja, R.; Spillane, C.; Robinson, S.J.; Links, M.; Clarke, C.; et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014, 5, 3706. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; He, J.; Liu, L.; Xie, R.; Qiu, L.; Li, X.; Yuan, W.; Chen, K.; Yin, Y.; Kyaw, M.M.M.; et al. A convenient, rapid and efficient method for establishing transgenic lines of Brassica napus. Plant Methods 2020, 16, 43. [Google Scholar] [CrossRef]
- Olhoft, P.M.; Flagel, L.E.; Donovan, C.M.; Somers, D.A. Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 2003, 216, 723–735. [Google Scholar] [CrossRef]
- Chu, U.C.; Kumar, S.; Sigmund, A.; Johnson, K.; Li, Y.; Vongdeuane, P.; Jones, T.J. Genotype-Independent Transformation and Genome Editing of Brassica napus Using a Novel Explant Material. Front. Plant Sci. 2020, 11, 579524. [Google Scholar] [CrossRef]
- Lu, C.; Kang, J. Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 2008, 27, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Silva, J.E.; Podicheti, R.; Macrander, J.; Yang, W.; Nazarenus, T.J.; Nam, J.-W.; Jaworski, J.G.; Lu, C.; Scheffler, B.E.; et al. Camelina seed transcriptome: A tool for meal and oil improvement and translational research. Plant Biotechnol. J. 2013, 11, 759–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Harrison, S.J.; Mott, E.K.; Parsley, K.; Aspinall, S.; Gray, J.C.; Cottage, A. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2006, 2, 19. [Google Scholar] [CrossRef] [Green Version]
- Belt, K.; Van Aken, O.; Murcha, M.; Millar, A.H.; Huang, S. An Assembly Factor Promotes Assembly of Flavinated SDH1 into the Succinate Dehydrogenase Complex. Plant Physiol. 2018, 177, 1439–1452. [Google Scholar] [CrossRef] [Green Version]
- Broda, M.; Aken, O.V. Studying retrograde signaling in plants. In Plant Programmed Cell Death; Springer: Berlin/Heidelberg, Germany, 2018; pp. 73–85. [Google Scholar]
- Miettinen, K.; Dong, L.; Navrot, N.; Schneider, T.; Burlat, V.; Pollier, J.; Woittiez, L.; Van Der Krol, S.; Lugan, R.; Ilc, T.; et al. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014, 5, 3606. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-H.; Sheng, M.; Chen, G.; Aldrich, J.R.; Chauhan, K.R. Iridodial: A powerful attractant for the green lacewing, Chrysopa septempunctata (Neuroptera: Chrysopidae). Die Naturwissenschaften 2006, 93, 461–465. [Google Scholar] [CrossRef]
- Sitther, V.; Tabatabai, B.; Enitan, O.; Dhekney, S. Agrobacterium-mediated transformation of Camelina sativa for production of transgenic plants. J. Biol. Methods 2018, 5, e83. [Google Scholar] [CrossRef] [Green Version]
- Sitther, V.; Tabatabai, B.; Enitan, O.; Fathabad, S.G.; Dhekney, S. Production of Transgenic Camelina sativa Plants via Agrobacterium-Mediated Transformation of Shoot Apical Meristems. Am. J. Plant Sci. 2019, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Van Aken, O.; Whelan, J.; Van Breusegem, F. Prohibitins: Mitochondrial partners in development and stress response. Trends Plant Sci. 2010, 15, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Miki, B.; McHugh, S. Selectable marker genes in transgenic plants: Applications, alternatives and biosafety. J. Biotechnol. 2004, 107, 193–232. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.-H.; Wang, H.-L.; Ding, B.-J.; Svensson, G.P.; Jarl-Sunesson, C.; Cahoon, E.B.; Hofvander, P.; Löfstedt, C. Green Chemistry Production of Codlemone, the Sex Pheromone of the Codling Moth (Cydia pomonella), by Metabolic Engineering of the Oilseed Crop Camelina (Camelina sativa). J. Chem. Ecol. 2021, 47, 950–967. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xie, L.; Chen, G.Q.; Lee, M.Y.; Loque, D.; Scheller, H.V. A transgene design for enhancing oil content in Arabidopsis and Camelina seeds. Biotechnol. Biofuels 2018, 11, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| GES | LP | GACGATTTGGGTACTGCTAAGG |
| RP | CTATTCCCTCCCTTCCTTCACT | |
| G80 | LP | AATCGGCAGAGGAAAAACAATA |
| RP | GCGAGGAATTAAGAAGGGAACT | |
| ACT2 | LP | AAGAGCAGCTCTTCAGTTGA |
| RP | AACCTCAGGACAACGGAATC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ontiveros-Cisneros, A.; Moss, O.; Van Moerkercke, A.; Van Aken, O. Evaluation of Antibiotic-Based Selection Methods for Camelina sativa Stable Transformants. Cells 2022, 11, 1068. https://doi.org/10.3390/cells11071068
Ontiveros-Cisneros A, Moss O, Van Moerkercke A, Van Aken O. Evaluation of Antibiotic-Based Selection Methods for Camelina sativa Stable Transformants. Cells. 2022; 11(7):1068. https://doi.org/10.3390/cells11071068
Chicago/Turabian StyleOntiveros-Cisneros, Abraham, Oliver Moss, Alex Van Moerkercke, and Olivier Van Aken. 2022. "Evaluation of Antibiotic-Based Selection Methods for Camelina sativa Stable Transformants" Cells 11, no. 7: 1068. https://doi.org/10.3390/cells11071068
APA StyleOntiveros-Cisneros, A., Moss, O., Van Moerkercke, A., & Van Aken, O. (2022). Evaluation of Antibiotic-Based Selection Methods for Camelina sativa Stable Transformants. Cells, 11(7), 1068. https://doi.org/10.3390/cells11071068






