Fission Yeast Autophagy Machinery
Abstract
:1. Introduction
2. Physiological Roles of Autophagy in Fission Yeast
3. Autophagy Induction Conditions in Fission Yeast
4. Methods to Monitor Autophagy in Fission Yeast
5. The Molecular Machinery for Autophagosome Assembly in Fission Yeast
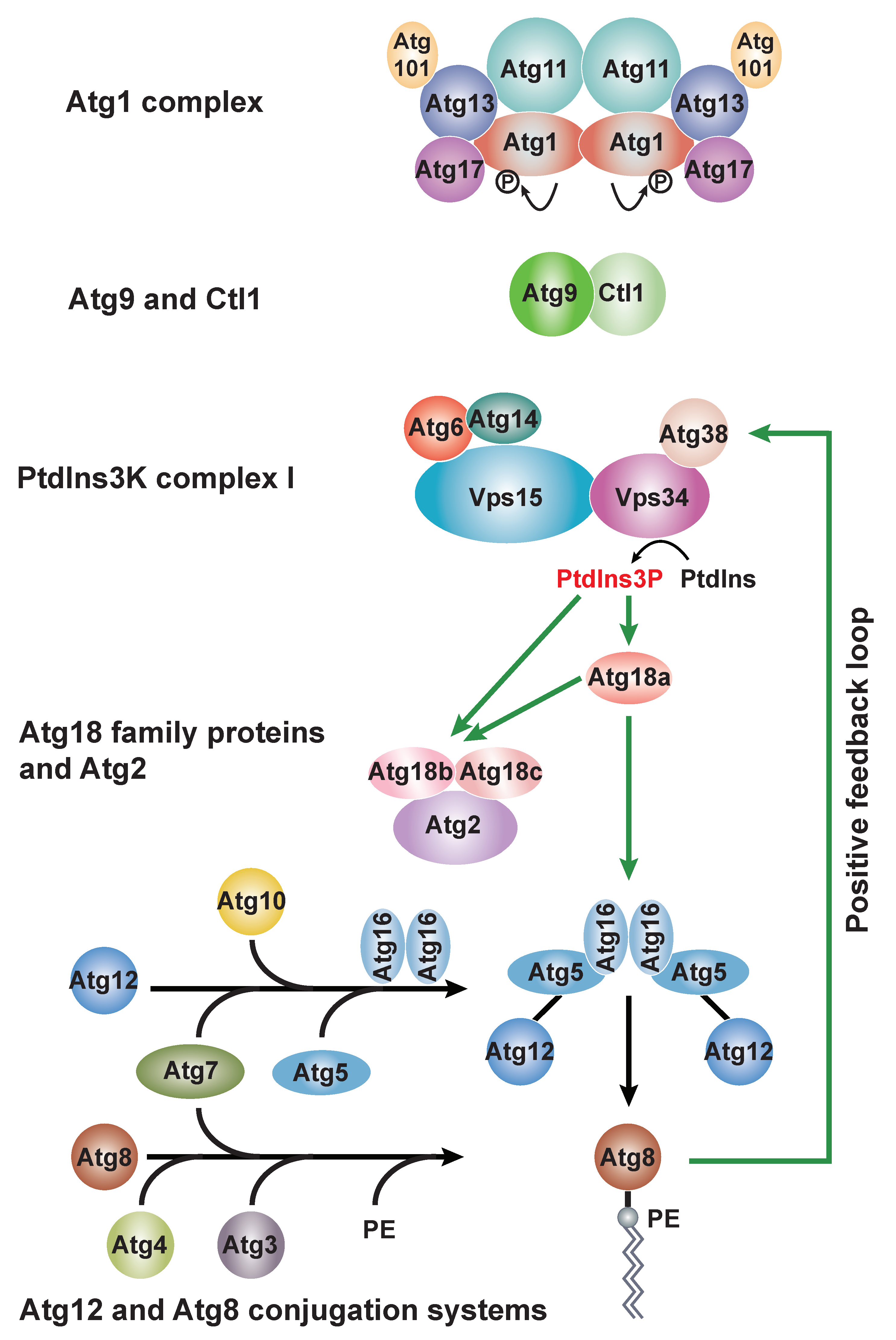
5.1. The Atg1 Protein Kinase Complex
5.2. Atg9 and Ctl1
5.3. PtdIns3K Complexes
5.4. Atg18 Family Proteins and Atg2
5.5. Two Ubiquitin-like Conjugation Systems
6. Selective Autophagy Receptors in S. pombe
7. Unanswered Questions
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klionsky, D.J.; Emr, S.D. Autophagy as a Regulated Pathway of Cellular Degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Klionsky, D.J. Autophagic Processes in Yeast: Mechanism, Machinery and Regulation. Genetics 2013, 194, 341–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Klionsky, D.J. Autophagosome Formation: Core Machinery and Adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef]
- Ohsumi, Y. Historical Landmarks of Autophagy Research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and Diversity in Autophagy Mechanisms: Lessons from Yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Kubota, Y.; Sekito, T.; Ohsumi, Y. Hierarchy of Atg Proteins in Pre-Autophagosomal Structure Organization. Genes Cells 2007, 12, 209–218. [Google Scholar] [CrossRef]
- Klionsky, D.J. The Importance of Diversity. Autophagy 2007, 3, 83–84. [Google Scholar] [CrossRef] [Green Version]
- King, J.S. Autophagy across the Eukaryotes: Is S. Cerevisiae the Odd One Out? Autophagy 2012, 8, 1159–1162. [Google Scholar] [CrossRef]
- Shen, X.-X.; Steenwyk, J.L.; LaBella, A.L.; Opulente, D.A.; Zhou, X.; Kominek, J.; Li, Y.; Groenewald, M.; Hittinger, C.T.; Rokas, A. Genome-Scale Phylogeny and Contrasting Modes of Genome Evolution in the Fungal Phylum Ascomycota. Sci. Adv. 2020, 6, eabd0079. [Google Scholar] [CrossRef]
- Mizushima, N. The Role of the Atg1/ULK1 Complex in Autophagy Regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kaizuka, T.; Mizushima, N.; Noda, N.N. Structure of the Atg101-Atg13 Complex Reveals Essential Roles of Atg101 in Autophagy Initiation. Nat. Struct. Mol. Biol. 2015, 22, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Nanji, T.; Liu, X.; Chew, L.H.; Li, F.K.; Biswas, M.; Yu, Z.-Q.; Lu, S.; Dong, M.-Q.; Du, L.-L.; Klionsky, D.J.; et al. Conserved and Unique Features of the Fission Yeast Core Atg1 Complex. Autophagy 2017, 13, 2018–2027. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-L.; Li, M.; Suo, F.; Liu, X.-M.; Shen, E.-Z.; Yang, B.; Dong, M.-Q.; He, W.-Z.; Du, L.-L. Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors. PLoS Genet. 2013, 9, e1003715. [Google Scholar] [CrossRef] [Green Version]
- Pan, Z.-Q.; Shao, G.-C.; Liu, X.-M.; Chen, Q.; Dong, M.-Q.; Du, L.-L. Atg1 Kinase in Fission Yeast Is Activated by Atg11-Mediated Dimerization and Cis-Autophosphorylation. eLife 2020, 9, e58073. [Google Scholar] [CrossRef] [PubMed]
- Kraft, C.; Peter, M.; Hofmann, K. Selective Autophagy: Ubiquitin-Mediated Recognition and Beyond. Nat. Cell Biol. 2010, 12, 836–841. [Google Scholar] [CrossRef]
- Liu, X.-M.; Sun, L.-L.; Hu, W.; Ding, Y.-H.; Dong, M.-Q.; Du, L.-L. ESCRTs Cooperate with a Selective Autophagy Receptor to Mediate Vacuolar Targeting of Soluble Cargos. Mol. Cell 2015, 59, 1035–1042. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-Y.; Zhang, J.; Liu, X.-M.; Li, Y.; Sui, J.; Dong, M.-Q.; Ye, K.; Du, L.-L. Molecular and Structural Mechanisms of ZZ Domain-Mediated Cargo Selection by Nbr1. EMBO J. 2021, 40, e107497. [Google Scholar] [CrossRef]
- Kohda, T.A.; Tanaka, K.; Konomi, M.; Sato, M.; Osumi, M.; Yamamoto, M. Fission Yeast Autophagy Induced by Nitrogen Starvation Generates a Nitrogen Source That Drives Adaptation Processes. Genes Cells 2007, 12, 155–170. [Google Scholar] [CrossRef]
- Mukaiyama, H.; Kajiwara, S.; Hosomi, A.; Giga-Hama, Y.; Tanaka, N.; Nakamura, T.; Takegawa, K. Autophagy-Deficient Schizosaccharomyces Pombe Mutants Undergo Partial Sporulation during Nitrogen Starvation. Microbiology 2009, 155, 3816–3826. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.-Q.; Sun, L.-L.; Jiang, Z.-D.; Liu, X.-M.; Zhao, D.; Wang, H.-T.; He, W.-Z.; Dong, M.-Q.; Du, L.-L. Atg38-Atg8 Interaction in Fission Yeast Establishes a Positive Feedback Loop to Promote Autophagy. Autophagy 2020, 16, 2036–2051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Liu, X.-M.; Yu, Z.-Q.; Sun, L.-L.; Xiong, X.; Dong, M.-Q.; Du, L.-L. Atg20- and Atg24-Family Proteins Promote Organelle Autophagy in Fission Yeast. J. Cell Sci. 2016, 129, 4289–4304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.-M.; Yamasaki, A.; Du, X.-M.; Coffman, V.C.; Ohsumi, Y.; Nakatogawa, H.; Wu, J.-Q.; Noda, N.N.; Du, L.-L. Lipidation-Independent Vacuolar Functions of Atg8 Rely on Its Noncanonical Interaction with a Vacuole Membrane Protein. eLife 2018, 7, e41237. [Google Scholar] [CrossRef]
- Fukuda, T.; Ebi, Y.; Saigusa, T.; Furukawa, K.; Yamashita, S.-I.; Inoue, K.; Kobayashi, D.; Yoshida, Y.; Kanki, T. Atg43 Tethers Isolation Membranes to Mitochondria to Promote Starvation-Induced Mitophagy in Fission Yeast. eLife 2020, 9, e61245. [Google Scholar] [CrossRef]
- Zhao, D.; Zou, C.-X.; Liu, X.-M.; Jiang, Z.-D.; Yu, Z.-Q.; Suo, F.; Du, T.-Y.; Dong, M.-Q.; He, W.; Du, L.-L. A UPR-Induced Soluble ER-Phagy Receptor Acts with VAPs to Confer ER Stress Resistance. Mol. Cell 2020, 79, 963–977.e3. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 Mediates Direct Lipid Transfer between Membranes for Autophagosome Formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 Is a Lipid Scramblase That Mediates Autophagosomal Membrane Expansion. Nat. Struct. Mol. Biol. 2020, 27, 1185–1193. [Google Scholar] [CrossRef]
- Tsukada, M.; Ohsumi, Y. Isolation and Characterization of Autophagy-Defective Mutants of Saccharomyces Cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Funakoshi, T.; Matsuura, A.; Noda, T.; Ohsumi, Y. Analyses of APG13 Gene Involved in Autophagy in Yeast, Saccharomyces Cerevisiae. Gene 1997, 192, 207–213. [Google Scholar] [CrossRef]
- Deutschbauer, A.M.; Williams, R.M.; Chu, A.M.; Davis, R.W. Parallel Phenotypic Analysis of Sporulation and Postgermination Growth in Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 2002, 99, 15530–15535. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, A.; Hasegawa, T.; Mori, S.; Ueno, M.; Tanaka, S.; Ushimaru, T.; Sato, S.; Uritani, M. A Starvation-Specific Serine Protease Gene, Isp6+, Is Involved in Both Autophagy and Sexual Development in Schizosaccharomyces Pombe. Curr. Genet. 2006, 49, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Takeshige, K.; Baba, M.; Tsuboi, S.; Noda, T.; Ohsumi, Y. Autophagy in Yeast Demonstrated with Proteinase-Deficient Mutants and Conditions for Its Induction. J. Cell Biol. 1992, 119, 301–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, T.; Ohsumi, Y. Tor, a Phosphatidylinositol Kinase Homologue, Controls Autophagy in Yeast. J. Biol. Chem. 1998, 273, 3963–3966. [Google Scholar] [CrossRef] [Green Version]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-Mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamada, Y.; Yoshino, K.; Kondo, C.; Kawamata, T.; Oshiro, N.; Yonezawa, K.; Ohsumi, Y. Tor Directly Controls the Atg1 Kinase Complex to Regulate Autophagy. Mol. Cell Biol. 2010, 30, 1049–1058. [Google Scholar] [CrossRef] [Green Version]
- Takahara, T.; Maeda, T. TORC1 of Fission Yeast Is Rapamycin-Sensitive. Genes Cells 2012, 17, 698–708. [Google Scholar] [CrossRef]
- Rallis, C.; Codlin, S.; Bähler, J. TORC1 Signaling Inhibition by Rapamycin and Caffeine Affect Lifespan, Global Gene Expression, and Cell Proliferation of Fission Yeast. Aging Cell 2013, 12, 563–573. [Google Scholar] [CrossRef] [Green Version]
- Otsubo, Y.; Nakashima, A.; Yamamoto, M.; Yamashita, A. TORC1-Dependent Phosphorylation Targets in Fission Yeast. Biomolecules 2017, 7, 50. [Google Scholar] [CrossRef] [Green Version]
- Mikawa, T.; Kanoh, J.; Ishikawa, F. Fission Yeast Vps1 and Atg8 Contribute to Oxidative Stress Resistance. Genes Cells 2010, 15, 229–242. [Google Scholar] [CrossRef]
- Shimasaki, T.; Okamoto, K.; Ohtsuka, H.; Aiba, H. Sulfur Depletion Induces Autophagy through Ecl1 Family Genes in Fission Yeast. Genes Cells 2020, 25, 825–830. [Google Scholar] [CrossRef]
- Xie, Z.; Nair, U.; Klionsky, D.J. Atg8 Controls Phagophore Expansion during Autophagosome Formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A Ubiquitin-like System Mediates Protein Lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Kirisako, T.; Baba, M.; Ishihara, N.; Miyazawa, K.; Ohsumi, M.; Yoshimori, T.; Noda, T.; Ohsumi, Y. Formation Process of Autophagosome Is Traced with Apg8/Aut7p in Yeast. J. Cell Biol. 1999, 147, 435–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The Reversible Modification Regulates the Membrane-Binding State of Apg8/Aut7 Essential for Autophagy and the Cytoplasm to Vacuole Targeting Pathway. J. Cell Biol. 2000, 151, 263–276. [Google Scholar] [CrossRef]
- Shintani, T.; Klionsky, D.J. Cargo Proteins Facilitate the Formation of Transport Vesicles in the Cytoplasm to Vacuole Targeting Pathway. J. Biol. Chem. 2004, 279, 29889–29894. [Google Scholar] [CrossRef] [Green Version]
- Morigasaki, S.; Shimada, K.; Ikner, A.; Yanagida, M.; Shiozaki, K. Glycolytic Enzyme GAPDH Promotes Peroxide Stress Signaling through Multistep Phosphorelay to a MAPK Cascade. Mol. Cell 2008, 30, 108–113. [Google Scholar] [CrossRef] [Green Version]
- Noda, T.; Matsuura, A.; Wada, Y.; Ohsumi, Y. Novel System for Monitoring Autophagy in the Yeast Saccharomyces Cerevisiae. Biochem. Biophys. Res. Commun. 1995, 210, 126–132. [Google Scholar] [CrossRef]
- Klionsky, D.J. Monitoring Autophagy in Yeast: The Pho8Delta60 Assay. Methods Mol. Biol. 2007, 390, 363–371. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Membrane Protein Sorting: Biosynthesis, Transport and Processing of Yeast Vacuolar Alkaline Phosphatase. EMBO J. 1989, 8, 2241–2250. [Google Scholar] [CrossRef]
- Backues, S.K.; Chen, D.; Ruan, J.; Xie, Z.; Klionsky, D.J. Estimating the Size and Number of Autophagic Bodies by Electron Microscopy. Autophagy 2014, 10, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Kabeya, Y.; Kamada, Y.; Baba, M.; Takikawa, H.; Sasaki, M.; Ohsumi, Y. Atg17 Functions in Cooperation with Atg1 and Atg13 in Yeast Autophagy. Mol. Biol. Cell 2005, 16, 2544–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragusa, M.J.; Stanley, R.E.; Hurley, J.H. Architecture of the Atg17 Complex as a Scaffold for Autophagosome Biogenesis. Cell 2012, 151, 1501–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papinski, D.; Kraft, C. Regulation of Autophagy By Signaling Through the Atg1/ULK1 Complex. J. Mol. Biol. 2016, 428, 1725–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yorimitsu, T.; Klionsky, D.J. Atg11 Links Cargo to the Vesicle-Forming Machinery in the Cytoplasm to Vacuole Targeting Pathway. Mol. Biol. Cell 2005, 16, 1593–1605. [Google Scholar] [CrossRef] [Green Version]
- Fujioka, Y.; Suzuki, S.W.; Yamamoto, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structural Basis of Starvation-Induced Assembly of the Autophagy Initiation Complex. Nat. Struct. Mol. Biol. 2014, 21, 513–521. [Google Scholar] [CrossRef]
- Yamamoto, H.; Fujioka, Y.; Suzuki, S.W.; Noshiro, D.; Suzuki, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ando, T.; Noda, N.N.; et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell 2016, 38, 86–99. [Google Scholar] [CrossRef] [Green Version]
- Kamber, R.A.; Shoemaker, C.J.; Denic, V. Receptor-Bound Targets of Selective Autophagy Use a Scaffold Protein to Activate the Atg1 Kinase. Mol. Cell 2015, 59, 372–381. [Google Scholar] [CrossRef] [Green Version]
- Torggler, R.; Papinski, D.; Brach, T.; Bas, L.; Schuschnig, M.; Pfaffenwimmer, T.; Rohringer, S.; Matzhold, T.; Schweida, D.; Brezovich, A.; et al. Two Independent Pathways within Selective Autophagy Converge to Activate Atg1 Kinase at the Vacuole. Mol. Cell 2016, 64, 221–235. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.H.; Jun, C.B.; Ro, S.-H.; Kim, Y.-M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.-H. ULK-Atg13-FIP200 Complexes Mediate MTOR Signaling to the Autophagy Machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [Green Version]
- Ganley, I.G.; Lam, D.H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1.ATG13.FIP200 Complex Mediates MTOR Signaling and Is Essential for Autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-Dependent MTORC1 Association with the ULK1-Atg13-FIP200 Complex Required for Autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.-Q.; Liu, X.-M.; Zhao, D.; Xu, D.-D.; Du, L.-L. Visual Detection of Binary, Ternary and Quaternary Protein Interactions in Fission Yeast Using a Pil1 Co-Tethering Assay. J. Cell Sci. 2021, 134, jcs258774. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kamada, Y.; Stromhaug, P.E.; Guan, J.; Hefner-Gravink, A.; Baba, M.; Scott, S.V.; Ohsumi, Y.; Dunn, W.A.; Klionsky, D.J. Cvt9/Gsa9 Functions in Sequestering Selective Cytosolic Cargo Destined for the Vacuole. J. Cell Biol. 2001, 153, 381–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, T.; Takamura, A.; Kishi, C.; Iemura, S.-I.; Natsume, T.; Guan, J.-L.; Mizushima, N. FIP200, a ULK-Interacting Protein, Is Required for Autophagosome Formation in Mammalian Cells. J. Cell Biol. 2008, 181, 497–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papinski, D.; Schuschnig, M.; Reiter, W.; Wilhelm, L.; Barnes, C.A.; Maiolica, A.; Hansmann, I.; Pfaffenwimmer, T.; Kijanska, M.; Stoffel, I.; et al. Early Steps in Autophagy Depend on Direct Phosphorylation of Atg9 by the Atg1 Kinase. Mol. Cell 2014, 53, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Wandelmer, J.; Kriegenburg, F.; Rohringer, S.; Schuschnig, M.; Gómez-Sánchez, R.; Zens, B.; Abreu, S.; Hardenberg, R.; Hollenstein, D.; Gao, J.; et al. Atg4 Proteolytic Activity Can Be Inhibited by Atg1 Phosphorylation. Nat. Commun. 2017, 8, 295. [Google Scholar] [CrossRef]
- Wold, M.S.; Lim, J.; Lachance, V.; Deng, Z.; Yue, Z. ULK1-Mediated Phosphorylation of ATG14 Promotes Autophagy and Is Impaired in Huntington’s Disease Models. Mol. Neurodegener. 2016, 11, 76. [Google Scholar] [CrossRef] [Green Version]
- Pengo, N.; Agrotis, A.; Prak, K.; Jones, J.; Ketteler, R. A Reversible Phospho-Switch Mediated by ULK1 Regulates the Activity of Autophagy Protease ATG4B. Nat. Commun. 2017, 8, 294. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Ma, K.; Gao, R.; Mu, C.; Chen, L.; Liu, Q.; Luo, Q.; Feng, D.; Zhu, Y.; Chen, Q. Regulation of MATG9 Trafficking by Src- and ULK1-Mediated Phosphorylation in Basal and Starvation-Induced Autophagy. Cell Res. 2017, 27, 184–201. [Google Scholar] [CrossRef] [Green Version]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.-Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.-L. ULK1 Induces Autophagy by Phosphorylating Beclin-1 and Activating VPS34 Lipid Kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [Green Version]
- Noda, T.; Kim, J.; Huang, W.P.; Baba, M.; Tokunaga, C.; Ohsumi, Y.; Klionsky, D.J. Apg9p/Cvt7p Is an Integral Membrane Protein Required for Transport Vesicle Formation in the Cvt and Autophagy Pathways. J. Cell Biol. 2000, 148, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Tucker, K.A.; Stromhaug, P.E.; Klionsky, D.J. The Atg1-Atg13 Complex Regulates Atg9 and Atg23 Retrieval Transport from the Pre-Autophagosomal Structure. Dev. Cell 2004, 6, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Lang, T.; Reiche, S.; Straub, M.; Bredschneider, M.; Thumm, M. Autophagy and the Cvt Pathway Both Depend on AUT9. J. Bacteriol. 2000, 182, 2125–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen, W.-L.; Legakis, J.E.; Nair, U.; Klionsky, D.J. Atg27 Is Required for Autophagy-Dependent Cycling of Atg9. Mol. Biol. Cell 2007, 18, 581–593. [Google Scholar] [CrossRef] [Green Version]
- Backues, S.K.; Orban, D.P.; Bernard, A.; Singh, K.; Cao, Y.; Klionsky, D.J. Atg23 and Atg27 Act at the Early Stages of Atg9 Trafficking in S. Cerevisiae. Traffic 2015, 16, 172–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Tito, S.; Hervás, J.H.; van Vliet, A.R.; Tooze, S.A. The Golgi as an Assembly Line to the Autophagosome. Trends Biochem. Sci. 2020, 45, 484–496. [Google Scholar] [CrossRef]
- Young, A.R.J.; Chan, E.Y.W.; Hu, X.W.; Köchl, R.; Crawshaw, S.G.; High, S.; Hailey, D.W.; Lippincott-Schwartz, J.; Tooze, S.A. Starvation and ULK1-Dependent Cycling of Mammalian Atg9 between the TGN and Endosomes. J. Cell Sci. 2006, 119, 3888–3900. [Google Scholar] [CrossRef] [Green Version]
- Guardia, C.M.; Tan, X.-F.; Lian, T.; Rana, M.S.; Zhou, W.; Christenson, E.T.; Lowry, A.J.; Faraldo-Gómez, J.D.; Bonifacino, J.S.; Jiang, J.; et al. Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery. Cell Rep. 2020, 31, 107837. [Google Scholar] [CrossRef]
- Lai, L.T.F.; Yu, C.; Wong, J.S.K.; Lo, H.S.; Benlekbir, S.; Jiang, L.; Lau, W.C.Y. Subnanometer Resolution Cryo-EM Structure of Arabidopsis Thaliana ATG9. Autophagy 2020, 16, 575–583. [Google Scholar] [CrossRef]
- Maeda, S.; Yamamoto, H.; Kinch, L.N.; Garza, C.M.; Takahashi, S.; Otomo, C.; Grishin, N.V.; Forli, S.; Mizushima, N.; Otomo, T. Structure, Lipid Scrambling Activity and Role in Autophagosome Formation of ATG9A. Nat. Struct. Mol. Biol. 2020, 27, 1194–1201. [Google Scholar] [CrossRef]
- Itakura, E.; Mizushima, N. Characterization of Autophagosome Formation Site by a Hierarchical Analysis of Mammalian Atg Proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two Distinct Vps34 Phosphatidylinositol 3-Kinase Complexes Function in Autophagy and Carboxypeptidase Y Sorting in Saccharomyces Cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 Forms Two Distinct Phosphatidylinositol 3-Kinase Complexes with Mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–5372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-Binding Proteins, Atg14L and Rubicon, Reciprocally Regulate Autophagy at Different Stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef]
- Araki, Y.; Ku, W.-C.; Akioka, M.; May, A.I.; Hayashi, Y.; Arisaka, F.; Ishihama, Y.; Ohsumi, Y. Atg38 Is Required for Autophagy-Specific Phosphatidylinositol 3-Kinase Complex Integrity. J. Cell Biol. 2013, 203, 299–313. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; He, L.; Behrends, C.; Araki, M.; Araki, K.; Jun Wang, Q.; Catanzaro, J.M.; Friedman, S.L.; Zong, W.-X.; Fiel, M.I.; et al. NRBF2 Regulates Autophagy and Prevents Liver Injury by Modulating Atg14L-Linked Phosphatidylinositol-3 Kinase III Activity. Nat. Commun. 2014, 5, 3920. [Google Scholar] [CrossRef]
- Kimura, K.; Miyake, S.; Makuuchi, M.; Morita, R.; Usui, T.; Yoshida, M.; Horinouchi, S.; Fukui, Y. Phosphatidylinositol-3 Kinase in Fission Yeast: A Possible Role in Stress Responses. Biosci. Biotechnol. Biochem. 1995, 59, 678–682. [Google Scholar] [CrossRef] [Green Version]
- Takegawa, K.; DeWald, D.B.; Emr, S.D. Schizosaccharomyces Pombe Vps34p, a Phosphatidylinositol-Specific PI 3-Kinase Essential for Normal Cell Growth and Vacuole Morphology. J. Cell Sci. 1995, 108 Pt 12, 3745–3756. [Google Scholar] [CrossRef]
- Mukaiyama, H.; Nakase, M.; Nakamura, T.; Kakinuma, Y.; Takegawa, K. Autophagy in the Fission Yeast Schizosaccharomyces Pombe. FEBS Lett. 2010, 584, 1327–1334. [Google Scholar] [CrossRef] [Green Version]
- Birgisdottir, Å.B.; Mouilleron, S.; Bhujabal, Z.; Wirth, M.; Sjøttem, E.; Evjen, G.; Zhang, W.; Lee, R.; O’Reilly, N.; Tooze, S.A.; et al. Members of the Autophagy Class III Phosphatidylinositol 3-Kinase Complex I Interact with GABARAP and GABARAPL1 via LIR Motifs. Autophagy 2019, 15, 1333–1355. [Google Scholar] [CrossRef] [Green Version]
- Krick, R.; Henke, S.; Tolstrup, J.; Thumm, M. Dissecting the Localization and Function of Atg18, Atg21 and Ygr223c. Autophagy 2008, 4, 896–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, H.; Meiling-Wesse, K.; Epple, U.D.; Thumm, M. Autophagy and the Cytoplasm to Vacuole Targeting Pathway Both Require Aut10p. FEBS Lett. 2001, 508, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Guan, J.; Stromhaug, P.E.; George, M.D.; Habibzadegah-Tari, P.; Bevan, A.; Dunn, W.A.; Klionsky, D.J. Cvt18/Gsa12 Is Required for Cytoplasm-to-Vacuole Transport, Pexophagy, and Autophagy in Saccharomyces Cerevisiae and Pichia Pastoris. Mol. Biol. Cell 2001, 12, 3821–3838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, H.; Meiling-Wesse, K.; Epple, U.D.; Thumm, M. Mai1p Is Essential for Maturation of Proaminopeptidase I but Not for Autophagy. FEBS Lett. 2002, 512, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Obara, K.; Sekito, T.; Niimi, K.; Ohsumi, Y. The Atg18-Atg2 Complex Is Recruited to Autophagic Membranes via Phosphatidylinositol 3-Phosphate and Exerts an Essential Function. J. Biol. Chem. 2008, 283, 23972–23980. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Suzuki, K.; Ohsumi, Y. Autophagosome Formation Can Be Achieved in the Absence of Atg18 by Expressing Engineered PAS-Targeted Atg2. FEBS Lett. 2012, 586, 2473–2478. [Google Scholar] [CrossRef]
- Juris, L.; Montino, M.; Rube, P.; Schlotterhose, P.; Thumm, M.; Krick, R. PI3P Binding by Atg21 Organises Atg8 Lipidation. EMBO J. 2015, 34, 955–973. [Google Scholar] [CrossRef] [Green Version]
- Proikas-Cezanne, T.; Waddell, S.; Gaugel, A.; Frickey, T.; Lupas, A.; Nordheim, A. WIPI-1alpha (WIPI49), a Member of the Novel 7-Bladed WIPI Protein Family, Is Aberrantly Expressed in Human Cancer and Is Linked to Starvation-Induced Autophagy. Oncogene 2004, 23, 9314–9325. [Google Scholar] [CrossRef] [Green Version]
- Bakula, D.; Müller, A.J.; Zuleger, T.; Takacs, Z.; Franz-Wachtel, M.; Thost, A.-K.; Brigger, D.; Tschan, M.P.; Frickey, T.; Robenek, H.; et al. WIPI3 and WIPI4 β-Propellers Are Scaffolds for LKB1-AMPK-TSC Signalling Circuits in the Control of Autophagy. Nat. Commun. 2017, 8, 15637. [Google Scholar] [CrossRef]
- Dooley, H.C.; Razi, M.; Polson, H.E.J.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12-5-16L1. Mol. Cell. 2014, 55, 238–252. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.-X.; Li, Y.; Ding, Y.-H.; Liu, J.-J.; Zhang, M.-J.; Dong, M.-Q.; Wang, H.-W.; Yu, L. Architecture of the ATG2B-WDR45 Complex and an Aromatic Y/HF Motif Crucial for Complex Formation. Autophagy 2017, 13, 1870–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, S.; Otomo, C.; Leitner, A.; Ohashi, K.; Aebersold, R.; Lander, G.C.; Otomo, T. Insights into Autophagosome Biogenesis from Structural and Biochemical Analyses of the ATG2A-WIPI4 Complex. Proc. Natl. Acad. Sci. USA 2018, 115, E9792–E9801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krick, R.; Tolstrup, J.; Appelles, A.; Henke, S.; Thumm, M. The Relevance of the Phosphatidylinositolphosphat-Binding Motif FRRGT of Atg18 and Atg21 for the Cvt Pathway and Autophagy. FEBS Lett. 2006, 580, 4632–4638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krick, R.; Busse, R.A.; Scacioc, A.; Stephan, M.; Janshoff, A.; Thumm, M.; Kühnel, K. Structural and Functional Characterization of the Two Phosphoinositide Binding Sites of PROPPINs, a β-Propeller Protein Family. Proc. Natl. Acad. Sci. USA 2012, 109, E2042–E2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baskaran, S.; Ragusa, M.J.; Boura, E.; Hurley, J.H. Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy. Mol. Cell 2012, 47, 339–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, Y.; Kobayashi, T.; Yamamoto, H.; Hoshida, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structure-Based Analyses Reveal Distinct Binding Sites for Atg2 and Phosphoinositides in Atg18. J. Biol. Chem. 2012, 287, 31681–31690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, N.; Nishimura, T.; Sakamaki, Y.; Koyama-Honda, I.; Yamamoto, H.; Mizushima, N. Differential Requirement for ATG2A Domains for Localization to Autophagic Membranes and Lipid Droplets. FEBS Lett. 2017, 591, 3819–3830. [Google Scholar] [CrossRef] [Green Version]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 Complex Tethers Pre-Autophagosomal Membranes to the Endoplasmic Reticulum for Autophagosome Formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Akioka, M.; Kondo-Kakuta, C.; Yamamoto, H.; Ohsumi, Y. Fine Mapping of Autophagy-Related Proteins during Autophagosome Formation in Saccharomyces Cerevisiae. J. Cell Sci. 2013, 126, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Sánchez, R.; Rose, J.; Guimarães, R.; Mari, M.; Papinski, D.; Rieter, E.; Geerts, W.J.; Hardenberg, R.; Kraft, C.; Ungermann, C.; et al. Atg9 Establishes Atg2-Dependent Contact Sites between the Endoplasmic Reticulum and Phagophores. J. Cell Biol. 2018, 217, 2743–2763. [Google Scholar] [CrossRef]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 Transports Lipids to Promote Autophagosome Biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osawa, T.; Ishii, Y.; Noda, N.N. Human ATG2B Possesses a Lipid Transfer Activity Which Is Accelerated by Negatively Charged Lipids and WIPI4. Genes Cells 2020, 25, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A Protein Conjugation System Essential for Autophagy. Nature 1998, 395, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Mizushima, N.; Ogawa, Y.; Matsuura, A.; Noda, T.; Ohsumi, Y. Apg10p, a Novel Protein-Conjugating Enzyme Essential for Autophagy in Yeast. EMBO J. 1999, 18, 5234–5241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Noda, T.; Ohsumi, Y. Apg16p Is Required for the Function of the Apg12p-Apg5p Conjugate in the Yeast Autophagy Pathway. EMBO J. 1999, 18, 3888–3896. [Google Scholar] [CrossRef] [Green Version]
- Kuma, A.; Mizushima, N.; Ishihara, N.; Ohsumi, Y. Formation of the Approximately 350-KDa Apg12-Apg5.Apg16 Multimeric Complex, Mediated by Apg16 Oligomerization, Is Essential for Autophagy in Yeast. J. Biol. Chem. 2002, 277, 18619–18625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef] [Green Version]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L Complex Specifies the Site of LC3 Lipidation for Membrane Biogenesis in Autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef] [Green Version]
- Lynch-Day, M.A.; Klionsky, D.J. The Cvt Pathway as a Model for Selective Autophagy. FEBS Lett. 2010, 584, 1359–1366. [Google Scholar] [CrossRef] [Green Version]
- Scott, S.V.; Guan, J.; Hutchins, M.U.; Kim, J.; Klionsky, D.J. Cvt19 Is a Receptor for the Cytoplasm-to-Vacuole Targeting Pathway. Mol. Cell 2001, 7, 1131–1141. [Google Scholar] [CrossRef] [Green Version]
- Kirkin, V.; Lamark, T.; Sou, Y.-S.; Bjørkøy, G.; Nunn, J.L.; Bruun, J.-A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A Role for NBR1 in Autophagosomal Degradation of Ubiquitinated Substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, T.-C.; Chen, C.-T.; Baron, D.; Onder, T.T.; Loewer, S.; Almeida, S.; Weismann, C.M.; Xu, P.; Houghton, J.-M.; Gao, F.-B.; et al. Midbody Accumulation through Evasion of Autophagy Contributes to Cellular Reprogramming and Tumorigenicity. Nat. Cell Biol. 2011, 13, 1214–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deosaran, E.; Larsen, K.B.; Hua, R.; Sargent, G.; Wang, Y.; Kim, S.; Lamark, T.; Jauregui, M.; Law, K.; Lippincott-Schwartz, J.; et al. NBR1 Acts as an Autophagy Receptor for Peroxisomes. J. Cell Sci. 2013, 126, 939–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo Recognition and Degradation by Selective Autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef]
- Birgisdottir, Å.B.; Lamark, T.; Johansen, T. The LIR Motif-Crucial for Selective Autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef] [Green Version]
- Takeda, K.; Yoshida, T.; Kikuchi, S.; Nagao, K.; Kokubu, A.; Pluskal, T.; Villar-Briones, A.; Nakamura, T.; Yanagida, M. Synergistic Roles of the Proteasome and Autophagy for Mitochondrial Maintenance and Chronological Lifespan in Fission Yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 3540–3545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-Mediated Selective Autophagy Degrades the Endoplasmic Reticulum and the Nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef]
- Khaminets, A.; Heinrich, T.; Mari, M.; Grumati, P.; Huebner, A.K.; Akutsu, M.; Liebmann, L.; Stolz, A.; Nietzsche, S.; Koch, N.; et al. Regulation of Endoplasmic Reticulum Turnover by Selective Autophagy. Nature 2015, 522, 354–358. [Google Scholar] [CrossRef]
- Fumagalli, F.; Noack, J.; Bergmann, T.J.; Cebollero, E.; Pisoni, G.B.; Fasana, E.; Fregno, I.; Galli, C.; Loi, M.; Soldà, T.; et al. Translocon Component Sec62 Acts in Endoplasmic Reticulum Turnover during Stress Recovery. Nat. Cell Biol. 2016, 18, 1173–1184. [Google Scholar] [CrossRef] [Green Version]
- Grumati, P.; Morozzi, G.; Hölper, S.; Mari, M.; Harwardt, M.-L.I.; Yan, R.; Müller, S.; Reggiori, F.; Heilemann, M.; Dikic, I. Full Length RTN3 Regulates Turnover of Tubular Endoplasmic Reticulum via Selective Autophagy. eLife 2017, 6, e25555. [Google Scholar] [CrossRef]
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 Is a Non-Canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell 2018, 44, 217–232.e11. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Ordureau, A.; Paulo, J.A.; Shoemaker, C.J.; Denic, V.; Harper, J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol. Cell 2019, 74, 891–908.e10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xiao, Y.; Chai, P.; Zheng, P.; Teng, J.; Chen, J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr. Biol. 2019, 29, 846–855.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chino, H.; Hatta, T.; Natsume, T.; Mizushima, N. Intrinsically Disordered Protein TEX264 Mediates ER-Phagy. Mol. Cell 2019, 74, 909–921.e6. [Google Scholar] [CrossRef] [PubMed]
- Nthiga, T.M.; Kumar Shrestha, B.; Sjøttem, E.; Bruun, J.-A.; Bowitz Larsen, K.; Bhujabal, Z.; Lamark, T.; Johansen, T. CALCOCO1 Acts with VAMP-Associated Proteins to Mediate ER-Phagy. EMBO J. 2020, 39, e103649. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Kanki, T. Mechanisms and Physiological Roles of Mitophagy in Yeast. Mol. Cells 2018, 41, 35–44. [Google Scholar] [CrossRef]
- Pickles, S.; Vigié, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The Ubiquitin Kinase PINK1 Recruits Autophagy Receptors to Induce Mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, K.; Kondo-Okamoto, N.; Ohsumi, Y. Mitochondria-Anchored Receptor Atg32 Mediates Degradation of Mitochondria via Selective Autophagy. Dev. Cell 2009, 17, 87–97. [Google Scholar] [CrossRef] [Green Version]
- Kanki, T.; Wang, K.; Cao, Y.; Baba, M.; Klionsky, D.J. Atg32 Is a Mitochondrial Protein That Confers Selectivity during Mitophagy. Dev. Cell 2009, 17, 98–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| S. pombe Autophagy Factor | Function in S. pombe | S. cerevisiae Homolog Involved in Autophagy | Human Homolog Involved in Autophagy |
|---|---|---|---|
| The Atg1 complex | |||
| Atg1 | Serine/threonine kinase [15] | Atg1 | ULK1 |
| Atg11 | Dimerizing Atg1 and activating Atg1 activity [15] | Atg11 | FIP200 |
| Atg17 | Component of the Atg1 complex [13] | Atg17 | − |
| Atg13 | Component of the Atg1 complex [13] | Atg13 | ATG13 |
| Atg101 | Stabilizing Atg13 [12,13] | − | ATG101 |
| Atg9 and Ctl1 | |||
| Atg9 | Lipid scramblase [27] | Atg9 | ATG9A/B |
| Ctl1 | Promoting proper localization of Atg9 [14] | − | − |
| The PtdIns3K complex I | |||
| Vps34 | PtdIns 3-kinase | Vps34 | VPS34 |
| Vps15 | Protein kinase required for Vps34 activity | Vps15 | VPS15 |
| Atg14 | Autophagy specific subunit of PtdIns3K complex I [14] | Atg14 | ATG14 |
| Atg6 | Forming a subcomplex with Atg14 [14] | Atg6 | Beclin 1 |
| Atg38 | Interacting with Atg8 to establish a positive feedback loop [21] | Atg38 | NRBF2 |
| Atg18 family proteins and Atg2 | |||
| Atg2 | Membrane tethering and lipid transfer [26] | Atg2 | ATG2A/B |
| Atg18a | Targeting the Atg12-Atg5·Atg16 complex to the PAS [14] | Atg21 | WIPI2 |
| Atg18b | Acting with Atg18c and Atg2 [14] | Atg18 | WIPI4 |
| Atg18c | Acting with Atg18b and Atg2 [14] | ||
| Atg12 and Atg8 conjugation systems | |||
| Atg12 | Ubiquitin-like protein conjugated to Atg5 [14,20] | Atg12 | ATG12 |
| Atg7 | E1 enzyme for Atg12 and Atg8 [14,20] | Atg7 | ATG7 |
| Atg10 | E2 enzyme for Atg12 [14] | Atg10 | ATG10 |
| Atg5 | Acting with Atg12 as the E3 enzyme for Atg8 [14,20] | Atg5 | ATG5 |
| Atg16 | Promoting the PAS localization of Atg12-Atg5 [14] | Atg16 | ATG16L1/2 |
| Atg8 | Ubiquitin-like protein conjugated to PE [14,20] | Atg8 | LC3 and GABARAP |
| Atg4 | Cysteine protease for Atg8 C-terminal processing and delipidation [14,20] | Atg4 | ATG4A-D |
| Atg3 | E2 enzyme for Atg8 [14,20] | Atg3 | ATG3 |
| Proteins involved in selective autophagy | |||
| Nbr1 | Targeting cytosolic hydrolases to the vacuole [17,18] | Atg19 | NBR1 |
| Atg20 | Atg20 family protein promoting ER and mitochondrial autophagy [22] | Atg20 | − |
| Atg24 and Atg24b | Atg24 family proteins promoting ER and mitochondrial autophagy [22] | Atg24 | − |
| Epr1 | ER-phagy receptor [25] | − | − |
| Atg43 | Mitophagy receptor [24] | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.-D.; Du, L.-L. Fission Yeast Autophagy Machinery. Cells 2022, 11, 1086. https://doi.org/10.3390/cells11071086
Xu D-D, Du L-L. Fission Yeast Autophagy Machinery. Cells. 2022; 11(7):1086. https://doi.org/10.3390/cells11071086
Chicago/Turabian StyleXu, Dan-Dan, and Li-Lin Du. 2022. "Fission Yeast Autophagy Machinery" Cells 11, no. 7: 1086. https://doi.org/10.3390/cells11071086






