Modulating the Ubiquitin–Proteasome System: A Therapeutic Strategy for Autoimmune Diseases
Abstract
1. Introduction
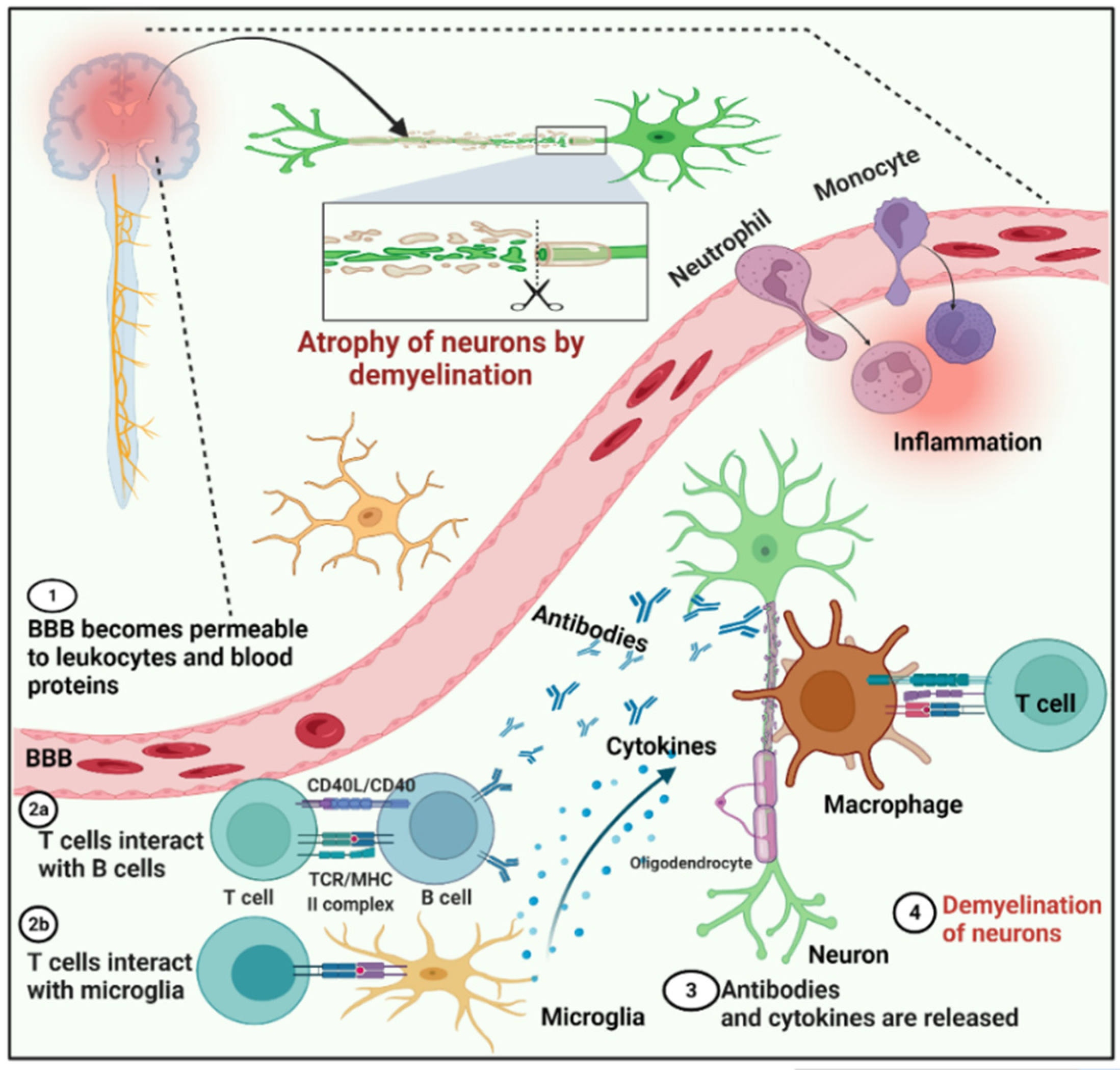
2. Multiple Sclerosis (MS)
3. Proteinopathies
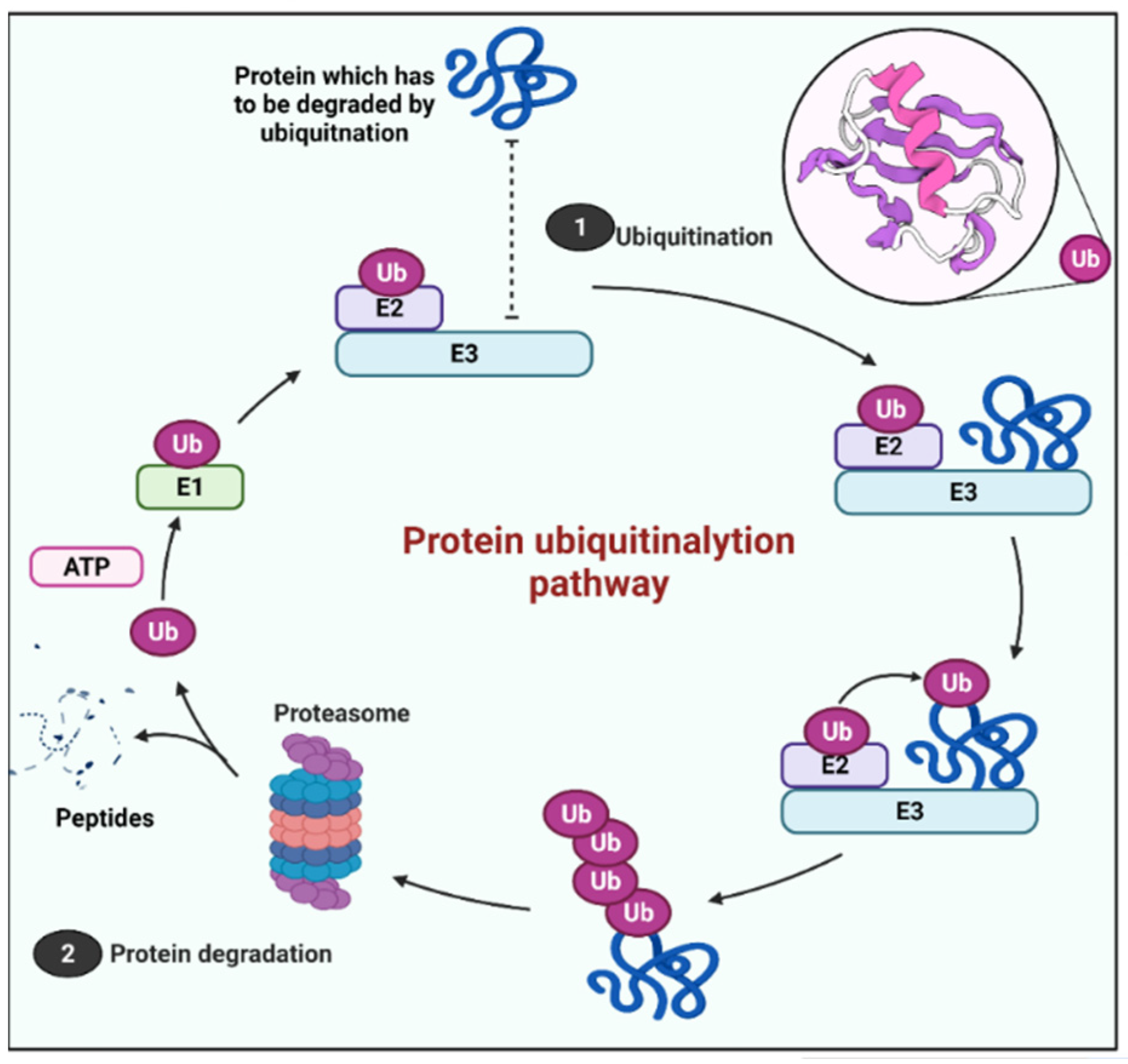
4. The UPS
5. Therapeutic Targets in UPS
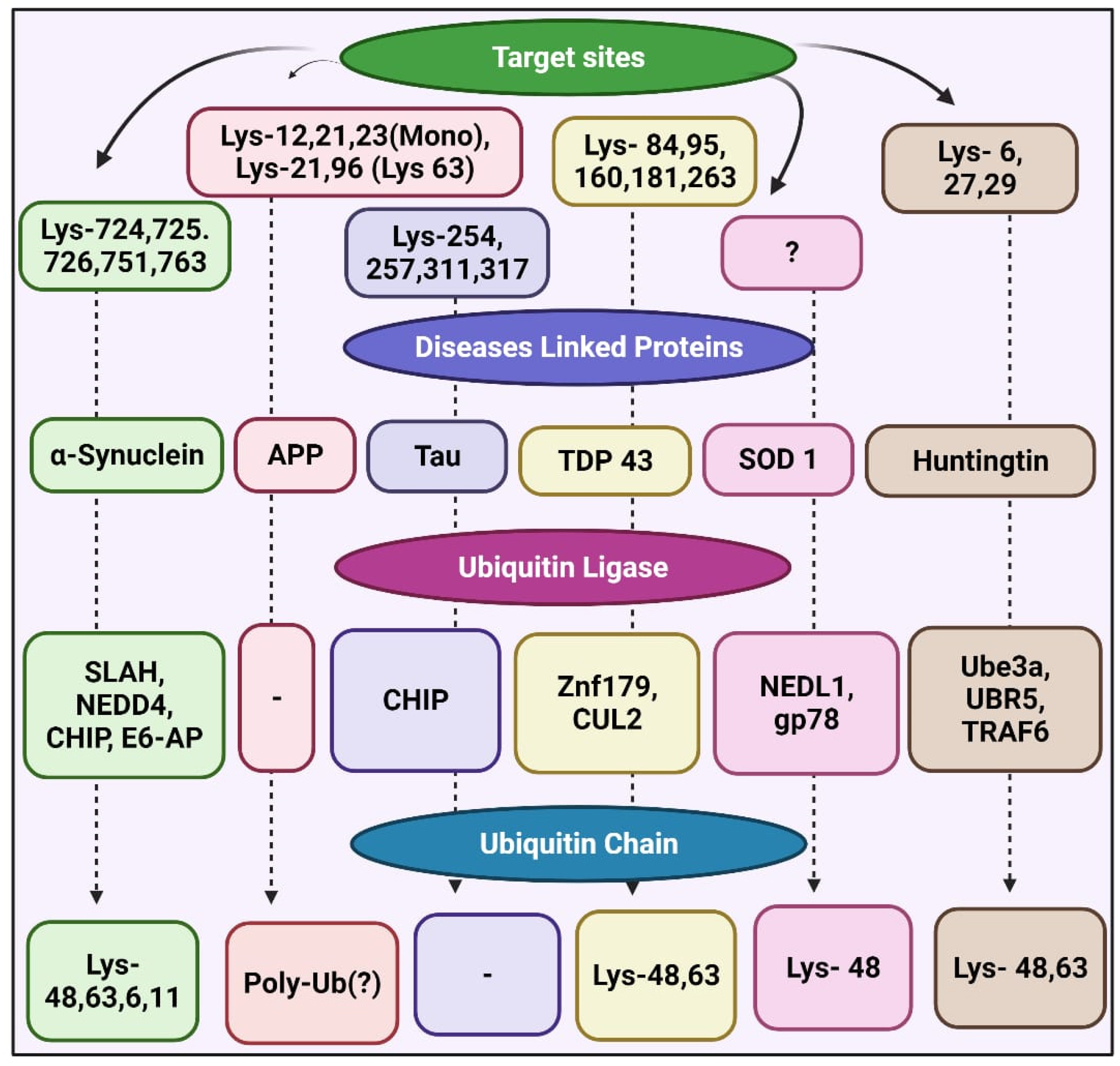
5.1. Targeting E3 in Proteinopathies
5.2. Potential Therapeutic Targets in MS
5.3. UBE2L3 as a Potential Target for Autoimmune Diseases
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roberts-Thomson, P.J.; Jackson, M.W.; Gordon, T.P. A seminal monograph: Mackay and Burnet's Autoimmune diseases. Med. J. Aust. 2012, 196, 74–76. [Google Scholar] [PubMed]
- Zinngrebe, J.; Montinaro, A.; Peltzer, N.; Walczak, H. Ubiquitin in the immune system. EMBO Rep. 2014, 15, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Belogurov, A., Jr.; Kudriaeva, A.; Kuzina, E.; Smirnov, I.; Bobik, T.; Ponomarenko, N.; Kravtsova-Ivantsiv, Y.; Ciechanover, A.; Gabibov, A. Multiple sclerosis autoantigen myelin basic protein escapes control by ubiquitination during proteasomal degradation. J. Biol. Chem. 2014, 289, 17758–17766. [Google Scholar] [CrossRef] [PubMed]
- Wootla, B.; Eriguchi, M.; Rodriguez, M. Is multiple sclerosis an autoimmune disease? Autoimmune Dis. 2012, 1, 969657. [Google Scholar] [CrossRef]
- Stys, P.K. Multiple sclerosis: Autoimmune disease or autoimmune reaction? Can. J. Neurol. Sci. 2010, 37 (Suppl. 2), S16–S23. [Google Scholar] [CrossRef]
- Huang, W.J.; Chen, W.W.; Zhang, X. Multiple sclerosis: Pathology, diagnosis and treatments (review). Exp. Ther. Med. 2017, 13, 3163–3166. [Google Scholar] [CrossRef]
- Ruan, J.; Schluter, D.; Wang, X. Deubiquitinating enzymes (DUBs): DoUBle-edged swords in CNS autoimmunity. J. Neuroinflammation 2020, 17, 102. [Google Scholar] [CrossRef]
- Dobson, R.; Giovannoni, G. Multiple sclerosis—A review. Eur. J. Neurol. 2019, 26, 27–40. [Google Scholar] [CrossRef]
- Anuppalle, M.; Maddirevula, S.; Huh, T.-L.; Rhee, M. Ubiquitin proteasome system networks in the neurological disorders. Animal Cells Syst. 2013, 17, 383–387. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar]
- Bui, Q.T.; Hong, J.H.; Kwak, M.; Lee, J.Y.; Lee, P.C. Ubiquitin-Conjugating Enzymes in Cancer. Cells 2021, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhao, H.; Yu, C.; Kang, Y.; Yang, X. Targeting Ubiquitin-Specific Protease 7 (USP7) in Cancer: A New Insight to Overcome Drug Resistance. Front. Pharmacol. 2021, 12, 648491. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Travaglio, M.; Popovic, R.; Leal, N.S.; Martins, L.M. Alzheimer’s and Parkinson’s Diseases Predict Different COVID-19 Outcomes: A UK Biobank Study. Geriatrics 2021, 6, 10. [Google Scholar] [PubMed]
- Momtaz, S.; Memariani, Z.; El-Senduny, F.F.; Sanadgol, N.; Golab, F.; Katebi, M.; Abdolghaffari, A.H.; Farzaei, M.H.; Abdollahi, M. Targeting Ubiquitin-Proteasome Pathway by Natural Products: Novel Therapeutic Strategy for Treatment of Neurodegenerative Diseases. Front. Physiol. 2020, 11, 361. [Google Scholar] [CrossRef]
- Bäuerlein, F.J.B.; Fernández-Busnadiego, R.; Baumeister, W. Investigating the Structure of Neurotoxic Protein Aggregates Inside Cells. Trends Cell Biol. 2020, 30, 951–966. [Google Scholar]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Körner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef]
- Bhatwa, A.; Wang, W.; Hassan, Y.I.; Abraham, N.; Li, X.-Z.; Zhou, T. Challenges Associated With the Formation of Recombinant Protein Inclusion Bodies in Escherichia coli and Strategies to Address Them for Industrial Applications. Front. Bioeng. Biotechnol. 2021, 9, 65. [Google Scholar] [CrossRef]
- Voges, D.; Zwickl, P.; Baumeister, W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999, 68, 1015–1068. [Google Scholar]
- Wang, J.; Maldonado, M.A. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases—PubMed. Cell. Mol. Immunol. 2006, 3, 255–261. [Google Scholar]
- Shin, J.Y.; Muniyappan, S.; Tran, N.N.; Park, H.; Lee, S.B.; Lee, B.H. Deubiquitination reactions on the proteasome for proteasome versatility. Int. J. Mol. Sci. 2020, 21, 5312. [Google Scholar] [CrossRef]
- Chondrogianni, N.; Gonos, E.S. Structure and function of the ubiquitin-proteasome system: Modulation of components. Prog. Mol. Biol. Transl. Sci. 2012, 109, 41–74. [Google Scholar] [CrossRef] [PubMed]
- Verbrugge, S.E.; Scheper, R.J.; Lems, W.F.; de Gruijl, T.D.; Jansen, G. Proteasome inhibitors as experimental therapeutics of autoimmune diseases. Arthritis Res. Ther. 2015, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, H.T.; Conley, T.; Muchamuel, T.; Jiang, J.; Lee, S.; Owen, T.; Barnard, J.; Nevarez, S.; Goldman, B.I.; Kirk, C.J.; et al. Beneficial effect of novel proteasome inhibitors in murine lupus via dual inhibition of type I interferon and autoantibody-secreting cells. Arthritis Rheum. 2012, 64, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Piedade, W.P.; Famulski, J.K. E3 ubiquitin ligase-mediated regulation of vertebrate ocular development; new insights into the function of SIAH enzymes. Biochem. Soc. Trans. 2021, 49, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Sherman, D.J.; Li, J. Proteasome Inhibitors: Harnessing Proteostasis to Combat Disease. Molecules 2020, 25, 671. [Google Scholar] [CrossRef]
- Benameur, T.; Soleti, R.; Panaro, M.A.; La Torre, M.E.; Monda, V.; Messina, G.; Porro, C. Curcumin as Prospective Anti-Aging Natural Compound: Focus on Brain. Molecules 2021, 26, 4794. [Google Scholar] [CrossRef]
- Mpakali, A.; Stratikos, E. The Role of Antigen Processing and Presentation in Cancer and the Efficacy of Immune Checkpoint Inhibitor Immunotherapy. Cancers 2021, 13, 134. [Google Scholar] [CrossRef]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar]
- George, A.J.; Hoffiz, Y.C.; Charles, A.J.; Zhu, Y.; Mabb, A.M. A comprehensive atlas of E3 ubiquitin ligase mutations in neurological disorders. Front. Genet. 2018, 9, 29. [Google Scholar]
- Ishida, T.; Ciulli, A. E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS Discov. Adv. Life Sci. R D 2021, 26, 484–502. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Gombart, A.F.; Pierre, A.; Maggini, S. A Review of Micronutrients and the Immune System-Working in Harmony to Reduce the Risk of Infection. Nutrients 2020, 12, 236. [Google Scholar] [CrossRef]
- Goru, S.K.; Pandey, A.; Gaikwad, A.B. E3 ubiquitin ligases as novel targets for inflammatory diseases. Pharmacol. Res. 2016, 106, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sui, P.; Li, R.; Zhang, Y.; Tan, C.; Garg, A.; Verheyden, J.M.; Sun, X. E3 ubiquitin ligase MDM2 acts through p53 to control respiratory progenitor cell number and lung size. Development 2019, 146, dev179820. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ma, L.; Wang, B.; Liu, J.; Wei, W. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 2017, 36, 683–702. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Argiles-Castillo, D.; Kane, E.I.; Zhou, A.; Spratt, D.E. HECT E3 ubiquitin ligases—Emerging insights into their biological roles and disease relevance. J. Cell Sci. 2020, 133, jcs228072. [Google Scholar] [CrossRef]
- Weber, J.; Polo, S.; Maspero, E. HECT E3 ligases: A tale with multiple facets. Front. Physiol. 2019, 10, 370. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 47–60. [Google Scholar] [CrossRef]
- Liu, L.; Wong, C.C.; Gong, B.; Yu, J. Functional significance and therapeutic implication of ring-type E3 ligases in colorectal cancer. Oncogene 2018, 37, 148–159. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A.P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Walden, H.; Rittinger, K. RBR ligase-mediated ubiquitin transfer: A tale with many twists and turns. Nat. Struct. Mol. Biol. 2018, 25, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Dai, X.; Jiang, W.; Li, Y.; Wei, W. RBR E3 ubiquitin ligases in tumorigenesis. Semin. Cancer Biol. 2020, 67, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Alquezar, C.; Arya, S.; Kao, A.W. Tau Post-translational Modifications: Dynamic Transformers of Tau Function, Degradation, and Aggregation. Front. Neurol. 2021, 11, 595532. [Google Scholar] [CrossRef] [PubMed]
- Munari, F.; Barracchia, C.G.; Franchin, C.; Parolini, F.; Capaldi, S.; Romeo, A.; Bubacco, L.; Assfalg, M.; Arrigoni, G.; D'Onofrio, M. Semisynthetic and Enzyme-Mediated Conjugate Preparations Illuminate the Ubiquitination-Dependent Aggregation of Tau Protein. Angew. Chem. Int. Ed. 2020, 59, 6607–6611. [Google Scholar]
- Guo, T.; Zhang, D.; Zeng, Y.; Huang, T.Y.; Xu, H.; Zhao, Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 40. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Li, J.-D. The roles of post-translational modifications on α-synuclein in the pathogenesis of Parkinson’s diseases. Front. Neurosci. 2019, 13, 381. [Google Scholar]
- Amer-Sarsour, F.; Kordonsky, A.; Berdichevsky, Y.; Prag, G.; Ashkenazi, A. Deubiquitylating enzymes in neuronal health and disease. Cell Death Dis. 2021, 12, 120. [Google Scholar] [CrossRef]
- Watanabe, Y.; Taguchi, K.; Tanaka, M. Ubiquitin, autophagy and neurodegenerative diseases. Cells 2020, 9, 2022. [Google Scholar]
- Walden, H.; Muqit, M.M.K. Ubiquitin and Parkinson’s disease through the looking glass of genetics. Biochem. J. 2017, 474, 1439–1451. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Huang, W.-C.; Lin, J.-H.; Kao, T.-J.; Lin, H.-C.; Lee, K.-H.; Lin, H.-C.; Shen, C.-K.J.; Chang, W.-C.; Huang, C.-C. Znf179 E3 ligase-mediated TDP-43 polyubiquitination is involved in TDP-43-ubiquitinated inclusions (UBI)(+)-related neurodegenerative pathology. J. Biomed. Sci. 2018, 25, 1–17. [Google Scholar]
- Peggion, C.; Massimino, M.L.; Stella, R.; Bortolotto, R.; Agostini, J.; Maldi, A.; Sartori, G.; Tonello, F.; Bertoli, A.; Lopreiato, R. Nucleolin rescues TDP-43 toxicity in yeast and human cell models. Front. Cell. Neurosci. 2021, 15, 115. [Google Scholar]
- Bi, M.; Du, X.; Jiao, Q.; Chen, X.; Jiang, H. Expanding the role of proteasome homeostasis in Parkinson's disease: Beyond protein breakdown. Cell Death Dis. 2021, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Gurfinkel, Y.; Polain, N.; Lamont, W.; Lyn Rea, S. Molecular Mechanisms Underlying TDP-43 Pathology in Cellular and Animal Models of ALS and FTLD. Int. J. Mol. Sci. 2021, 22, 4705. [Google Scholar] [CrossRef]
- Kang, J.A.; Jeon, Y.J. How Is the Fidelity of Proteins Ensured in Terms of Both Quality and Quantity at the Endoplasmic Reticulum? Mechanistic Insights into E3 Ubiquitin Ligases. Int. J. Mol. Sci. 2021, 22, 2078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xu, Y.; Liu, Y.; Ye, Y. gp78 functions downstream of Hrd1 to promote degradation of misfolded proteins of the endoplasmic reticulum. Mol. Biol. Cell 2015, 26, 4438–4450. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.M.; Toonen, L.J.A.; van Roon-Mom, W.M.C. Ataxin-3 protein and RNA toxicity in spinocerebellar ataxia type 3: Current insights and emerging therapeutic strategies. Mol. Neurobiol. 2014, 49, 1513–1531. [Google Scholar] [CrossRef][Green Version]
- Joshi, V.; Upadhyay, A.; Kumar, A.; Mishra, A. Gp78 E3 Ubiquitin Ligase: Essential Functions and Contributions in Proteostasis. Front. Cell. Neurosci. 2017, 11. [Google Scholar] [CrossRef]
- Sap, K.A.; Reits, E.A. Strategies to Investigate Ubiquitination in Huntington's Disease. Front. Chem. 2020, 8, 485. [Google Scholar] [CrossRef]
- Bhat, K.P.; Yan, S.; Wang, C.-E.; Li, S.; Li, X.-J. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. USA 2014, 111, 5706–5711. [Google Scholar]
- Baker, H.A.; Bernardini, J.P. It’s not just a phase; ubiquitination in cytosolic protein quality control. Biochem. Soc. Trans. 2021, 49, 365–377. [Google Scholar] [CrossRef]
- Li, J.; Liu, N.; Tang, L.; Yan, B.; Chen, X.; Zhang, J.; Peng, C. The relationship between TRAF6 and tumors. Cancer Cell Int. 2020, 20, 429. [Google Scholar] [CrossRef] [PubMed]
- Tracz, M.; Bialek, W. Beyond K48 and K63: Non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021, 26, 1. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Ng, J.; Sivaraman, J. Exploring the ”Other” subfamily of HECT E3-ligases for therapeutic intervention. Pharmacol. Ther. 2021, 224, 107809. [Google Scholar] [CrossRef] [PubMed]
- Anilkumar, A.C.; Foris, L.A.; Tadi, P. Acute Disseminated Encephalomyelitis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Minagar, A.; Barnett, M.H.; Benedict, R.H.; Pelletier, D.; Pirko, I.; Sahraian, M.A.; Frohman, E.; Zivadinov, R. The thalamus and multiple sclerosis: Modern views on pathologic, imaging, and clinical aspects. Neurology 2013, 80, 210–219. [Google Scholar]
- Minagar, A. Current and future therapies for multiple sclerosis. Scientifica 2013, 2013, 11. [Google Scholar] [CrossRef]
- Paramonova, N.; Kalnina, J.; Dokane, K.; Dislere, K.; Trapina, I.; Sjakste, T.; Sjakste, N. Genetic variations in the PSMA6 and PSMC6 proteasome genes are associated with multiple sclerosis and response to interferon-β therapy in Latvians. Exp. Ther. Med. 2021, 21, 478. [Google Scholar] [CrossRef]
- Innao, V.; Rizzo, V.; Allegra, A.G.; Musolino, C.; Allegra, A. Promising Anti-Mitochondrial Agents for Overcoming Acquired Drug Resistance in Multiple Myeloma. Cells 2021, 10, 439. [Google Scholar] [CrossRef]
- Honke, N.; Shaabani, N.; Zhang, D.-E.; Hardt, C.; Lang, K.S. Multiple functions of USP18. Cell Death Dis. 2016, 7, e2444. [Google Scholar] [CrossRef]
- Malhotra, S.; Morcillo-Suarez, C.; Nurtdinov, R.; Rio, J.; Sarro, E.; Moreno, M.; Castillo, J.; Navarro, A.; Montalban, X.; Comabella, M. Roles of the ubiquitin peptidase USP18 in multiple sclerosis and the response to interferon-beta treatment. Eur. J. Neurol. 2013, 20, 1390–1397. [Google Scholar] [CrossRef]
- Yang, W.; Lee, Y.H.; Jones, A.E.; Woolnough, J.L.; Zhou, D.; Dai, Q.; Wu, Q.; Giles, K.E.; Townes, T.M.; Wang, H. The histone H2A deubiquitinase Usp16 regulates embryonic stem cell gene expression and lineage commitment. Nat. Commun. 2014, 5, 3818. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, R.-B.; Cao, Q.; Fan, K.-Q.; Huang, L.-J.; Yu, J.-S.; Gao, Z.-J.; Huang, T.; Zhong, J.-Y.; Mao, X.-T.; et al. USP16-mediated deubiquitination of calcineurin A controls peripheral T cell maintenance. J. Clin. Investig. 2019, 129, 2856–2871. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Tan, X.; Shi, Y.; Xu, G.; Mao, R.; Gu, X.; Fan, Y.; Yu, Y.; Burlingame, S.; Zhang, H.; et al. USP11 negatively regulates TNFalpha-induced NF-kappaB activation by targeting on IkappaBalpha. Cell Signal. 2010, 22, 386–394. [Google Scholar] [CrossRef]
- Schweitzer, K.; Bozko, P.M.; Dubiel, W.; Naumann, M. CSN controls NF-κB by deubiquitinylation of IκBα. EMBO J. 2007, 26, 1532–1541. [Google Scholar]
- Antao, A.M.; Tyagi, A.; Kim, K.S.; Ramakrishna, S. Advances in deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers 2020, 12, 1579. [Google Scholar]
- Rusilowicz-Jones, E.V.; Jardine, J.; Kallinos, A.; Pinto-Fernandez, A.; Guenther, F.; Giurrandino, M.; Barone, F.G.; McCarron, K.; Burke, C.J.; Murad, A.; et al. USP30 sets a trigger threshold for PINK1-PARKIN amplification of mitochondrial ubiquitylation. Life Sci. Alliance 2020, 3, e202000768. [Google Scholar] [CrossRef]
- Liang, J.-R.; Martinez, A.; Lane, J.D.; Mayor, U.; Clague, M.J.; Urbé, S. USP30 deubiquitylates mitochondrial Parkin substrates and restricts apoptotic cell death. EMBO Rep. 2015, 16, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Perga, S.; Montarolo, F.; Martire, S.; Bonaldo, B.; Bono, G.; Bertolo, J.; Magliozzi, R.; Bertolotto, A. Overexpression of the ubiquitin-editing enzyme A20 in the brain lesions of Multiple Sclerosis patients: Moving from systemic to central nervous system inflammation. Brain Pathol. 2021, 31, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Alpi, A.F.; Chaugule, V.; Walden, H. Mechanism and disease association of E2-conjugating enzymes: Lessons from UBE2T and UBE2L3. Biochem. J. 2016, 473, 3401–3419. [Google Scholar] [CrossRef]
- Wang, X.; Mulas, F.; Yi, W.; Brunn, A.; Nishanth, G.; Just, S.; Waisman, A.; Brück, W.; Deckert, M.; Schlüter, D. OTUB1 inhibits CNS autoimmunity by preventing IFN-γ-induced hyperactivation of astrocytes. EMBO J. 2019, 38, e100947. [Google Scholar] [CrossRef]
- Budroni, V.; Versteeg, G.A. Negative Regulation of the Innate Immune Response through Proteasomal Degradation and Deubiquitination. Viruses 2021, 13, 584. [Google Scholar]
- Severa, M.; Farina, C.; Salvetti, M.; Coccia, E.M. Three Decades of Interferon-β in Multiple Sclerosis: Can We Repurpose This Information for the Management of SARS-CoV2 Infection? Front. Immunol. 2020, 11, 1459. [Google Scholar] [CrossRef] [PubMed]
- John, L.; Krauth, M.T.; Podar, K.; Raab, M.-S. Pathway-Directed Therapy in Multiple Myeloma. Cancers 2021, 13, 1668. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Liu, L.; Gan, W. The Roles of Post-Translational Modifications on mTOR Signaling. Int. J. Mol. Sci. 2021, 22, 1784. [Google Scholar] [CrossRef] [PubMed]
- Popova, N.V.; Jücker, M. The role of mTOR signaling as a therapeutic target in cancer. Int. J. Mol. Sci. 2021, 22, 1743. [Google Scholar] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Bingol, B.; Tea, J.S.; Phu, L.; Reichelt, M.; Bakalarski, C.E.; Song, Q.; Foreman, O.; Kirkpatrick, D.S.; Sheng, M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 2014, 510, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Crosas, B. Deubiquitinating enzyme inhibitors and their potential in cancer therapy. Curr. Cancer Drug Targets 2014, 14, 506–516. [Google Scholar] [CrossRef] [PubMed]
- De Jager, P.L.; Jia, X.; Wang, J.; De Bakker, P.I.; Ottoboni, L.; Aggarwal, N.T.; Piccio, L.; Raychaudhuri, S.; Tran, D.; Aubin, C. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009, 41, 776–782. [Google Scholar] [PubMed]
- Torre, S.; Polyak, M.J.; Langlais, D.; Fodil, N.; Kennedy, J.M.; Radovanovic, I.; Berghout, J.; Leiva-Torres, G.A.; Krawczyk, C.M.; Ilangumaran, S.; et al. USP15 regulates type I interferon response and is required for pathogenesis of neuroinflammation. Nat. Immunol. 2017, 18, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Cocco, E.; Marrosu, M.G. The current role of mitoxantrone in the treatment of multiple sclerosis. Expert Rev. Neurother. 2014, 14, 607–616. [Google Scholar]
- Gandhi, R.; Laroni, A.; Weiner, H.L. Role of the innate immune system in the pathogenesis of multiple sclerosis. J. Neuroimmunol. 2010, 221, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Adrianto, I.; Wiley, G.B.; Lessard, C.J.; Kelly, J.A.; Adler, A.J.; Glenn, S.B.; Williams, A.H.; Ziegler, J.T.; Comeau, M.E.; et al. A functional haplotype of UBE2L3 confers risk for systemic lupus erythematosus. Genes Immun. 2012, 13, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.J.; Vyse, S.; Shields, A.M.; Boeltz, S.; Gordon, P.A.; Spector, T.D.; Lehner, P.J.; Walczak, H.; Vyse, T.J. UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am. J. Hum. Genet. 2015, 96, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Lavanya, V.; Jamal, S.; Ahmed, N. Ubiquitin Mediated Posttranslational Modification of Proteins Involved in Various Signaling Diseases. In Protein Modificomics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 109–130. [Google Scholar]
- Bulatov, E.; Khaiboullina, S.; dos Reis, H.J.; Palotas, A.; Venkataraman, K.; Vijayalakshmi, M.; Rizvanov, A. Ubiquitin-proteasome system: Promising therapeutic targets in autoimmune and neurodegenerative diseases. Bionanoscience 2016, 6, 341–344. [Google Scholar] [CrossRef]
- Moser, E.K.; Oliver, P.M. Regulation of autoimmune disease by the E3 ubiquitin ligase Itch. Cell. Immunol. 2019, 340, 103916. [Google Scholar] [CrossRef]
- Giordana, M.T.; Richiardi, P.; Trevisan, E.; Boghi, A.; Palmucci, L. Abnormal ubiquitination of axons in normally myelinated white matter in multiple sclerosis brain. Neuropathol. Appl. Neurobiol. 2002, 28, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Z.; Gu, L.; Yin, R.; Li, H.; Zhang, X.; Cao, T.; Jiang, C. Toxic effects of decabromodiphenyl ether (BDE-209) on human embryonic kidney cells. Front. Genet. 2014, 5, 118. [Google Scholar]
- Minagar, A.; Ma, W.; Zhang, X.; Wang, X.; Zhang, K.; Alexander, J.S.; Gonzalez-Toledo, E.; Albitar, M. Plasma ubiquitin-proteasome system profile in patients with multiple sclerosis: Correlation with clinical features, neuroimaging, and treatment with interferon-beta-1b. Neurol. Res. 2012, 34, 611–618. [Google Scholar] [CrossRef]
| Types | Functional Domain | Members | Reference |
|---|---|---|---|
| HECTs | N and C lobes and flexible lobes in between | NEDD4, ITCH, SMURF1, SMURF2, WWP1, WWP2, UBR5 HERC1, HERC2, HERC3, HERC4, E6AP | [35,36,37] |
| RINGs | RING folded structure with or without zinc binding domain | c-CBL, E4B, cIAP, CHIP, Mdm2-MdmX, SCF, CRL2s, CRL3s, CRL4s, CRL5s, Cullin7/FBXW8, APC/C | [38,39,40] |
| RBRs | Two ring domains on terminal with one internal ring domain | HHARI, ARIH2/TRIAD1, NF14/TRIAD2, RNF216/TRIAD3, PARC/ CUL9, ANKIB1, PAPKIN, HOIL-1L, HOIP | [41,42] |
| USP | Nature | Characteristics/Signaling | Therapeutic Target | Ref. |
|---|---|---|---|---|
| USP30 | Deubiquitinating enzyme with a transmembrane domain | Mitochondria-anchored DUBs; PINK1/Parkin-mediated mitophagy in cells | Potential target for neuro-autoimmune disease | [87] |
| USP18 | Deubiquitinating enzyme | Acts as a negative regulator of type-I interferon (IFN) signaling; involved in IFN-β signaling | Low level of USP18 Expression is directly related to the severity of MS | [70,88] |
| USP16 | Deubiquitinating enzyme | Deubiquitination of PLK1 and histone H2A to control chromosome function | Specific USP16 inhibitors may be effective in treating MS caused by T cells. | [72] |
| A20 | Deubiquitinating and E3 ligase domains | Encoded through TNFAIP3 gene; crucial gatekeeper of immune homeostasis/ involved in NF-κB signaling | Mutation in TNFAIP3 gene leads to autoimmune diseases including MS | [70,89] |
| USP15 | Deubiquitinating enzyme | Regulates type-I interferon response; activation of the transcription factor NF-κB and regulation of its inhibitor IκBα | Potential target for neurodegenerative diseases | [90] |
| USP11 | Deubiquitinating enzyme | Suppresses TNFα-and stimulates activation of NF-κB by targeting IκBα | DUB inhibitor targets the USP11 and acts as an immunosuppressive drug to protect against multiple sclerosis | [91] |
| OTUB1 | Deubiquitinating enzyme | It inhibits IFN-γ-activated JAK2-STAT1 signaling via Lys48 deubiquitinating and stabilizing SOCS1, the JAK2 inhibitor. | Potent target as T and NK cells are important mediators in MS and OTUB1 hinders the activation of T cells and NK cells | [80,92] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, D.; Lee, J.Y.; Puranik, N.; Chauhan, P.S.; Chavda, V.; Jin, J.-O.; Lee, P.C.W. Modulating the Ubiquitin–Proteasome System: A Therapeutic Strategy for Autoimmune Diseases. Cells 2022, 11, 1093. https://doi.org/10.3390/cells11071093
Yadav D, Lee JY, Puranik N, Chauhan PS, Chavda V, Jin J-O, Lee PCW. Modulating the Ubiquitin–Proteasome System: A Therapeutic Strategy for Autoimmune Diseases. Cells. 2022; 11(7):1093. https://doi.org/10.3390/cells11071093
Chicago/Turabian StyleYadav, Dhananjay, Ji Yeon Lee, Nidhi Puranik, Pallavi S. Chauhan, Vishal Chavda, Jun-O. Jin, and Peter C. W. Lee. 2022. "Modulating the Ubiquitin–Proteasome System: A Therapeutic Strategy for Autoimmune Diseases" Cells 11, no. 7: 1093. https://doi.org/10.3390/cells11071093
APA StyleYadav, D., Lee, J. Y., Puranik, N., Chauhan, P. S., Chavda, V., Jin, J.-O., & Lee, P. C. W. (2022). Modulating the Ubiquitin–Proteasome System: A Therapeutic Strategy for Autoimmune Diseases. Cells, 11(7), 1093. https://doi.org/10.3390/cells11071093









