Single-Cell Molecular Characterization to Partition the Human Glioblastoma Tumor Microenvironment Genetic Background
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Glioblastoma Tissue Collection
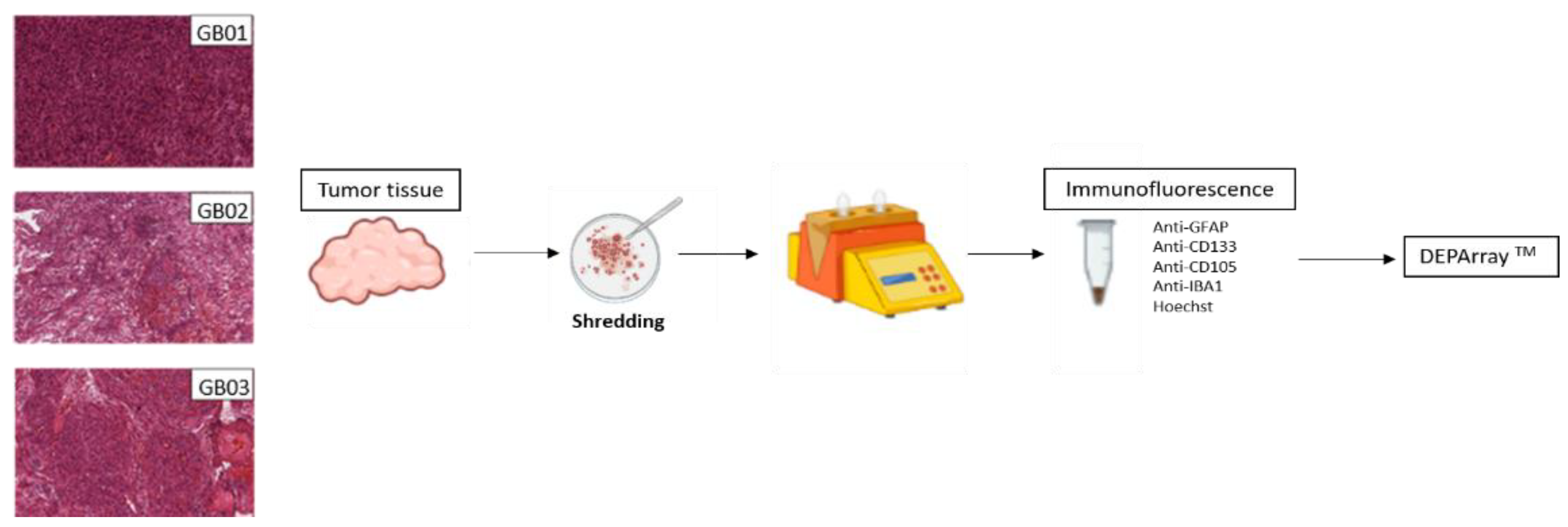
2.2. Tumor Dissociation to Single-Cell Suspensions
2.3. Immunofluorescence of Single-Cell Suspensions
2.4. Single-Cell Isolation by DEPArrayTM NxT
2.5. Immunofluorescence of GB Tissues
2.6. DNA Extraction from Fresh Tissues
2.7. Ampli1™ Whole Genome Amplification and Low Pass Analysis
2.8. CNA Calling
3. Results
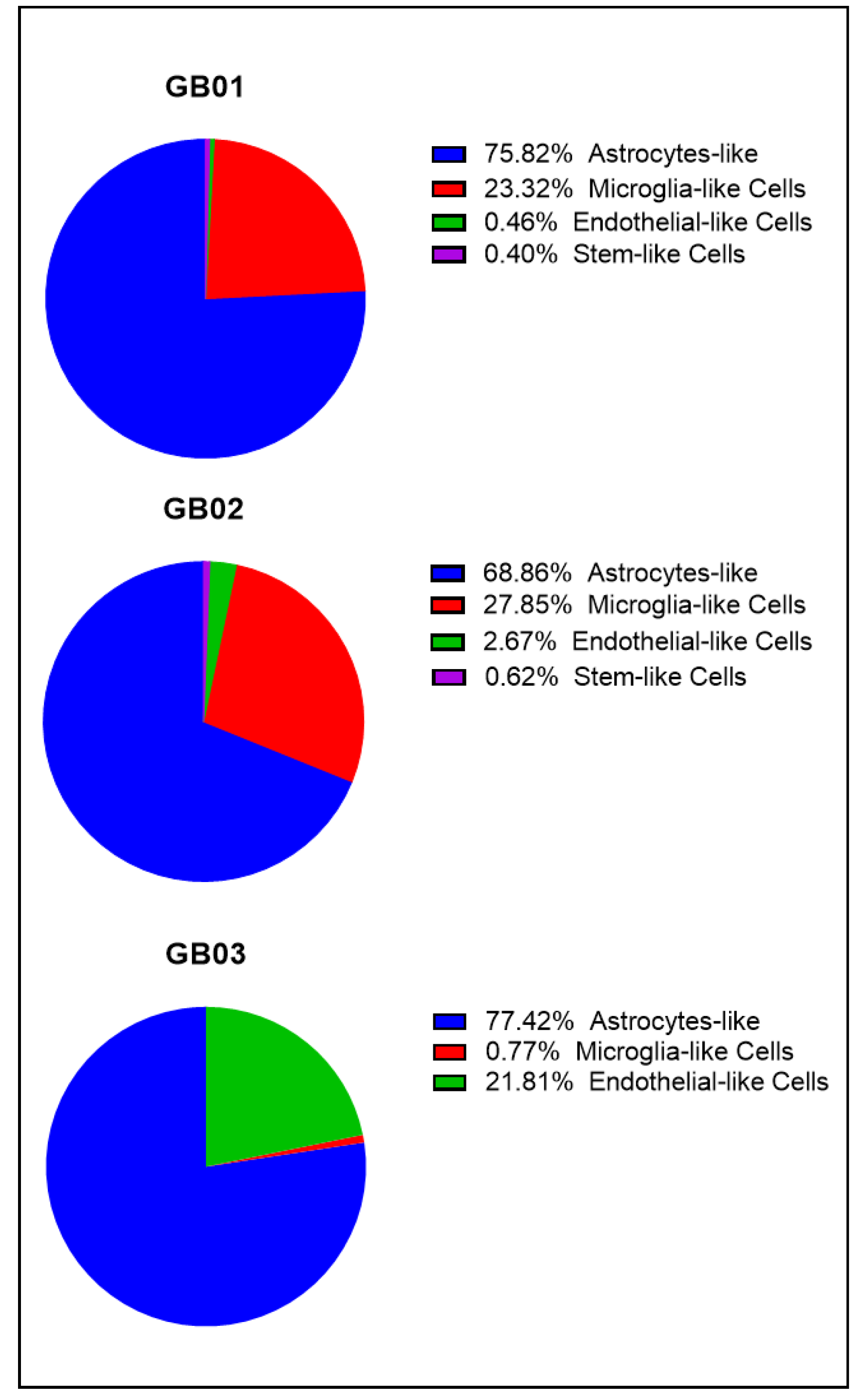
3.1. Isolation of Single-Cells from GB Fresh Tissues with DEPArrayTM NxT
Cell Populations in GB01, GB02, and GB03
3.2. Copy Number Aberrations (CNAs) Analysis
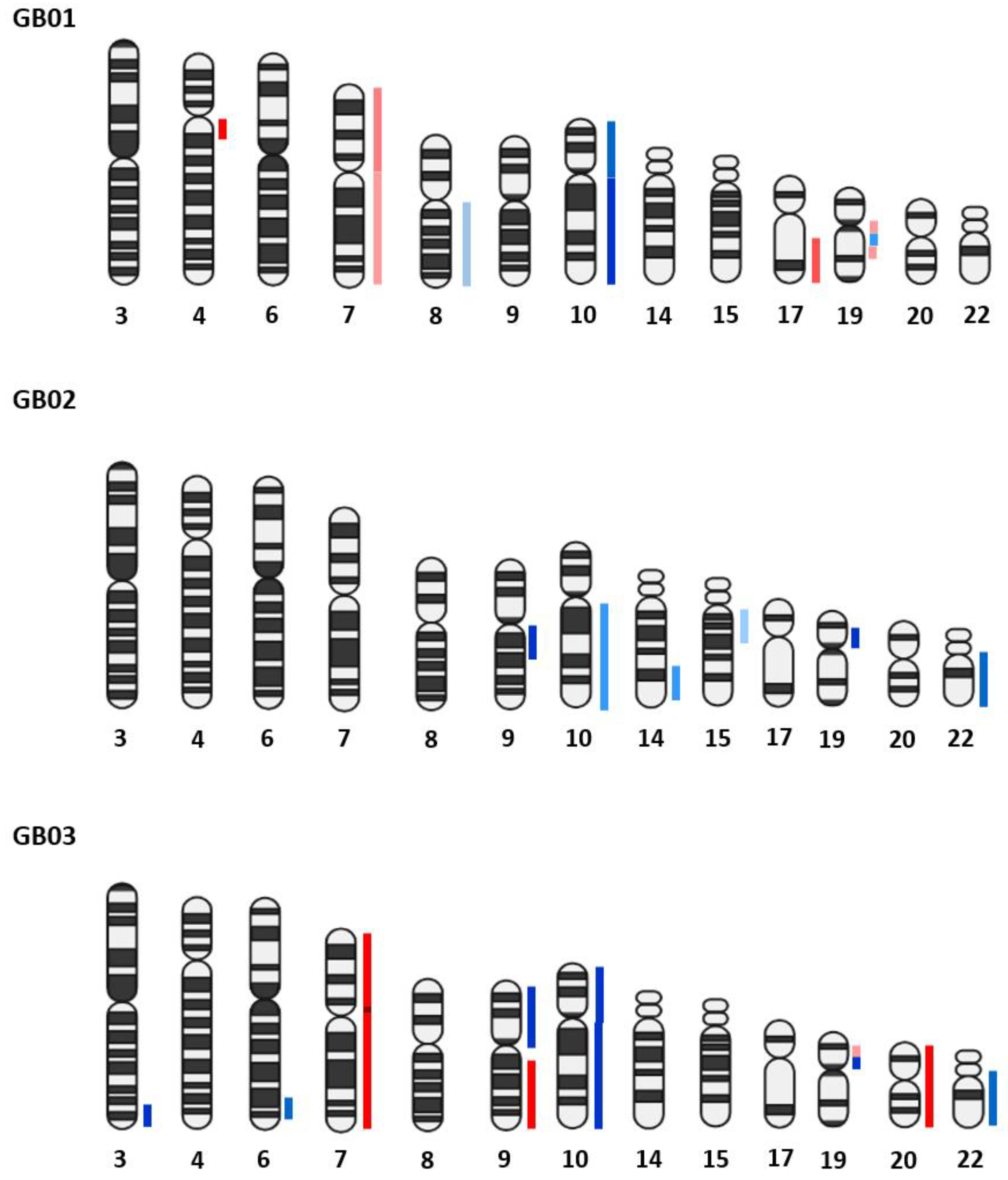
3.2.1. GB Bulk Tissues
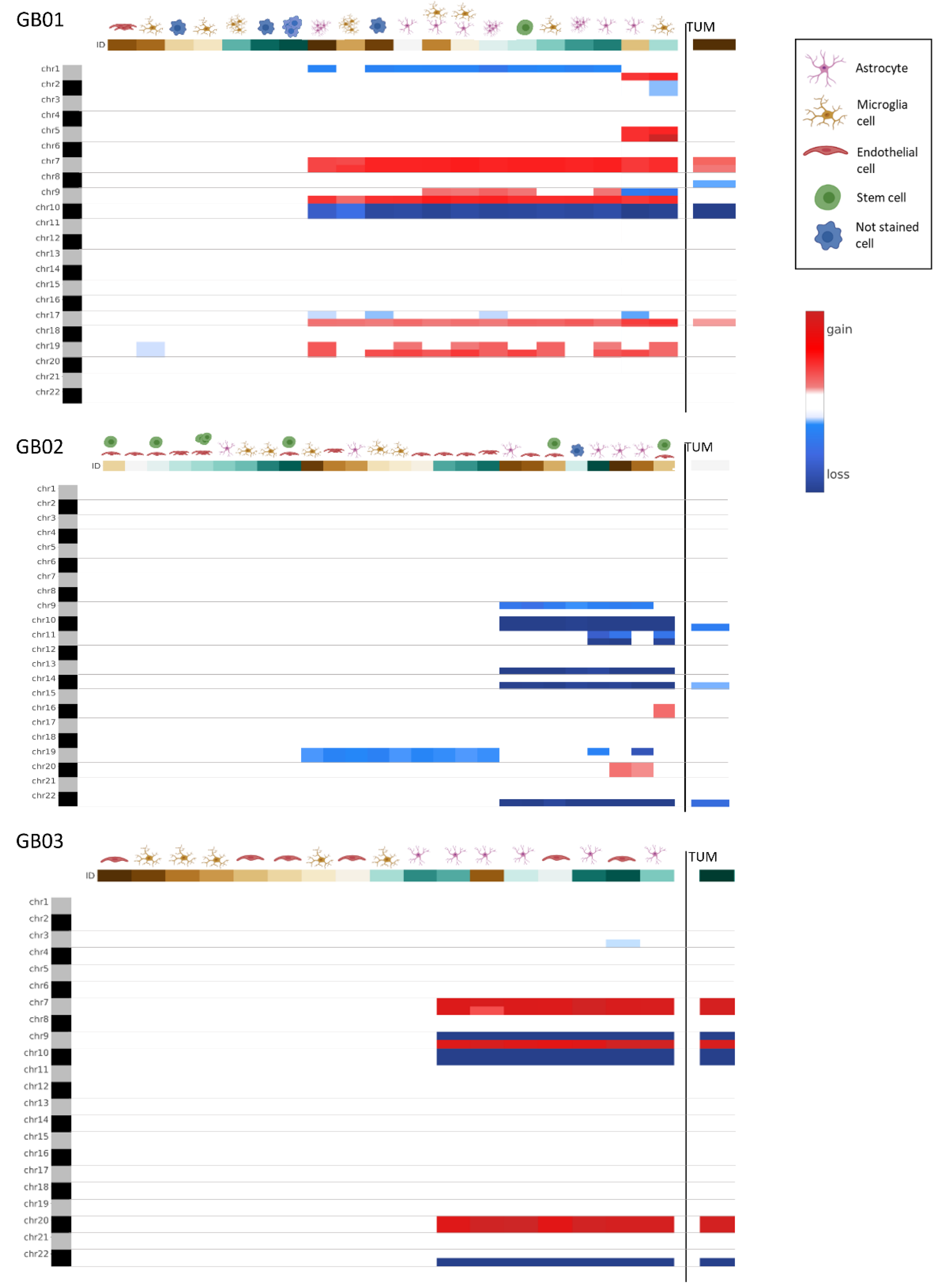
3.2.2. GB Single Cells
3.2.3. Comparison between Bulk Tissues and Single Cells
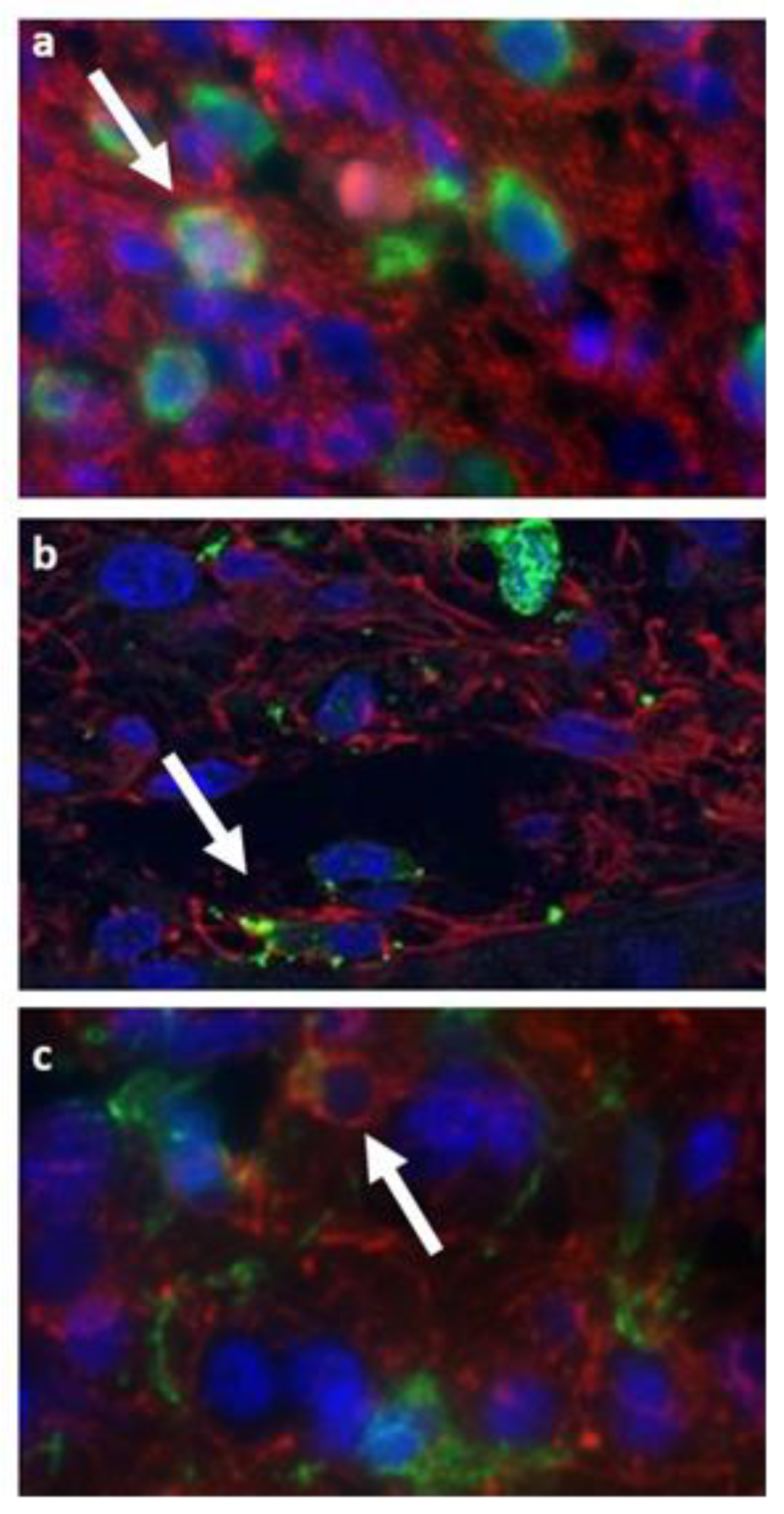
3.3. Double Staining Cells Immunofluorescence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Zhang, X.; Ding, K.; Wang, J.; Li, X.; Zhao, P. Chemoresistance caused by the microenvironment of glioblastoma and the corresponding solutions. Biomed. Pharmacother. 2019, 109, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Hambardzumyan, D.; Bergers, G. Glioblastoma: Defining Tumor Niches. Trends Cancer 2015, 1, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Parada, L.F. The molecular and genetic basis of neurological tumours. Nat. Rev. Cancer 2002, 2, 616–626. [Google Scholar] [CrossRef]
- Yap, T.A.; Gerlinger, M.; Futreal, P.A.; Pusztai, L.; Swanton, C. Intratumor Heterogeneity: Seeing the Wood for the Trees. Sci. Transl. Med. 2012, 4, 127ps10. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.; Reimand, J.; Lan, X.; Head, R.; Zhu, X.; Kushida, M.; Bayani, J.; Pressey, J.C.; Lionel, A.C.; Clarke, I.D.; et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. USA 2015, 112, 851–856. [Google Scholar] [CrossRef] [Green Version]
- Shlush, L.I.; Hershkovitz, D. Clonal evolution models of tumor heterogeneity. Am. Soc. Clin. Oncol. Educ. Book. Am. Soc. Clin. Oncol. Annu. Meet. 2015, 35, e662–e665. [Google Scholar] [CrossRef]
- Albertson, D.G.; Collins, C.; McCormick, F.; Gray, J.W. Chromosome aberrations in solid tumors. Nat. Genet. 2003, 34, 369–376. [Google Scholar] [CrossRef]
- Shlien, A.; Malkin, D. Copy number variations and cancer. Genome Med. 2009, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLendon, R.; Friedman, A.; Bigner, D.; Van Meir, E.G.; Brat, D.J.; Mastrogianakis, G.M.; Olson, J.J.; Mikkelsen, T.; Lehman, N.; Aldape, K.; et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Assem, M.; Sibenaller, Z.; Agarwal, S.; Al-Keilani, M.S.; Alqudah, M.A.Y.; Ryken, T.C. Enhancing Diagnosis, Prognosis, and Therapeutic Outcome Prediction of Gliomas Using Genomics. Omics A J. Integr. Biol. 2012, 16, 113–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hänggi, D.; Wick, W.; et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676. [Google Scholar] [CrossRef] [Green Version]
- Laks, E.; McPherson, A.; Zahn, H.; Lai, D.; Steif, A.; Brimhall, J.; Biele, J.; Wang, B.; Masud, T.; Ting, J.; et al. Clonal Decomposition and DNA Replication States Defined by Scaled Single-Cell Genome Sequencing. Cell 2019, 179, 1207. [Google Scholar] [CrossRef]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Franch-Expósito, S.; Bassaganyas, L.; Vila-Casadesús, M.; Hernández-Illán, E.; Esteban-Fabró, R.; Díaz-Gay, M.; Lozano, J.J.; Castells, A.; Llovet, J.M.; Castellví-Bel, S.; et al. CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications. Elife 2020, 9, e50267. [Google Scholar] [CrossRef]
- Fan, F.; Zhang, H.; Dai, Z.; Zhang, Y.; Xia, Z.; Cao, H.; Yang, K.; Hu, S.; Guo, Y.; Ding, F.; et al. A comprehensive prognostic signature for glioblastoma patients based on transcriptomics and single cell sequencing. Cell. Oncol. (Dordr) 2021, 44, 917–935. [Google Scholar] [CrossRef]
- Bailey, P.; Cushing, H. A Classification of the Tumors of the Glioma Group on a Histogenetic Basis with a Correlated Study of Prognosis. J. Am. Med. Assoc. 1926, 87, 268. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Castro, L.N.G.; Tirosh, I.; Suvà, M.L. Decoding Cancer Biology One Cell at a Time. Cancer Discov. 2021, 11, 960. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couturier, C.P.; Ayyadhury, S.; Le, P.U.; Nadaf, J.; Monlong, J.; Riva, G.; Allache, R.; Baig, S.; Yan, X.; Bourgey, M.; et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 2020, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An integrative model of cellular states, plasticity and genetics for glioblastoma. Cell 2019, 178, 835. [Google Scholar] [CrossRef] [PubMed]
- Reemst, K.; Noctor, S.C.; Lucassen, P.J.; Hol, E.M. The Indispensable Roles of Microglia and Astrocytes during Brain Development. Front. Hum. Neurosci. 2016, 10, 566. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a Cancer Stem Cell in Human Brain Tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J.E.; Kahng, J.Y.; Kim, S.H.; Park, J.S.; Yoon, S.J.; Um, J.Y.; Kim, W.K.; Lee, J.K.; Park, J.; et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 2018, 560, 243–247. [Google Scholar] [CrossRef]
- Matias, D.; Balça-Silva, J.; da Graça, G.C.; Wanjiru, C.M.; Macharia, L.W.; Nascimento, C.P.; Roque, N.R.; Coelho-Aguiar, J.M.; Pereira, C.M.; Dos Santos, M.F.; et al. Microglia/astrocytes–glioblastoma crosstalk: Crucial molecular mechanisms and microenvironmental factors. Front. Cell. Neurosci. 2018, 12, 235. [Google Scholar] [CrossRef] [Green Version]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Keane, L.; Cheray, M.; Blomgren, K.; Joseph, B. Multifaceted microglia-key players in primary brain tumour heterogeneity. Nat. Rev. Neurol. 2021, 17, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Graeber, M.B.; Scheithauer, B.W.; Kreutzberg, G.W. Microglia in brain tumors. Glia 2002, 40, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Leite, D.M.; Zvar Baskovic, B.; Civita, P.; Neto, C.; Gumbleton, M.; Pilkington, G.J. A human co-culture cell model incorporating microglia supports glioblastoma growth and migration, and confers resistance to cytotoxics. FASEB J. 2020, 34, 1710–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, S.A.; Boddeke, H.W.G.M.; Kettenmann, H. Microglia in Physiology and Disease. Annu. Rev. Physiol. 2017, 79, 619–643. [Google Scholar] [CrossRef] [PubMed]
- Bowman, R.L.; Klemm, F.; Akkari, L.; Pyonteck, S.M.; Sevenich, L.; Quail, D.F.; Dhara, S.; Simpson, K.; Gardner, E.E.; Iacobuzio-Donahue, C.A.; et al. Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies. Cell Rep. 2016, 17, 2445–2459. [Google Scholar] [CrossRef] [Green Version]
- Maas, S.L.N.; Abels, E.R.; Van De Haar, L.L.; Zhang, X.; Morsett, L.; Sil, S.; Guedes, J.; Sen, P.; Prabhakar, S.; Hickman, S.E.; et al. Glioblastoma hijacks microglial gene expression to support tumor growth. J. Neuroinflamm. 2020, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Daneman, R.; Prat, A. The blood–brain barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; He, L.; Lugano, R.; Zhang, Y.; Cao, H.; He, Q.; Chao, M.; Liu, B.; Cao, Q.; Wang, J.; et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI Insight 2021, 6, e150861. [Google Scholar] [CrossRef]
- Chouleur, T.; Tremblay, M.L.; Bikfalvi, A. Mechanisms of invasion in glioblastoma. Curr. Opin. Oncol. 2020, 32, 631–639. [Google Scholar] [CrossRef]
- Charalambous, C.; Hofman, F.M.; Chen, T.C. Functional and phenotypic differences between glioblastoma multiforme-derived and normal human brain endothelial cells. J. Neurosurg. 2005, 102, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Miebach, S.; Grau, S.; Hummel, V.; Rieckmann, P.; Tonn, J.C.; Goldbrunner, R.H. Isolation and culture of microvascular endothelial cells from gliomas of different WHO grades. J. Neurooncol. 2006, 76, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Luissint, A.C.; Artus, C.; Glacial, F.; Ganeshamoorthy, K.; Couraud, P.O. Tight junctions at the blood brain barrier: Physiological architecture and disease-associated dysregulation. Fluids Barriers CNS 2012, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Chadalavada, K.; Wilshire, J.; Kowalik, U.; Hovinga, K.E.; Geber, A.; Fligelman, B.; Leversha, M.; Brennan, C.; Tabar, V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature 2010, 468, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Pallini, R.; Biffoni, M.; Todaro, M.; Invernici, G.; Cenci, T.; Maira, G.; Parati, E.A.; Stassi, G.; Larocca, L.M.; et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 2010, 468, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Reduzzi, C.; Vismara, M.; Gerratana, L.; Silvestri, M.; De Braud, F.; Raspagliesi, F.; Verzoni, E.; Di Cosimo, S.; Locati, L.D.; Cristofanilli, M.; et al. The curious phenomenon of dual-positive circulating cells: Longtime overlooked tumor cells. Semin. Cancer Biol. 2020, 60, 344–350. [Google Scholar] [CrossRef]
- Fais, S. Cannibalism: A way to feed on metastatic tumors. Cancer Lett. 2007, 258, 155–164. [Google Scholar] [CrossRef]
- Coopman, P.J.; Do, M.T.; Thompson, E.W.; Mueller, S.C. Phagocytosis of cross-linked gelatin matrix by human breast carcinoma cells correlates with their invasive capacity. Clin. Cancer Res. 1998, 4, 507–515. [Google Scholar]
- Persson, A.; Englund, E. The glioma cell edge—Winning by engulfing the enemy? Med. Hypotheses 2009, 73, 336–337. [Google Scholar] [CrossRef]
- Chang, G.H.F.; Barbaro, N.M.; Pieper, R.O. Phosphatidylserine-dependent phagocytosis of apoptotic glioma cells by normal human microglia, astrocytes, and glioma cells. Neuro Oncol. 2000, 2, 174–183. [Google Scholar] [CrossRef]
- Huysentruyt, L.C.; Seyfried, T.N. Perspectives on the mesenchymal origin of metastatic cancer. Cancer Metastasis Rev. 2010, 29, 695–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelmsson, U.; Eliasson, C.; Bjerkvig, R.; Pekny, M. Loss of GFAP expression in high-grade astrocytomas does not contribute to tumor development or progression. Oncogene 2003, 22, 3407–3411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hieronymus, H.; Murali, R.; Tin, A.; Yadav, K.; Abida, W.; Moller, H.; Berney, D.; Scher, H.; Carver, B.; Scardino, P.; et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 2018, 7, e37294. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Palacios-Corona, R.; Trevino, V. Integration and comparison of different genomic data for outcome prediction in cancer. BioData Min. 2015, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Roh, W.; Chen, P.L.; Reuben, A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Gopalakrishnan, V.; Wang, F.; Cooper, Z.A.; Reddy, S.M.; et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017, 9, eaah3560. [Google Scholar] [CrossRef] [Green Version]


| Cases | Age | Sex | Primary or Recurrence | Brain Location | IDH1/IDH2 | Pathology Report | Therapy Administered |
|---|---|---|---|---|---|---|---|
| GB01 | 30 | M | Primary | parietal lobe | WT | Glioblastoma (Grade IV WHO) (GFAP+, MKI67-20%) | Levetiracetam, Soldesam, Lansoprazole |
| GB02 | 47 | M | Primary | right temporal lobe | WT | Glioblastoma (Grade IV WHO) (GFAP+, MKI67-30%) | Levetiracetam, Dexamethasone, Omeprazole |
| GB03 | 65 | M | Primary | right frontal lobe | WT | Glioblastoma (Grade IV WHO) (GFAP+, MKI67-20%) | Levetiracetam, Lansoprazole, Dexamethasone (Mannitol pre-op) |
| GB01 | |
|---|---|
| Single Cells Collected | CNA |
| Group of endothelial-like cells | WT |
| Microglia-like cell | 19- |
| Microglia-like cell | WT |
| Microglia-like cell | 1p-, 7+, 9q+, 10-, 17q+, 19+ |
| Microglia-like cell | 1q+, 2-, 5+, 7+, 9p-, 9q+, 10-, 17q+, 19+ |
| Group of microglia-like cells | WT |
| Group of microglia-like cells | 7+, 9q+, 10-, 17q+ |
| Astrocyte-like | 1p-, 7+, 9q+, 10-, 17q+, 19+ |
| Astrocyte-like | 1p-, 7+, 9q+, 10-, 17q+, 19+ |
| Astrocyte-like | 1q+, 5+, 7+, 9p-, 9q+, 10-, 17p-, 17q+, 19q+ |
| Group of astrocytes-like | 1p-, 7+, 9q+, 10-, 17p-, 17q+, 19+ |
| Group of astrocytes-like | 1p-, 7+, 9+, 10-, 17p-, 17q+, 19+ |
| Group of astrocytes-like | 1p-, 7+, 9q+, 10-, 17q+ |
| Stem-like cell | 1p-, 7+, 9+, 10-, 17q+, 19q+ |
| Astrocyte/microglia-like cell | 1p-, 7+, 9+, 10-, 17q+, 19q+ |
| Astrocyte/microglia-like cell | 1p-, 7+, 9+, 10-, 17q+, 19+ |
| Not stained cell | WT |
| Not stained cell | WT |
| Not stained cell | 1p-, 7+, 9q+, 10-, 17p-, 17q+, 19q+ |
| Group of not stained cells | WT |
| GB02 | |
|---|---|
| Single Cells Collected | CNA |
| Endothelial-like cell | WT |
| Endothelial-like cell | 19- |
| Endothelial-like cell | 19- |
| Endothelial-like cell | 19- |
| Endothelial-like cell | 9p-, 10-, 13q-, 14q-, 22q- |
| Group of endothelial-like cells | WT |
| Group of endothelial-like cells | 19- |
| Group of endothelial-like cells | 19- |
| Astrocyte-like | WT |
| Astrocyte-like | 19- |
| Astrocyte-like | 9p-, 10-, 13q-, 14q-, 22q- |
| Astrocyte-like | 9p-, 10-, 11- 13q-, 14q-, 19p-, 22q- |
| Astrocyte-like | 9p-, 10-, 11- 13q-, 14q-, 20+, 22q- |
| Astrocyte-like | 9p-, 10-, 13q-, 14q-, 19p-, 20+ 22q- |
| Microglia-like cell | WT |
| Microglia-like cell | WT |
| Microglia-like cell | 19- |
| Microglia-like cell | 19- |
| Microglia-like cell | 19- |
| Endothelial/stem-like cell | WT |
| Endothelial/stem-like cell | WT |
| Endothelial/stem-like cell | WT |
| Endothelial/stem-like cell | 9p-, 10-, 13q-, 14q-, 22q- |
| Endothelial/stem-like cell | 10-, 11-, 13q-, 14q-, 16+, 22q- |
| Group of endothelial/stem-like cells | WT |
| Not stained cell | 9p-, 10-, 13q-, 14q-, 22q- |
| GB03 | |
|---|---|
| Single Cells Collected | CNA |
| Endothelial-like cell | WT |
| Endothelial-like cell | WT |
| Endothelial-like cell | WT |
| Endothelial-like cell | WT |
| Endothelial-like cell | 7+, 9p-, 9q+, 10-, 20+, 22q- |
| Endothelial-like cell | 3q-, 7+, 9p-, 9q+, 10-, 20+, 22q- |
| Microglia-like cell | WT |
| Microglia-like cell | WT |
| Microglia-like cell | WT |
| Microglia-like cell | WT |
| Microglia-like cell | WT |
| Astrocyte-like | WT |
| Astrocyte-like | 7+, 9p-, 9q+, 10-, 20+, 22q- |
| Astrocyte-like | 7+, 9p-, 9q+, 10-, 20+, 22q- |
| Astrocyte-like | 7+, 9p-, 9q+, 10-, 20+, 22q- |
| Astrocyte-like | 7+, 9p-, 9q+, 10-, 20+, 22q- |
| Astrocyte-like | 7+, 9p-, 9q+, 10-, 20+, 22q- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lessi, F.; Franceschi, S.; Morelli, M.; Menicagli, M.; Pasqualetti, F.; Santonocito, O.; Gambacciani, C.; Pieri, F.; Aquila, F.; Aretini, P.; et al. Single-Cell Molecular Characterization to Partition the Human Glioblastoma Tumor Microenvironment Genetic Background. Cells 2022, 11, 1127. https://doi.org/10.3390/cells11071127
Lessi F, Franceschi S, Morelli M, Menicagli M, Pasqualetti F, Santonocito O, Gambacciani C, Pieri F, Aquila F, Aretini P, et al. Single-Cell Molecular Characterization to Partition the Human Glioblastoma Tumor Microenvironment Genetic Background. Cells. 2022; 11(7):1127. https://doi.org/10.3390/cells11071127
Chicago/Turabian StyleLessi, Francesca, Sara Franceschi, Mariangela Morelli, Michele Menicagli, Francesco Pasqualetti, Orazio Santonocito, Carlo Gambacciani, Francesco Pieri, Filippo Aquila, Paolo Aretini, and et al. 2022. "Single-Cell Molecular Characterization to Partition the Human Glioblastoma Tumor Microenvironment Genetic Background" Cells 11, no. 7: 1127. https://doi.org/10.3390/cells11071127
APA StyleLessi, F., Franceschi, S., Morelli, M., Menicagli, M., Pasqualetti, F., Santonocito, O., Gambacciani, C., Pieri, F., Aquila, F., Aretini, P., & Mazzanti, C. M. (2022). Single-Cell Molecular Characterization to Partition the Human Glioblastoma Tumor Microenvironment Genetic Background. Cells, 11(7), 1127. https://doi.org/10.3390/cells11071127









